1. Introduction
Atherosclerosis is the main cause of morbidity and mortality in developed countries. It is a complex, progressive disease of the arterial wall, resulting in life-threatening thrombotic episodes such as acute coronary syndromes. More than 75% of plaques that result in acute myocardial infarction and/or death are non-stenotic [
1]. Numerous studies based on histology and intravascular imaging techniques have tried to detect the type of atherosclerotic plaque that results in an acute coronary syndrome, whose most frequent mechanism is plaque rupture [
1,
2]. This prone-to-rupture type of plaque is characterized as “high risk” and has specific morphologic features: a thin fibrous cap that covers a large superficial lipid pool and often exhibits local signs of inflammation [
2,
3,
4].
Optical coherence tomography (OCT, intravascular OCT) is a light-based medical imaging modality. It was first demonstrated by Huang D et al. in 1991 [
5]. In intravascular imaging, OCT has been established as the frequency or Fourier domain (FD—OCT) system due to the advantages it presents (in terms of speed and sensitivity) [
6,
7]. In recent years, continuous improvements in the technology of OCT systems, mainly in terms of the Micro-Electro-Mechanical Systems (MEMS) [
8], have allowed their easy application in the study of biological tissues and the achievement of an axial resolution <2 μm.
In Intravascular Optical coherence tomography, the light is in the near infrared (NIR) range, typically with wavelengths of approximately 1.3 μm, which provides a 10-fold higher spatial resolution than that of intravascular ultrasound (IVUS), allowing the precise measurement of the fibrous cap thickness and detection of plaque components. OCT can be used to identify various stages of plaque morphology. The main limitation of intravascular OCT is the limited penetration depth in the artery wall, as it depends on tissue type and usually ranges from 0.1 to 2.0 mm using typical IVOCT NIR light.
Intravascular OCT can identify several types of atherosclerotic plaques. According to the published expert consensus documents, an atherosclerotic plaque (atheroma) in intravascular OCT images is defined as a mass lesion (focal thickening) or loss of a layered structure of the vessel wall. A fibrous plaque has high backscattering and a relatively homogeneous intravascular OCT signal. A calcific plaque contains intravascular OCT evidence of calcium that appears as a signal-poor or heterogeneous region with a sharply delineated border (leading, trailing, and/or lateral edges). This definition applies to larger calcifications. A lipid plaque by intravascular OCT is a signal-poor region with poorly delineated borders, a fast signal drop-off and little or no signal backscattering, covered by a fibrous cap. Mixed lesions are called heterogeneous plaques or mixed lesions [
9,
10].
The assessment of the degree of the development and the type of plaque is very important for identifying high-risk plaques. The use of OCT involves the collection of a large amount of imaging data (e.g., 100–300 cross-sectional images are collected for each artery), the examination of which requires a lot of time and great effort from a skilled medical specialist. Besides, it can be very hard to distinguish plaques even for experts on the field [
9,
11]. Therefore, an automated method is needed to detect and categorize the microstructures of interest and objectify the characterization. Athanasiou et al. [
12] demonstrated that automatic characterization methods can provide accurate results despite the propagation of error caused by the image formation, the intermediate steps, and the final classification phase of such methods.
The aim of this work is the development of a new deep learning method to detect atherosclerotic tissues in intravascular OCT images. Steps for the classification of detected plaques into four categories are proposed. The method is based on the classification of the A-lines into normal and abnormal (tissues) and the differentiation of the abnormal tissues into four plaque types. The main idea is to configure the input to a CNN architecture and to choose CNN properties and training options that offer better classification results.
The present paper is structured as follows: In
Section 2, we discuss related work. In
Section 3, we refer to the material of the study and describe the proposed method in detail.
Section 4 presents the obtained results. In
Section 5, we discuss the methodological approach and the outcomes of the study. Finally,
Section 6 presents the conclusions of the study.
2. Related Work
Several methods attempted to automatically detect specific plaque regions. Wang et al. [
13] proposed an automatic method for lumen segmentation and calcified plaque detection. The first task is achieved by a dynamic programming scheme and the latter by combining edge detection with active contour models. Other approaches apply machine learning techniques based on hand-crafted or automatically extracted features. Specifically, Shalev et al. [
14] used hand-crafted features to train Support Vector Machines (SVM) and Radial Basis Function (RBF) classifiers to discriminate calcifications, lipid plaques, and fibrous tissue/plaque (one grouped category). Similarly, Athanasiou et al. [
15] used hand-crafted features to train SVM, neural networks and Random Forests (RF) classifiers to classify the detected plaque into four types: calcified, lipid, mixed, and fibrous tissue. Random Forests provided the best classification results. It is not clear, however, if the last type refers to fibrous plaque or normal fibrous tissue. Prakash et al. [
16] proposed an unsupervised classification approach that uses the K-means clustering algorithm applied to statistical texture features to identify background, plaque, and normal tissue areas. Their method provided a rough estimation and was evaluated by visual inspection.
Instead of using hand-crafted features, other studies applied deep learning to OCT cross-sections or patches, attempting a semantic interpretation. Gessert et al. [
17] used ResNet and DenseNet to classify OCT images in three categories: normal with calcified plaque and with fibrous or lipid plaque. This method does not classify image regions but whole images. Oliveira et al. [
18] applied a fully connected CNN known as SegNet. The dataset consisted of 51 images of 13 patients. Patches of 100 × 100 pixels were input to SegNet, which produced a probability map with two classes: normal and calcified plaque. Probabilities derived from overlapping patches were combined in post-processing steps to classify each pixel of the image. The accuracy was 0.7 in this task. He et al. [
19] presented a method that applied an original CNN to 269 images of 22 patients. Images were divided into square patches of 51 × 51 pixels, which were used to train the network. They chose to classify tissue in 4 categories (neglecting fibrous plaque). The accuracy of the algorithm in detecting calcified plaque was too low, resulting in Dice = 0.22. Most of the existing methods are patch-based (pixel classification), having several limitations. Plaque types are not considered, e.g., fibrous plaque. Moreover, it is uncertain if these methods can generalize to other images because there is no reference to data handling and splitting into training, validation, and test sets. Generally, patch-based approaches have difficulty achieving tissue classification because some plaques present similar textures. In addition, the stratification of artery wall layers (intima, media, and adventitia), which is not depicted in patches, is more indicative of the presence of plaque structures.
Therefore, several methods were proposed for axial line (A-line) classification. A-line classification does not permit the delineation of plaque structures, which is attempted by patch-based methods. However, it permits the estimation of the extent of the plaque present on the arterial wall. Rico-Jimenez et al. [
20] modeled every axial line in IVOCT data as a linear combination of several depth profiles. After the estimation of these profiles through least-square optimization, they classified the tissue types based on their morphological features. While they examine four A-line types—Intimal-thickening, Fibrotic, Superficial-Lipid, and Fibrotic-Lipid—their method was tested in ex vivo data and was successful in discriminating lipid and non-lipid tissue (85%). Prabhu et al. [
21] applied dual binary classifiers and SVM to features extracted from A-lines to achieve 81.56% accuracy in three-way classification. Kolluru et al. [
22] applied deep learning combined with Conditional Random Fields to achieve 83.16% accuracy in the same task. Lee et al. [
23] built upon these methods and enhanced their performance (89% accuracy) by introducing lumen morphology features and active learning. Abdolminalfi et al. [
24] used a CNN to extract features from A-lines. These features were input to three classifiers (neural networks, random forests, SVM) and trained to distinguish intima and media in the arterial wall. The outcome of this method is related to the detection of plaques because the clear appearance of intima and media layers is indicative of healthy tissue. Therefore, Zahnd et al. [
25] proposed a method that, after detecting the layers of the arterial wall, determined if wall parts (series of A-lines) are healthy or diseased, and they had 0.91 median accuracy. Their dataset consisted of 260 images, but it was imbalanced (containing mostly healthy tissue).
One aspect of plaque detection and plaque calcification to be considered is the use of optical-transformed images instead of intensity images acquired by OCT systems. Boi et al. [
26] published a review of deep learning IVOCT approaches, including methods for the calculation of backscattering and the attenuation coefficients of tissue in OCT images. It is suggested that these methods may enhance performance in plaque detection and plaque classification tasks. One of these methods was the study of van Soest et al. [
27], who highlighted attenuation coefficient’s importance for tissue classification and modeled it with more accuracy. Foin et al. [
28] showed that the estimation of the attenuation coefficient leads to better manual segmentation by medical experts. Liu et al. [
29] expanded van Soest et al.’s work and presented a detailed method for the estimation of backscattering and attenuation coefficients. Liu et al.’s [
29] formulas were used in the present work.
3. Materials and Methods
3.1. Materials
We studied 183 intravascular FD-OCT images derived from 33 patients who underwent a clinically indicated cardiac catheterization. All imaging data were blindly acquired at Hippokration Hospital, Athens, Greece, General Hospital of Nikaia, Piraeus, Greece and New Tokyo Hospital, Chiba, Japan, using standardized image acquisition protocols.
The OCT acquisition was performed with a frequency-domain OCT imaging system (C7-XRT OCT Intravascular Imaging System, Westford, MA, USA) at a pullback speed of 20 mm/s, axial resolution of 15 μm, and maximum frame rate of 100 frames/s. Temporary blood clearance was achieved with contrast infusion. Expert cardiologists annotated 183 images with tissue and plaque types according to the standards of published consent of experts [
9]. Plaques that appeared in the annotated images were: 84 lipid, 80 calcified, 70 fibrous, και 42 mixed.
3.2. Methodology Overview
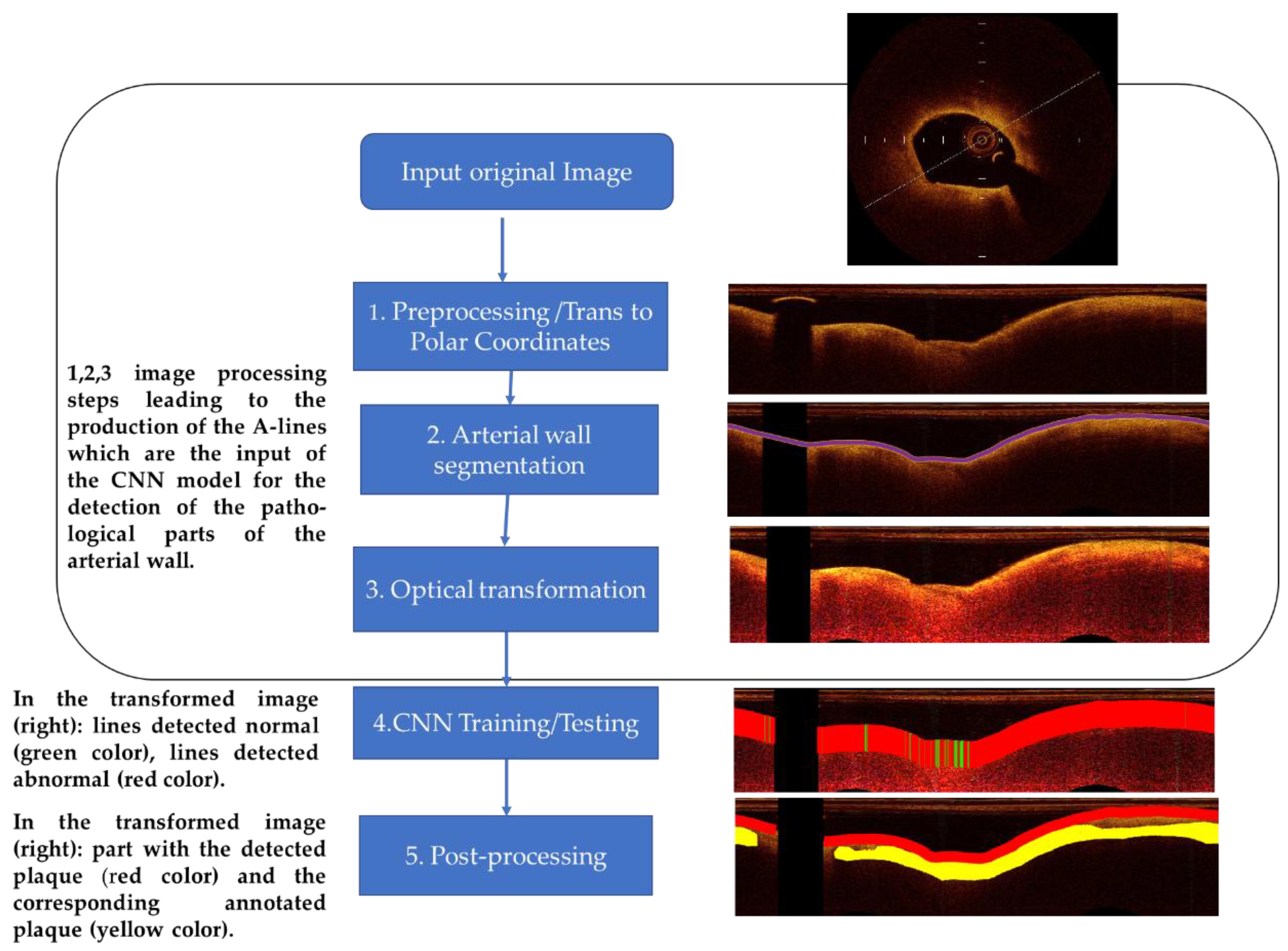
A-line is the propagation line of the light beam that starts from the catheter and reaches the depth of the biological tissue where it is completely attenuated. The method, which classifies A-lines to normal and plaque, consists of the following analysis steps: (1) preprocessing—image preparation, (2) arterial wall segmentation, (3) OCT-specific transformation based on the attenuation coefficient estimation, (outcome the A-lines that are input to the CNN), (4) CNN training—testing procedure (for the detection of the pathological tissue and classification of the different tissue types), and (5) post-processing based on the majority of the classifications. The employed image processing and CNN classification procedure workflow is illustrated in
Figure 1 and described below.
3.3. Preprocessing
Preprocessing steps included: (i) dataset “alignment” by image resizing, (ii) calibration markers’ noise removal, and (iii) polar coordinates transformation:
Due to the fixed size of the CNN input and to the examination of texture features, images were “aligned”, i.e., they were resized to have the same size and resolution. Most images in the dataset used had the size of 1024 × 1024, and we chose to oversample smaller images. Few images in our dataset had higher resolution than 2048 × 2048 pixels. We chose to downsample the latter because oversampling all other images of the dataset to their size would significantly alter the original data.
Calibration markers represent non-natural pixels in the images and were thus removed. The intensities of pixels in calibration markers’ positions were estimated by taking the median value of their region utilizing a 5 × 5 window.
The transformation of images to polar coordinates was proposed by several studies [
12,
13] both for the delineation of arterial wall-lumen border and for semantic analysis by deep learning techniques. In this work, this transformation was also an essential step to achieve these tasks and to extract A-lines that are input to the proposed deep neural network architecture. Given a pixel (x,y) in the Cartesian domain, its corresponding position (ρ,θ) in polar coordinates is given by [
30]:
where (Cx,Cy) is the image center coordinates (the central point of the catheter) in the Cartesian domain. We considered that we have enough information from the image in the Cartesian domain to represent a depth of 383 pixels and 1532 angles. Consequently, the size of the resulting images in polar coordinates was 383 × 1532 pixels.
3.4. Segmentation of the Arterial Wall
After preprocessing, the ARC-OCT algorithm was applied for the delineation of lumen-wall borders [
30]. ARC-OCT includes mainly OCT-specific (depth-resolved) transformation of images, thresholding, morphological operations and contour smoothing. The lumen-wall border was not defined in A-lines with guide-wire artifacts, but it is estimated by the neighboring parts of the border. However, image parts influenced by guide-wire artifacts were excluded from further examination in this work. In
Figure 2, the result of the lumen segmentation algorithm is presented.
The delineation of the lumen-wall border is equivalent to arterial wall segmentation with the assumption that what is deeper than the lumen-wall border and up to the point where there is no backscattered signal is considered the arterial wall. The arterial wall is the region where plaques and normal tissue can be found. Therefore, we applied ARC-OCT to produce one-dimensional A-line patches that start at the lumen-wall border and include pixels from the arterial wall. Each A-line patch acquired a tag, if at least one pixel is part of a manual delineated plaque. The tags were: fibrous plaque, lipid plaque, calcified plaque, mixed plaque, or normal tissue.
3.5. A-Line Size Selection
The penetration depth (how deeply within the tissue we can obtain OCT image data) usually does not exceed 2 mm in coronary arteries. At this depth, the light signal is usually completely attenuated. Medical experts suggested that 1.5 mm is the penetration depth on the arterial wall where information about tissue properties can be found in any case. This depth corresponds to 90 pixels in the dataset’s resolution [
9]. We carried out relative tests with A-lines of different sizes (120, 110, 100, 90, 80, 70, and 60 pixels) to verify that A-lines of a 1 × 90 size are a reasonable option. Finally, we chose the A-line size of 90 pixels as input in our CNN model.
3.6. Attenuation Coefficient Estimation
Backscattered light, which forms the OCT image, is attenuated in arterial wall parts. Consequently, intensities of the same tissue type, e.g., endothelial, diverge. However, it is preferable that regions of a certain tissue type share a similar appearance. The latter is attempted by estimating attenuation coefficient values in every pixel of an OCT image. The outcome image is more realistic, i.e., a specific tissue type is represented by a smaller range of intensities while maintaining texture information. To test the effect of using the attenuation coefficient in the classification of A-lines, transformations of OCT-images were carried out according to the work presented by Liu et al. [
29]. The attenuation μ in every pixel of an OCT image can be estimated with the OCT image intensities by using the formula [
29]:
where I[i,j] is the intensity in the original image; Δ is the physical size of a pixel (mm/pixel); i indicates the A-line number; j indicates the depth. In the rest of this document, transformed images are often referred to as attenuation images for brevity.
In
Figure 3, an example of these transformations is presented. In the
Section 4, we show that A-line patches extracted from images representing estimated attenuation coefficient values are classified with higher accuracy than A-line patches extracted from images with original intensity values.
3.7. Deep Learning
CNNs are neural networks with convolutional layers followed by dense layers. CNN-based methods outperformed other methods in many computer vision problems. Specifically, accurate methods in A-line classification in IVOCT images [
22,
23] are using CNNs. Therefore, in this work, various original (trained from scratch) and pretrained CNN architectures were tested.
Pretrained CNNs are fixed, open-source methods that were trained with natural images. They can be efficient in medical image analysis by fine-tuning their parameters [
31]. Fine-tuning the parameters leads to faster convergence during training than training from scratch. Such a pretrained network is AlexNet [
32], which consists of five convolutional layers with rectified linear unit (ReLU) activations and three dense layers. Some of its activations are followed by max-pooling layers. Although there are more efficient architectures, AlexNet is still a basis for the development of simple architectures that are more easily trained due to their small number of layers.
Having been trained with natural images that are included in large databases, pretrained networks may not be suitable for medical images that represent the interior of the human body. Besides, resizing images to fit these networks may significantly alter their original representation. More specialized networks are needed for medical image analysis. In medical image analysis, regions are segmented based on their texture. When there is no abundance of images, semantic methods are not efficient. Therefore, images are segmented to fix-sized patches. This introduces the need for CNN architectures with smaller input sizes than state-of-the-art architectures. To analyze CT images of lungs, Anthimopoulos et al. [
33] proposed an original CNN that is specialized in classifying small parts of the images. The main characteristics were: five convolutional and three dense layers. This network could be considered a miniature of AlexNet, but it does not use pooling layers.
Inspired by common architectures such as AlexNet and the more specialized miniature network by Anthimopoulos et al. [
33] and after experiments with different CNNs, in this work, we propose the use of a 1-D CNN that takes as input axial-lines with the corresponding attributes (
Figure 4. To test the nearly optimal depth of A-lines for classification, the input size varied from 1 × 60 × 3 and 1 × 120 × 3 by changing the size of the second dimension by 10 pixels each time. The resulting network we finally propose has input 1 × 90 × 3. It consisted of 3 convolutional layers, 2 dense layers, and an output layer. The size of the convolutional kernels was small to keep small the receptive field so that high spatial frequencies stimulate the kernels. Their exact size of 1 × 4, 1 × 2, 1 × 10 was chosen after experiments. In these experiments, the number of feature maps in each layer was also examined. The use of only one big dense layer accelerated convergence. A much smaller dense layer followed to capture A-line appearances that could be grouped in few categories. Overfitting was dealt with dropout layers after the dense layer and with L2 regularization (The L2 parameter was set equal to 0.0005). After experimentation with activations, leaky ReLU [
34] was selected for the convolution layers. Leaky ReLU activation is more robust when applied to the convolutional layers. The last dense layer was followed by a softmax activation. The Adam optimizer was used to reduce categorical cross-entropy [
35]. Hyper-parameter tuning was used to select the learning rate to be equal to 0.001. After 200 iterations without improvement of the training error, the training stopped.
With this architecture as a basis, deeper CNNs were examined, and their results were compared. These CNNs also converged fast and achieved a similar validation and test accuracy for each task. However, the architecture in
Figure 4 (and
Table 1) slightly surpasses, in most cases, the performance of other deeper CNNs. In the Results section, we refer to its performance except for a comparison with a deeper CNN. In the latter case, two more convolutional layers were added as dropout layers following the convolutional layers.
3.8. Post-Processing
The similar performance of the CNN architectures in the normal/abnormal classification case is attributed to the descriptive properties of the data. Many A-lines were correctly classified as normal or abnormal with different CNN architectures. On the other hand, a smaller percentage of A-lines are misclassified. The test set includes information about the adjacency of A-lines. Based on the minimum extent of plaques, which is 40 A-lines, and the fact that a single normal A-line cannot reside inside a plaque region, it is assumed that an individual A-line, which is not classified as belonging to the same class with most of a group of 20 adjacent A-lines, is misclassified. Therefore, a post-processing step to smooth classification results and correct misclassifications was introduced.
Specifically, let the notation of classification be: C(i) = 0 (normal) or 1 (plaque), where i is the A-line number. For i = 1, 21, 42, …, M (where M is the total number of A-lines in an image): C(i:i + 20) values are updated to 0 if Σ(C(i:i + 20) ≤ 10 and C(i:i + 20) are update to 1 if Σ(C(i:i + 20) > 10. This step decides the classification result of an A-line based on the dominant class of the classification results of adjacent A-lines. This produces a more realistic outcome, and it can improve test accuracy by 1–2% in most cases. An example of a post-processing result is shown in
Figure 5.
3.9. Experimental Setup
All experiments were performed on a laptop computer featuring an Intel Core i7-4720HQ 2.60 GHz CPU, 8 GB RAM memory and a 3GB NVIDIA GeForce RTX 1080 GPU. The algorithms were implemented in MatLab (2020a, MathWorks, Natick, MA, USA).
Regarding the evaluation of the method, five-fold cross-validation was used: from 183 cross-sectional images of coronary images of 33 patients, 6 groups were randomly selected. Each group consisted of images of 5 different patients, apart from one group that consisted of images of 3 patients. To assess the efficiency of the proposed pipeline, 5 rounds of tests were applied. In each run, one of the aforementioned groups was used as the validation set, one as the test set, and the remaining images were used as the training set. Training, validation, and test sets combined consisted of 196,580 A-lines (11,333 calcified, 39,956 fibrous, 17,724 mixed plaque, 40,183 lipid plaque, and 87,384 normal tissue).
5. Discussion
Most methods presented in
Section 2 do not consider all the plaque types that can be identified in OCT images by medical experts [
9]. More often, these methods do not distinguish fibrous plaque. Our study includes two more types of plaque than the previously mentioned works [
22,
23]. Specifically, we considered four plaque types: fibrous, lipid, calcific, and mixed. A-line classification methods were based on a quite arbitrary outer wall border segmentation to select the input (A-line patch) to the classifier. Alternatively, other methods used whole A-lines (from the catheter to maximal depth). We selected A-line patches that start from the arterial lumen-wall border and include pixels that are part of the arterial wall. Instead of choosing an endpoint pixel at a certain depth, we compared the classification results of our pipeline giving as input A-line patches of different sizes. In addition, inspired by the work of Boi et al. [
26] and using Liu et al. [
29] approach, we estimated the attenuation coefficient in every pixel of each image. The outcome images offered better A-line classification results than the images based on original intensities. Finally, the CNN architecture and the various training options we proposed were the result of testing, OCT-specific intuition, and adoption of ideas that we considered to fit the problem.
More common methods of splitting the image into square patches were not preferred. Instead, A-lines were chosen because the stratification of the tissue of the arterial wall is the major anatomic feature of the plaque. Concerning A-line segment selection, our estimation is that it is better to extract the more meaningful part that light can be backscattered than the whole A-line used in other studies. The co-occurrence of two plaques in the same wall part and the extent of plaques were not examined in this work since this adds high variability in classes. In the case of co-occurrence of plaques, the dataset is too small for reliable classification Further, the problem of OCT penetration in the vessel wall depending on the OCT technology used is an issue we need to take into account when developing four class classifiers.
The data examined are single frames, and after extended testing using ALEXnet, we achieved the results presented in this paper. Of course, a deep learning approach is desirable to be applied on a large set of data so that the evaluation of the deep learning approach can reach as high as possible performance metrics. Given the size of the data we used, we believe that the proposed architecture achieved the best possible evaluation metrics. Further, various data augmentation techniques are not a favorable option because of the specific nature of OCT A-lines. The generative adversarial network (GAN) for data augmentation may be a suitable option, and it can be employed in future work given the availability of more data. In this case, we believe that it is best to add in vivo images, which offer real and not synthetic variability, to the dataset. One other aspect of the data to be considered is that by transforming images to polar coordinates, images are represented with their original acquisition form. The method could be applied to raw data (possibly with better results since calibration markers and going back and forth to coordinates’ systems introduces errors).
One more important point to stress is that computer vision algorithms’ efficiency can be enhanced with specialized pre-processing relevant to the modality, especially in the case of medical images. The attenuation properties of the tissue were examined in this study. Backscattering properties could be examined too, but according to [
29], backscattering coefficients in homogeneous regions are linearly related to the attenuation coefficients. Experiments were carried out with the same setup for attenuation images and for original images. The setup of the attenuation images surpassed at least slightly the performance of the method in comparison to the setup of the original data. An interesting finding in this work is that in the classification of A-lines to fibrolipidic or fibrocalcific, the accuracy was enhanced from 74.94% to 83.4% by using attenuation coefficients estimation.
Concerning deep learning methods, possibly, there is room for improvement since the exhaustive use of original architectures and subsequent hyper-parameter tuning may lead to better classification. However, several tests were carried out with much different convolutional network set-ups that did not achieve significantly different results. Therefore, we believe that we have acquired the maximum of the descriptive features that can be acquired by this dataset.
It was chosen to split the dataset into three parts: training, validation, and testing. Each part was derived from a different random group of patients. The validation set was used in training for regularization, and the test set was not used at all in training, which stopped after several epochs with no training accuracy improvement. Therefore, this method has the potential to generalize new data if there is an availability of such data.
In conclusion, there are four key steps that are original in the method presented: initial image segmentation (A-lines are not considered whole but part of the arterial wall), the use of OCT-specific transformations, a specific CNN architecture for A-line classification and simple but efficient post-processing. Finally, the annotated dataset developed and used for the purposes of this study is a valuable resource for our scientific community. The results and especially the sensitivity in detecting abnormal A-lines can be found satisfactory since we considered more types of tissues than other studies: normal, lipid, fibrous, calcified, and mixed. Only Athanasiou et al. [
15] examined the same tissue types, but they used many heuristic and hand-crafted steps, and therefore, their method cannot easily generalize to new data. The method presented here is very slightly dependent on parameters and has the potential for a more complete semantic analysis of the intravascular OCT images.