Application and Future Prospective of Lactic Acid Bacteria as Natural Additives for Silage Production—A Review
Abstract
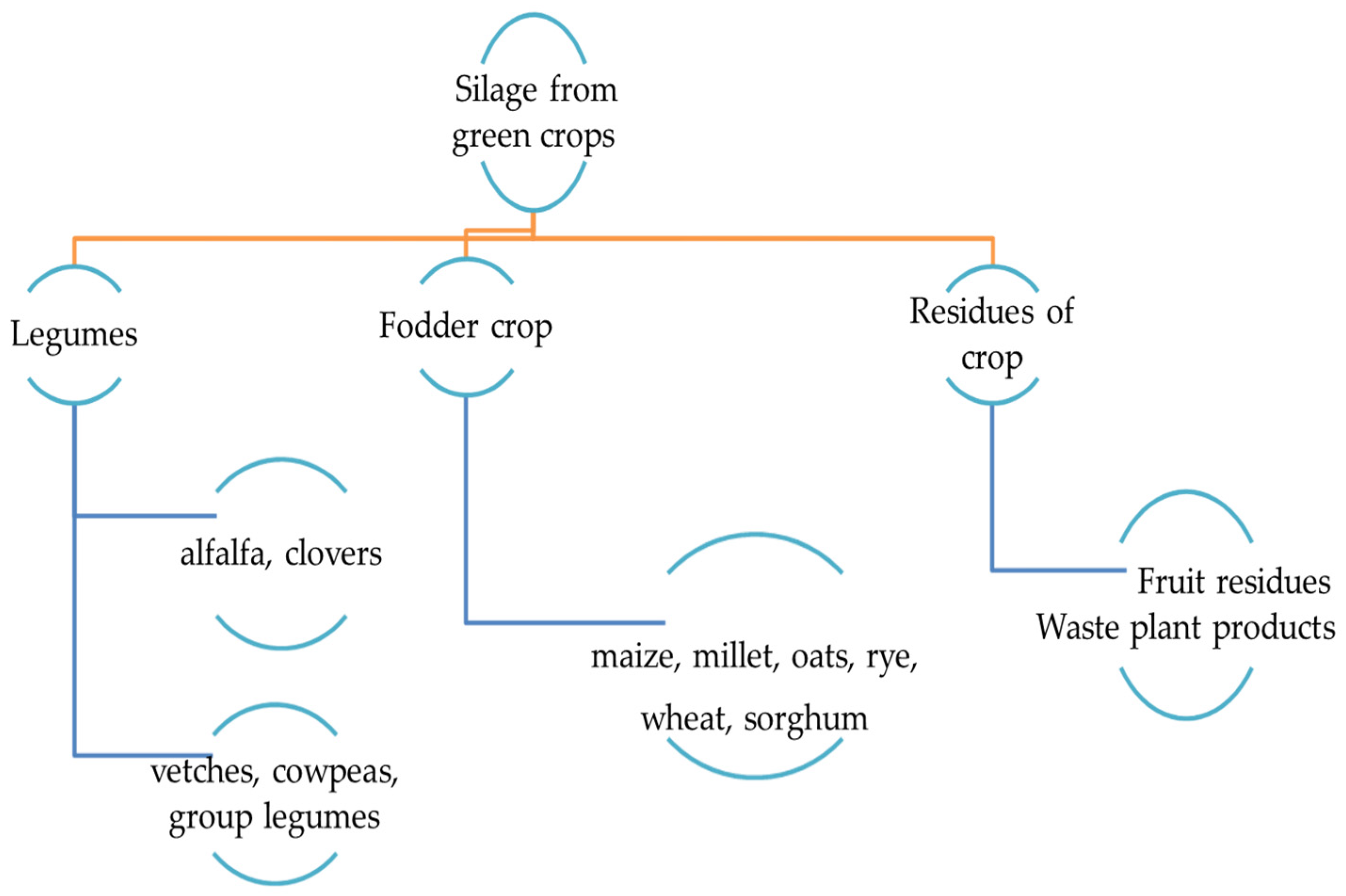
:1. Introduction
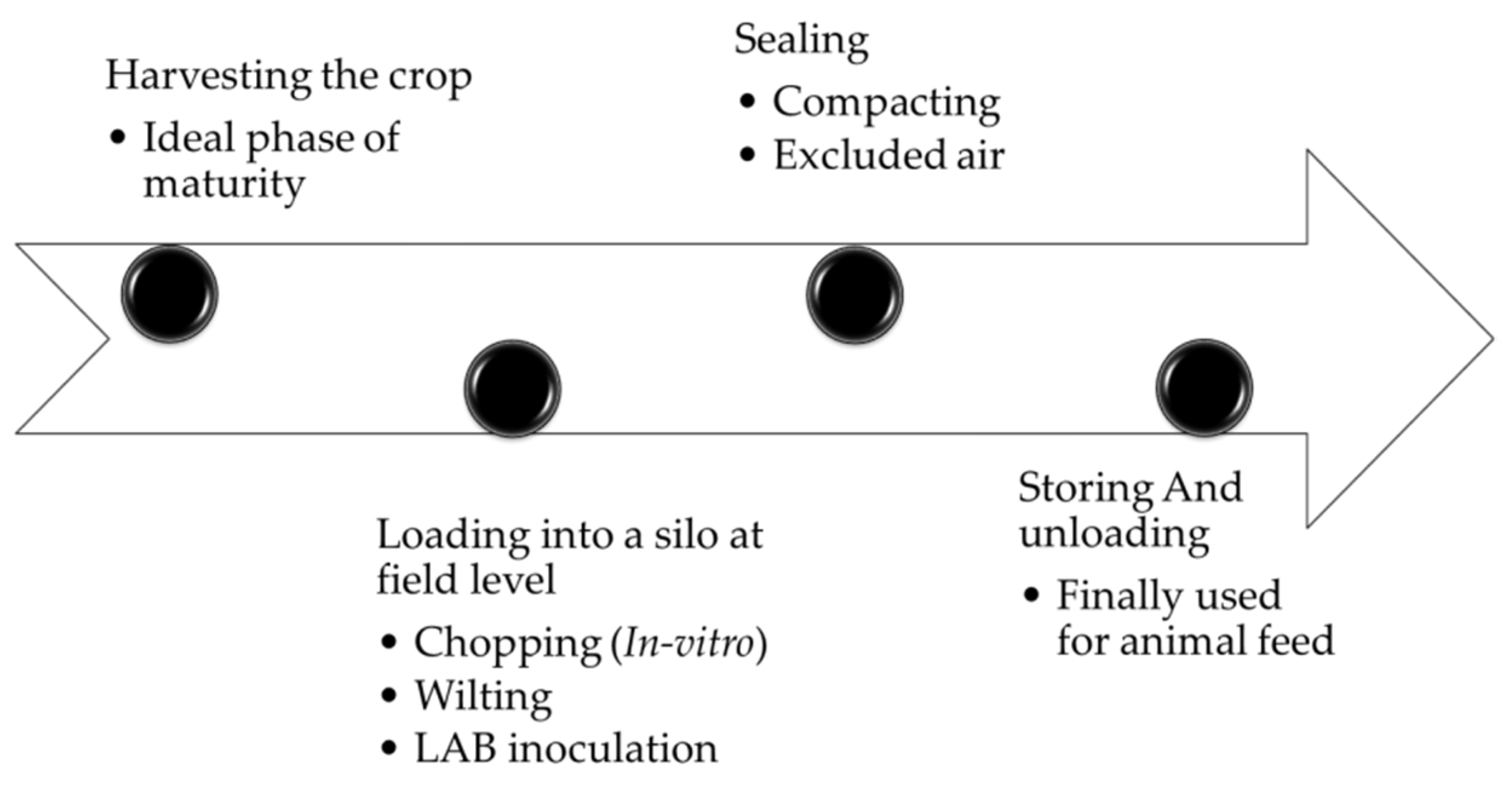
2. Features and the Procedure in Producing Silage
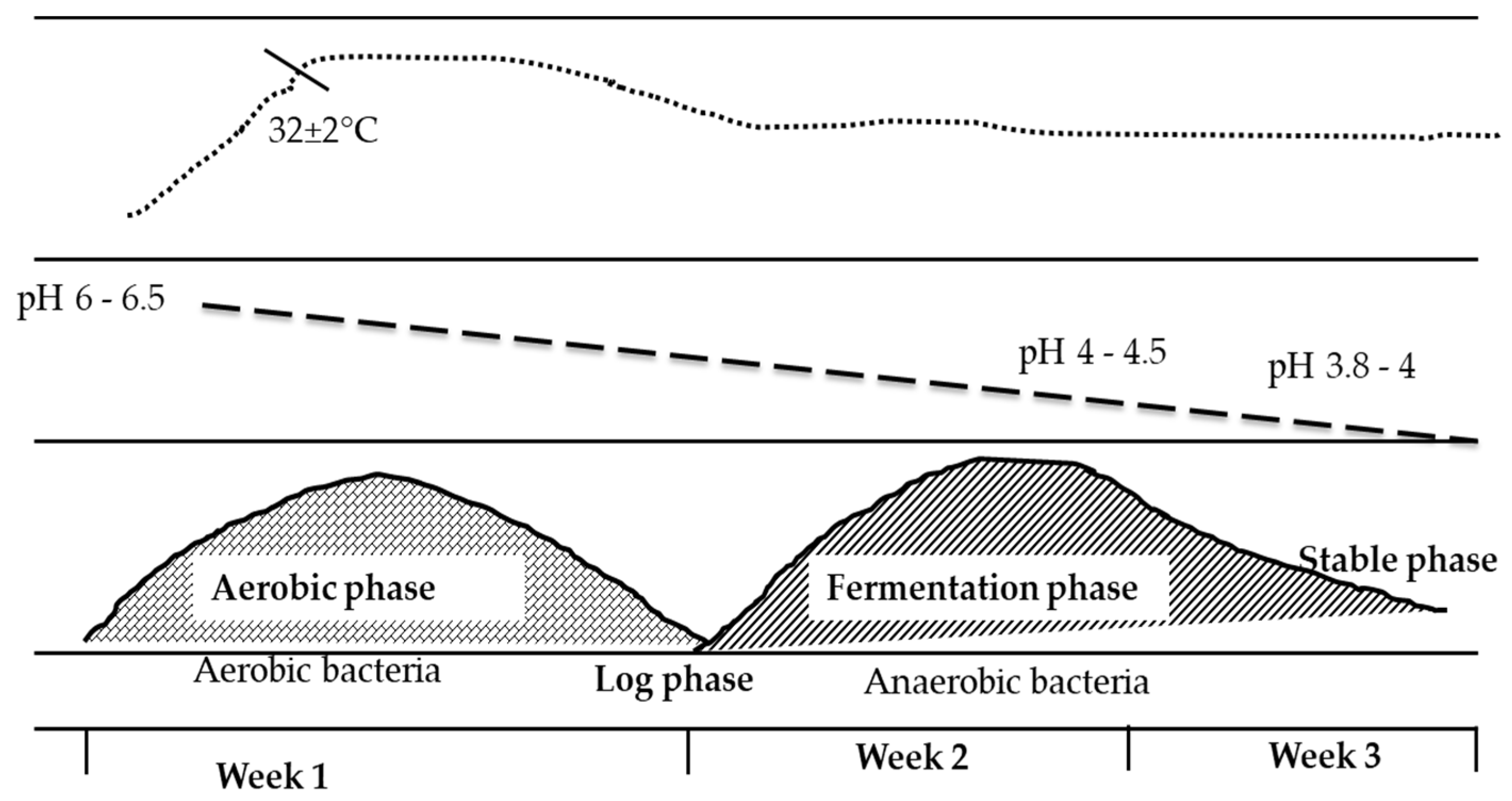
3. What Happens to Samples When They Are Ensiled at Different Stages?
4. Naturally Occurring Microbiota in Plant Samples
5. Beneficial Microbiota and Their Impact on Ensiled Silages
6. Undesirable Microbial Population and Its Complication in Ensiled Silages
6.1. Yeasts and Molds
6.2. Bacterial Species
6.3. Bacterial Synthesis of Biogenic Amines
7. Synthetic and Natural Additives for Silage Production
7.1. Synthetic Additives for Silage Production
7.2. Natural Additives for Silage Production
8. Changes in Fermented Silage by LAB
9. Insight on Homo- and Heterofermentative LAB
9.1. Homofermentative LAB
9.2. Heterofermentative LAB
10. Criteria for Selection of LAB for Silage Production
11. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Oliveira, A.S.; Weinberg, Z.G.; Ogunade, I.M.; Cervantes, A.A.P.; Arriola, K.G.; Jiang, Y.; Kim, D.; Li, X.; Goncalves, M.C.M.; Vyas, D.; et al. Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J. Dairy Sci. 2017, 100, 4587–4603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Dov, E.; Shapiro, O.H.; Siboni, N.; Kushmaro, A. Advantage of using inosine at the 3’ termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl. Environ. Microbiol. 2006, 72, 6902–6906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kung, L., Jr.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage review: Interpretation of chemical, microbial, and organoleptic components of silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef]
- Pahlow, G.; Muck, R.; Driehuis, F.; Oude Elferink, S.; Spoelstra, S.F. Microbiology of Ensiling. Silage Sci. Technol. 2003, 42, 31–93. [Google Scholar]
- Weinberg, Z.G.; Muck, R.E. New trends and opportunities in the development and use of inoculants for silage. FEMS Microbiol. Rev. 1996, 19, 53–68. [Google Scholar] [CrossRef]
- Weinberg, Z.G.; Ashbell, G.; Chen, Y.; Gamburg, M.; Sela, S. The effect of sewage irrigation on safety and hygiene of forage crops and silage. Anim. Feed Sci. Technol. 2004, 116, 271–280. [Google Scholar] [CrossRef]
- Wambacq, E.; Vanhoutte, I.; Audenaert, K.; De Gelder, L.; Haesaert, G. Occurrence, prevention and remediation of toxigenic fungi and mycotoxins in silage: A review. J. Sci. Food Agric. 2016, 96, 2284–2302. [Google Scholar] [CrossRef]
- Ávila, C.L.S.; Pinto, J.C.; Figueiredo, H.C.P.; Schwan, R.F. Effects of an indigenous and a commercial Lactobacillus buchneri strain on quality of sugar cane silage. Grass Forage Sci. 2009, 64, 384–394. [Google Scholar] [CrossRef]
- Santos, A.O.; Ávila, C.L.; Pinto, J.C.; Carvalho, B.F.; Dias, D.R.; Schwan, R.F. Fermentative profile and bacterial diversity of corn silages inoculated with new tropical lactic acid bacteria. J. Appl. Microbiol. 2016, 120, 266–279. [Google Scholar] [CrossRef]
- Ávila, C.L.S.; Carvalho, B.F. Silage fermentation—updates focusing on the performance of micro-organisms. J. Appl. Microbiol. 2020, 128, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Soundharrajan, I.; Kuppusamy, P.; Park, H.; Kim, J.; Kim, W.; Jung, J.; Choi, K. Lactic Acid Bacteria Mixture as Inoculants on Low Moisture Italian Ryegrass Silage Fermentation. J. Korean Soc. Grassl. Forage Sci. 2019, 39, 127–131. [Google Scholar] [CrossRef]
- Kuppusamy, P.; Soundharrajan, I.; Park, H.; Kim, J.; Kim, W.; Jung, J.; Choi, K. Effects of Lactic Acid Bacteria Inoculants on Fermentation of Low Moisture Fresh Rice Straw Silage at Different Storage Periods. J. Korean Soc. Grassl. Forage Sci. 2019, 39, 165–170. [Google Scholar] [CrossRef]
- Borreani, G.; Piano, S.; Tabacco, E. Aerobic stability of maize silage stored under plastic films with different oxygen permeability. J. Sci. Food Agric. 2014, 94, 2684–2690. [Google Scholar] [CrossRef]
- Borreani, G.; Tabacco, E.; Cavallarin, L. A new oxygen barrier film reduces aerobic deterioration in farm-scale corn silage. J. Dairy Sci. 2007, 90, 4701–4706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muck, R.E. Recent advances in silage microbiology. ARS USDA Submiss. 2013, 22, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Ogunade, I.M.; Jiang, Y.; Pech Cervantes, A.A.; Kim, D.H.; Oliveira, A.S.; Vyas, D.; Weinberg, Z.G.; Jeong, K.C.; Adesogan, A.T. Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: Effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 2018, 101, 2048–2059. [Google Scholar] [CrossRef]
- Liu, Y.W.; Liong, M.T.; Tsai, Y.C. New perspectives of Lactobacillus plantarum as a probiotic: The gut-heart-brain axis. J. Microbiol. 2018, 56, 601–613. [Google Scholar] [CrossRef]
- Jones, D.J.C. The Biochemistry of Silage (2nd edn), by p. McDonald, A. R. Henderson & S. J. E. Heron. 340 pp. Kingston, Kent: Chalcombe Publications (1991). £49.50 (UK) £55.00 (elsewhere) (hardback). ISBN 0 948617 22 5. J. Agric. Sci. 2009, 117, 386. [Google Scholar] [CrossRef]
- Vandamme, P.; De Bruyne, K.; Pot, B. Lactic Acid Bacteria: Biodiversity and Taxonomy; John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 31–44. [Google Scholar]
- Pang, H.; Zhang, M.; Qin, G.; Tan, Z.; Li, Z.; Wang, Y.; Cai, Y. Identification of lactic acid bacteria isolated from corn stovers. Anim. Sci. J. = Nihon Chikusan Gakkaiho 2011, 82, 642–653. [Google Scholar] [CrossRef]
- Napasirth, V.; Napasirth, P.; Sulinthone, T.; Phommachanh, K.; Cai, Y. Microbial population, chemical composition and silage fermentation of cassava residues. Anim. Sci. J. 2015, 86, 842–848. [Google Scholar] [CrossRef]
- Müller, T.; Ulrich, A.; Ott, E.-M.; Müller, M. Identification of plant-associated enterococci. J. Appl. Microbiol. 2001, 91, 268–278. [Google Scholar] [CrossRef]
- Wang, L.-T.; Kuo, H.-P.; Wu, Y.-C.; Tai, C.-J.; Lee, F.-L. Lactobacillus taiwanensis sp. nov., isolated from silage. Int. J. Syst. Evol. Microbiol. 2009, 59, 2064–2068. [Google Scholar] [CrossRef] [PubMed]
- McAllister, T.A.; Dunière, L.; Drouin, P.; Xu, S.; Wang, Y.; Munns, K.; Zaheer, R. Silage review: Using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 2018, 101, 4060–4074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O‘Brien, M.; O’Kiely, P.; Forristal, P.D.; Fuller, H. Fungi isolated from contaminated baled grass silage on farms in the Irish Midlands. FEMS Microbiol. Lett. 2005, 247, 131–135. [Google Scholar] [CrossRef] [Green Version]
- van Egmond, H.P. Natural toxins: Risks, regulations and the analytical situation in Europe. Anal. Bioanal. Chem. 2004, 378, 1152–1160. [Google Scholar] [CrossRef]
- Richard, E.; Heutte, N.; Sage, L.; Pottier, D.; Bouchart, V.; Lebailly, P.; Garon, D. Toxigenic fungi and mycotoxins in mature corn silage. Food Chem. Toxicol. 2007, 45, 2420–2425. [Google Scholar] [CrossRef]
- O’Brien, M.; Nielsen, K.F.; O’Kiely, P.; Forristal, P.D.; Fuller, H.T.; Frisvad, J.C. Mycotoxins and Other Secondary Metabolites Produced in Vitro by Penicillium paneum Frisvad and Penicillium roqueforti Thom Isolated from Baled Grass Silage in Ireland. J. Agric. Food Chem. 2006, 54, 9268–9276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driehuis, F.; Wikselaar, P. The occurrence and prevention of ethanol fermentation in high-dry-matter grass silage. J. Sci. Food Agric. 2000, 80, 711–718. [Google Scholar] [CrossRef]
- Orsi, R.B.; Corrêa, B.; Possi, C.R.; Schammass, E.A.; Nogueira, J.R.; Dias, S.M.C.; Malozzi, M.A.B. Mycoflora and occurrence of fumonisins in freshly harvested and stored hybrid maize. J. Stored Prod. Res. 2000, 36, 75–87. [Google Scholar] [CrossRef]
- Roigé, M.B.; Aranguren, S.M.; Riccio, M.B.; Pereyra, S.; Soraci, A.L.; Tapia, M.O. Mycobiota and mycotoxins in fermented feed, wheat grains and corn grains in Southeastern Buenos Aires Province, Argentina. Rev. Iberoam. De Micol. 2009, 26, 233–237. [Google Scholar] [CrossRef]
- Niderkorn, V.; Morgavi, D.P.; Pujos, E.; Tissandier, A.; Boudra, H. Screening of fermentative bacteria for their ability to bind and biotransform deoxynivalenol, zearalenone and fumonisins in an in vitro simulated corn silage model. Food Addit. Contam. 2007, 24, 406–415. [Google Scholar] [CrossRef] [Green Version]
- Murphy, P.A.; Hendrich, S.; Landgren, C.; Bryant, C.M. Food Mycotoxins: An Update. J. Food Sci. 2006, 71, R51–R65. [Google Scholar] [CrossRef]
- Cavallarin, L.; Tabacco, E.; Antoniazzi, S.; Borreani, G. Aflatoxin accumulation in whole crop maize silage as a result of aerobic exposure. J. Sci. Food Agric. 2011, 91, 2419–2425. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Riley, R.T. An historical overview of field disease outbreaks known or suspected to be caused by consumption of feeds contaminated with Fusarium toxins. Anim. Feed Sci. Technol. 2007, 137, 201–212. [Google Scholar] [CrossRef]
- Myllykoski, J.; Lindström, M.; Keto-Timonen, R.; Söderholm, H.; Jakala, J.; Kallio, H.; Sukura, A.; Korkeala, H. Type C bovine botulism outbreak due to carcass contaminated non-acidified silage. Epidemiol. Infect. 2009, 137, 284–293. [Google Scholar] [CrossRef] [Green Version]
- Mobashar, M.; Hummel, J.; Blank, R.; Südekum, K.-H. Ochratoxin A in Ruminants–A Review on Its Degradation by Gut Microbes and Effects on Animals. Toxins 2010, 2, 809–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudra, H. Mycotoxins: An insidious menacing factor for the quality of forages and the performances of the ruminants. Fourrages 2009, 199, 265–280. [Google Scholar]
- Vissers, M.M.M.; Te Giffel, M.C.; Driehuis, F.; De Jong, P.; Lankveld, J.M.G. Minimizing the Level of Bacillus cereus Spores in Farm Tank Milk. J. Dairy Sci. 2007, 90, 3286–3293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- te Giffel, M.C.; Wagendorp, A.; Herrewegh, A.; Driehuis, F. Bacterial spores in silage and raw milk. Antonie van Leeuwenhoek 2002, 81, 625–630. [Google Scholar] [CrossRef]
- Abee, T.; Groot, M.N.; Tempelaars, M.; Zwietering, M.; Moezelaar, R.; van der Voort, M. Germination and outgrowth of spores of Bacillus cereus group members: Diversity and role of germinant receptors. Food Microbiol. 2011, 28, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Aureli, P.; Franciosa, G.; Scalfaro, C. Pathogens in Milk | Clostridium spp. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 47–53. [Google Scholar] [CrossRef]
- Lindström, M.; Myllykoski, J.; Sivelä, S.; Korkeala, H. Clostridium botulinum in cattle and dairy products. Crit. Rev. Food Sci. Nutr. 2010, 50, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.L.; McAdams, S.C.; Whitlock, R.H. Type A botulism in horses in the United States: A review of the past ten years (1998–2008). J. Vet. Diagn. Investig. 2010, 22, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, L.; Mohammed, H.O.; McDonough, P.L. Farm-management and milking practices associated with the presence of Listeria monocytogenes in New York state dairy herds. Prev. Vet. Med. 2001, 51, 63–73. [Google Scholar] [CrossRef]
- Dhama, K.; Karthik, K.; Tiwari, R.; Shabbir, M.Z.; Barbuddhe, S.; Malik, S.V.S.; Singh, R.K. Listeriosis in animals, its public health significance (food-borne zoonosis) and advances in diagnosis and control: A comprehensive review. Vet. Q. 2015, 35, 211–235. [Google Scholar] [CrossRef]
- Schoder, D.; Melzner, D.; Schmalwieser, A.; Zangana, A.; Winter, P.; Wagner, M. Important vectors for Listeria monocytogenes transmission at farm dairies manufacturing fresh sheep and goat cheese from raw milk. J. Food Prot. 2011, 74, 919–924. [Google Scholar] [CrossRef]
- Vilar, M.J.; Yus, E.; Sanjuán, M.L.; Diéguez, F.J.; Rodríguez-Otero, J.L. Prevalence of and risk factors for Listeria species on dairy farms. J. Dairy Sci. 2007, 90, 5083–5088. [Google Scholar] [CrossRef]
- Cernicchiaro, N.; Pearl, D.L.; Ghimire, S.; Gyles, C.L.; Johnson, R.P.; LeJeune, J.T.; Ziebell, K.; McEwen, S.A. Risk factors associated with Escherichia coli O157:H7 in Ontario beef cow-calf operations. Prev. Vet. Med. 2009, 92, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Fenlon, D.R.; Wilson, J. Growth of Escherichia coli O157 in poorly fermented laboratory silage: A possible environmental dimension in the epidemiology of E. coli O157. Lett. Appl. Microbiol. 2000, 30, 118–121. [Google Scholar] [CrossRef]
- Dunière, L.; Gleizal, A.; Chaucheyras-Durand, F.; Chevallier, I.; Thévenot-Sergentet, D. Fate of Escherichia coli O26 in corn silage experimentally contaminated at ensiling, at silo opening, or after aerobic exposure, and protective effect of various bacterial inoculants. Appl. Environ. Microbiol. 2011, 77, 8696–8704. [Google Scholar] [CrossRef]
- Baines, D.; Erb, S.; Turkington, K.; Kuldau, G.; Juba, J.; Masson, L.; Mazza, A.; Roberts, R. Mouldy feed, mycotoxins and Shiga toxin - producing Escherichia colicolonization associated with Jejunal Hemorrhage Syndrome in beef cattle. BMC Vet. Res. 2011, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, M.V. Tuberculosis: A reemerging disease at the interface of domestic animals and wildlife. Curr. Top. Microbiol. Immunol. 2007, 315, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.M.; Srinivasan, V.; Murinda, S.E.; Oliver, S.P. Detection of Campylobacter jejuni in dairy farm environmental samples using SYBR Green real-time polymerase chain reaction. Foodborne Pathog. Dis. 2005, 2, 160–168. [Google Scholar] [CrossRef]
- Plym, F.L.; Wierup, M. Salmonella contamination: A significant challenge to the global marketing of animal food products. Rev. Sci. et Tech. 2006, 25, 541–554. [Google Scholar]
- Aschenbach, J.R.; Gäbel, G. Effect and absorption of histamine in sheep rumen: Significance of acidotic epithelial damage. J. Anim. Sci. 2000, 78, 464–470. [Google Scholar] [CrossRef]
- Steidlová, S.; Kalac, P. The effects of using lactic acid bacteria inoculants in maize silage on the formation of biogenic amines. Arch. fur Tierernahr. 2003, 57, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Knický, M.; Lingvall, P. Possibilities to avoid growth of clostridia and/or fungi in wilted silage by use of organic and inorganic salts. In Proceedings of the XIX International Grassland Congress Brazil, São pedro, Brazil, 11–21 February 2001; pp. 788–789. [Google Scholar]
- Biro, D.; Juracek, M.; Kacaniova, M.; Simko, M.; Galik, B.; Michalkova, J.; Gyongyova, E. Occurrence of microscopic fungi and mycotoxins in conserved high moisture corn from Slovakia. Ann. Agric. Environ. Med. 2009, 16, 227–232. [Google Scholar]
- Nascimento Agarussi, M.C.; Gomes Pereira, O.; Paula, R.A.d.; Silva, V.P.d.; Santos Roseira, J.P.; Fonseca e Silva, F. Novel lactic acid bacteria strains as inoculants on alfalfa silage fermentation. Sci. Rep. 2019, 9, 8007. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Yao, D.; Li, D.; Lin, Y.; Bureenok, S.; Ni, K.; Yang, F. Effects of Lactic Acid Bacteria Isolated From Rumen Fluid and Feces of Dairy Cows on Fermentation Quality, Microbial Community, and in vitro Digestibility of Alfalfa Silage. Front. Microbiol. 2020, 10, 2998. [Google Scholar] [CrossRef]
- Puntillo, M.; Gaggiotti, M.; Oteiza, J.M.; Binetti, A.; Massera, A.; Vinderola, G. Potential of Lactic Acid Bacteria Isolated From Different Forages as Silage Inoculants for Improving Fermentation Quality and Aerobic Stability. Front. Microbiol. 2020, 11, 3091. [Google Scholar] [CrossRef]
- Kuppusamy, P.; Kim, D.; Soundharrajan, I.; Park, H.S.; Jung, J.S.; Yang, S.H.; Choi, K.C. Low-Carbohydrate Tolerant LAB Strains Identified from Rumen Fluid: Investigation of Probiotic Activity and Legume Silage Fermentation. Microorganisms 2020, 8, 1044. [Google Scholar] [CrossRef]
- Ni, K.; Wang, Y.; Cai, Y.; Pang, H. Natural Lactic Acid Bacteria Population and Silage Fermentation of Whole-crop Wheat. Asian-Australas J. Anim. Sci. 2015, 28, 1123–1132. [Google Scholar] [CrossRef]
- Zhang, T.; Li, L.; Wang, X.-f.; Zeng, Z.-h.; Hu, Y.-g.; Cui, Z.-j. Effects of Lactobacillus buchneri and Lactobacillus plantarum on fermentation, aerobic stability, bacteria diversity and ruminal degradability of alfalfa silage. World J. Microbiol. Biotechnol. 2009, 25, 965–971. [Google Scholar] [CrossRef]
- Fabiszewska, A.U.; Zielińska, K.J.; Wróbel, B. Trends in designing microbial silage quality by biotechnological methods using lactic acid bacteria inoculants: A minireview. World J. Microbiol. Biotechnol. 2019, 35, 76. [Google Scholar] [CrossRef] [Green Version]
- Amaral, R.C.; Carvalho, B.F.; Costa, D.M.; Morenz, M.J.F.; Schwan, R.F.; Ávila, C.L.d.S. Novel lactic acid bacteria strains enhance the conservation of elephant grass silage cv. BRS Capiaçu. Anim. Feed Sci. Technol. 2020, 264, 114472. [Google Scholar] [CrossRef]
- Zhao, S.S.; Wang, Y.P.; Yang, F.Y.; Wang, Y.; Zhang, H. Screening a Lactobacillus plantarum strain for good adaption in alfalfa ensiling and demonstrating its improvement of alfalfa silage quality. J. Appl. Microbiol. 2020, 129, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.M.; Rinne, M. Highlights of progress in silage conservation and future perspectives. Grass Forage Sci. 2018, 73, 40–52. [Google Scholar] [CrossRef]
- Ávila, C.L.S.; Carvalho, B.F.; Pinto, J.C.; Duarte, W.F.; Schwan, R.F. The use of Lactobacillus species as starter cultures for enhancing the quality of sugar cane silage. J. Dairy Sci. 2014, 97, 940–951. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Shuai, Y.; Yan, Y.; Ran, Q.; Wang, X.; Li, D.; Cai, Y.; Zhang, X. Microbial Community and FermentationDynamics of Corn Silage Prepared withHeat-Resistant Lactic Acid Bacteria in a HotEnvironment. Microorganisms 2020, 8, 719. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Zhou, W.; Yang, F.-y.; Chen, X.-y.; Zhang, Q. Effects of Wilting and Lactobacillus plantarum Addition on the Fermentation Quality and Microbial Community of Moringa oleifera Leaf Silage. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.; Sun, L.; Chen, C.; Lin, J.; Yang, F.; Cai, Y. Exploring the microbial community structure and metabolic gene clusters during silage fermentation of paper mulberry, and developing the high-protein woody plant as ruminant feed. bioRxiv 2020, 275, 114766. [Google Scholar] [CrossRef]
- Pereira, G.A.; Santos, E.M.; Araújo, G.G.L.; Oliveira, J.S.; Pinho, R.M.A.; Zanine, A.d.M.; Souza, A.F.N.; Macedo, A.J.S.; Neto, J.M.C.; Nascimento, T.V.C. Isolation and identification of lactic acid bacteria in fresh plants and in silage from Opuntia and their effects on the fermentation and aerobic stability of silage. J. Agric. Sci. 2020, 157, 684–692. [Google Scholar] [CrossRef]
- Huyen, N.; Martinez, I.; Pellikaan, W. Using Lactic Acid Bacteria as Silage Inoculants or Direct-Fed Microbials to Improve In Vitro Degradability and Reduce Methane Emissions in Dairy Cows. Agronomy 2020, 10, 1482. [Google Scholar] [CrossRef]
- Broberg, A.; Jacobsson, K.; Ström, K.; Schnürer, J. Metabolite profiles of lactic acid bacteria in grass silage. Appl. Environ. Microbiol. 2007, 73, 5547–5552. [Google Scholar] [CrossRef] [Green Version]
- Lindqvist, H.; Nadeau, E.; Jensen, S.K. Alpha-tocopherol and β-carotene in legume–grass mixtures as influenced by wilting, ensiling and type of silage additive. Grass Forage Sci. 2012, 67, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.Q.; Wei, S.N.; Liu, C.; Kim, H.J.; Kim, J.G. Effect of harvest dates on β-carotene content and forage quality of rye (Secale cereale L.) silage and hay. J. Anim. Sci. Technol. 2021, 63, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Kung, L., Jr.; Santos, M.; DerBedrosian, M. The effect of feeding cows corn silage with or without L. buchneri 40788 and supplemented with or without Levucell SC; Lallemand Animal Nutrition Internal Report; Lallemand Animal Nutrition: Milwaukee, WI, USA, 2010. [Google Scholar]
- Weinberg, Z.G.; Ashbell, G.; Hen, Y.; Azrieli, A.; Szakacs, G.; Filya, I. Ensiling whole-crop wheat and corn in large containers with Lactobacillus plantarum and Lactobacillus buchneri. J. Ind. Microbiol. Biotechnol. 2002, 28, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Blajman, J.; Vinderola, G.; Paez, R.; Signorini, M. The role of homofermentative and heterofermentative lactic acid bacteria for alfalfa silage: A meta-analysis. J. Agric. Sci. 2020, 158, 1–12. [Google Scholar] [CrossRef]
- Kung, L. Silage Additives: Where are we going? In Proceedings of the XVII International Silage Conference, Piracicaba, Brazil, 1–3 July 2015. [Google Scholar]
- Guan, H.; Ke, W.; Yan, Y.; Shuai, Y.; Li, X.; Ran, Q.; Yang, Z.; Wang, X.; Cai, Y.; Zhang, X. Screening of natural lactic acid bacteria with potential effect on silage fermentation, aerobic stability and aflatoxin B1 in hot and humid area. J. Appl. Microbiol. 2020, 128, 1301–1311. [Google Scholar] [CrossRef]
- Xu, D.; Ding, W.; Ke, W.; Li, F.; Zhang, P.; Guo, X. Modulation of Metabolome and Bacterial Community in Whole Crop Corn Silage by Inoculating Homofermentative Lactobacillus plantarum and Heterofermentative Lactobacillus buchneri. Front. Microbiol. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Hammes, W.P.; Hertel, C. Genus Lactobacillus beijerinck 1901, 212AL. The Firmicutes. In Bergey’s Manualof Systematic Bacteriology, 2nd ed.; De Vos, G.M.G., Jones, D., Krieg, N.R., Ludwig, W., Rainey, F.A., Schleifer, K., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2009; pp. 465–490. [Google Scholar]
- Pot, B. Tsakalidou, Taxonomy and metabolism of Lactobacillus; Caister Acadedmic Press: Norfolk, UK, 2009; pp. 3–58. [Google Scholar]
- Ström, K.; Sjögren, J.; Broberg, A.; Schnürer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef] [Green Version]
- Oude Elferink, S.J.; Krooneman, J.; Gottschal, J.C.; Spoelstra, S.F.; Faber, F.; Driehuis, F. Anaerobic conversion of lactic acid to acetic acid and 1, 2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 2001, 67, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Sriramulu, D.D.; Liang, M.; Hernandez-Romero, D.; Raux-Deery, E.; Lünsdorf, H.; Parsons, J.B.; Warren, M.J.; Prentice, M.B. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J. Bacteriol. 2008, 190, 4559–4567. [Google Scholar] [CrossRef] [Green Version]
- Nsereko, V.L.; Smiley, B.K.; Rutherford, W.M.; Spielbauer, A.; Forrester, K.J.; Hettinger, G.H.; Harman, E.K.; Harman, B.R. Influence of inoculating forage with lactic acid bacterial strains that produce ferulate esterase on ensilage and ruminal degradation of fiber. Anim. Feed Sci. Technol. 2008, 145, 122–135. [Google Scholar] [CrossRef]
- Borreani, G.; Tabacco, E.; Schmidt, R.J.; Holmes, B.J.; Muck, R.E. Silage review: Factors affecting dry matter and quality losses in silages. J. Dairy Sci. 2018, 101, 3952–3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrero, F.; Piano, S.; Tabacco, E.; Borreani, G. Effects of conservation period and Lactobacillus hilgardii inoculum on the fermentation profile and aerobic stability of whole corn and sorghum silages. J. Sci. Food Agric. 2019, 99, 2530–2540. [Google Scholar] [CrossRef] [PubMed]
- Ganzorig, O.; Sumisa, F.; Batjargal, B.; Yoshida, T. Isolation and Identification of new Lactic Acid Bacteria with Potent Biological Activity and Yeasts in Airag, a Traditional Mongolian Fermented Beverage. Food Sci. Technol. Res. 2016, 22, 575–582. [Google Scholar] [CrossRef]
- Valerio, F.; Lavermicocca, P.; Pascale, M.; Visconti, A. Production of phenyllactic acid by lactic acid bacteria: An approach to the selection of strains contributing to food quality and preservation. FEMS Microbiol. Lett. 2004, 233, 289–295. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Hu, W.; Mills, J.A.; Kung, L., Jr. The development of lactic acid bacteria and Lactobacillus buchneri and their effects on the fermentation of alfalfa silage. J. Dairy Sci. 2009, 92, 5005–5010. [Google Scholar] [CrossRef] [Green Version]
- Johanningsmeier, S.D.; McFeeters, R.F. Metabolism of lactic acid in fermented cucumbers by Lactobacillus buchneri and related species, potential spoilage organisms in reduced salt fermentations. Food Microbiol. 2013, 35, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Reich, L.J.; Kung, L. Effects of combining Lactobacillus buchneri 40788 with various lactic acid bacteria on the fermentation and aerobic stability of corn silage. Anim. Feed Sci. Technol. 2010, 159, 105–109. [Google Scholar] [CrossRef]
- Driehuis, F.; Oude Elferink, S.J.W.H.; Van Wikselaar, P.G. Fermentation characteristics and aerobic stability of grass silage inoculated with Lactobacillus buchneri, with or without homofermentative lactic acid bacteria. Grass Forage Sci. 2001, 56, 330–343. [Google Scholar] [CrossRef]
- Addah, W.; Baah, J.; Okine, E.K.; McAllister, T.A. A third-generation esterase inoculant alters fermentation pattern and improves aerobic stability of barley silage and the efficiency of body weight gain of growing feedlot cattle1. J. Anim. Sci. 2012, 90, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Arriola, K.G.; Queiroz, O.C.M.; Romero, J.J.; Casper, D.; Muniz, E.; Hamie, J.; Adesogan, A.T. Effect of microbial inoculants on the quality and aerobic stability of bermudagrass round-bale haylage. J. Dairy Sci. 2015, 98, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yuan, X.; Dong, Z.; Li, J.; Guo, G.; Bai, Y.; Zhang, J.; Shao, T. Characteristics of isolated lactic acid bacteria and their effects on the silage quality. Asian-Australas J. Anim. Sci. 2017, 30, 819–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, B.F.; Sales, G.F.C.; Schwan, R.F.; Ávila, C.L.S. Criteria for lactic acid bacteria screening to enhance silage quality. J. Appl. Microbiol. 2021, 130, 341–355. [Google Scholar] [CrossRef]
- Soundharrajan, I.; Kim, D.H.; Srisesharam, S.; Kuppusamy, P.; Park, H.S.; Yoon, Y.H.; Kim, W.H.; Song, Y.G.; Choi, K.C. Application of customised bacterial inoculants for grass haylage production and its effectiveness on nutrient composition and fermentation quality of haylage. 3 Biotech 2017, 7, 321. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soundharrajan, I.; Park, H.S.; Rengasamy, S.; Sivanesan, R.; Choi, K.C. Application and Future Prospective of Lactic Acid Bacteria as Natural Additives for Silage Production—A Review. Appl. Sci. 2021, 11, 8127. https://doi.org/10.3390/app11178127
Soundharrajan I, Park HS, Rengasamy S, Sivanesan R, Choi KC. Application and Future Prospective of Lactic Acid Bacteria as Natural Additives for Silage Production—A Review. Applied Sciences. 2021; 11(17):8127. https://doi.org/10.3390/app11178127
Chicago/Turabian StyleSoundharrajan, Ilavenil, Hyung Soo Park, Sathya Rengasamy, Ravikumar Sivanesan, and Ki Choon Choi. 2021. "Application and Future Prospective of Lactic Acid Bacteria as Natural Additives for Silage Production—A Review" Applied Sciences 11, no. 17: 8127. https://doi.org/10.3390/app11178127






