Abstract
Significant growth in Electronic Health Records (EHR) over the last decade has provided an abundance of clinical text that is mostly unstructured and untapped. This huge amount of clinical text data has motivated the development of new information extraction and text mining techniques. Named Entity Recognition (NER) and Relationship Extraction (RE) are key components of information extraction tasks in the clinical domain. In this paper, we highlight the present status of clinical NER and RE techniques in detail by discussing the existing proposed NLP models for the two tasks and their performances and discuss the current challenges. Our comprehensive survey on clinical NER and RE encompass current challenges, state-of-the-art practices, and future directions in information extraction from clinical text. This is the first attempt to discuss both of these interrelated topics together in the clinical context. We identified many research articles published based on different approaches and looked at applications of these tasks. We also discuss the evaluation metrics that are used in the literature to measure the effectiveness of the two these NLP methods and future research directions.
1. Introduction
The amount of text generated every day is increasing drastically in different domains such as health care, news articles, scientific literature, and social media. Since 2010, the International Data Corporation (IDC) has predicted that the amount of data can potentially grow 50-fold to 40 billion terabytes by 2020 [1]. Textual data is very common in most domains, but automated comprehension is difficult due to its unstructured nature and has led to the design of several text mining (TM) techniques in the last decade.
TM refers to the extraction of interesting and nontrivial patterns or knowledge from text [2]. Common text mining tasks include text preprocessing, text classification, question-answering, clustering, and statistical techniques.
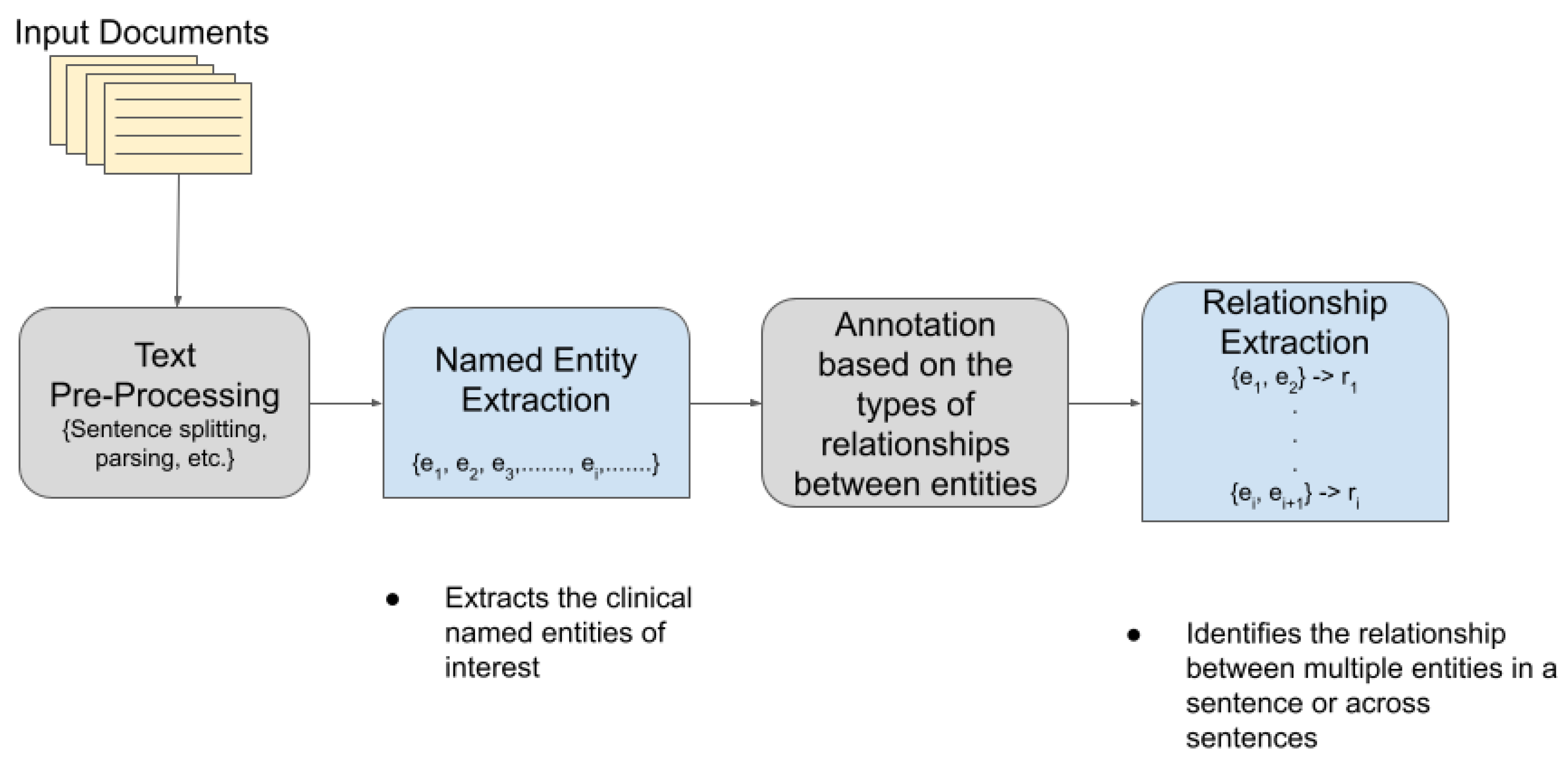
TM has become extremely popular and useful in the biomedical and healthcare domains. In healthcare, about 80% of the total medical data is unstructured and untapped after its creation [3]. This unstructured data from hospitals, healthcare clinics, or biomedical labs can come in many forms such as text, images, and signals. Out of the various text mining tasks and techniques, our goal in this paper is to review the current state-of-the-art in Clinical Named Entity Recognition (NER) and Relationship Extraction (RE)-based techniques. Clinical NER is a natural language processing (NLP) method used for extracting important medical concepts and events i.e., clinical NEs from the data [4]. Relationship Extraction (RE) is used for detecting and classifying the annotated semantic relationships between the recognized entities. Significant research on NER and RE has been carried out in the past both on clinical narratives and other types of text. For example, in the sentence, “Her white count remained elevated despite discontinuing her G-CSF”, the words in bold are the various entities in the sentence. After the entities are recognized, the relationship between two or more entities is extracted. In this case, “her white count” and “elevated” are found to be related to each other in a manner dissimilar to the nature of the relationship between “elevated” and “her G-CSF”. In the sentence “Atorvastatin is found to have therapeutic effects in breast cancer although no clinical trials are performed at present”, the NE of interest includes the name of the drug (atorvastatin) and the disease name (breast cancer), whereas the drug–disease relation (atorvastatin–breast cancer) is the relationship of interest. Figure 1 shows a pictorial representation of the association between NER and RE.
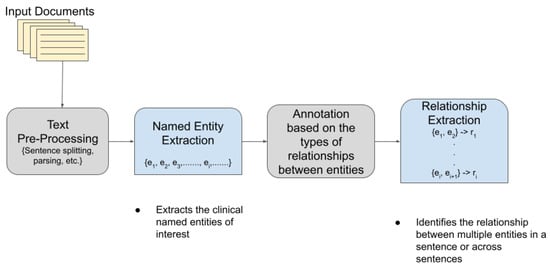
Figure 1.
Association between Named Entity Recognition and Relationship Extraction.
2. Background
Over the years, many toolkits and applications have been introduced to address different NLP tasks in the clinical domain, including NER and RE. The WEKA Data Mining Software [5] first came into existence in the late nineties. It was updated several times over the years to include NLP systems for language identification, tokenization, sentence boundary detection, and named entity recognition. Later on, the clinical NLP toolkit, CLAMP (Clinical Language Annotation, Modeling, and Processing) [6] was introduced in 2018 and provides a GUI-based state-of-the-art NLP system. CLAMP achieved good performance on NER and concept encoding and is also publicly available for research use. Comprehend Medical, a NER- and RE-related Web Service (2019) [7], is a very recent effort that introduces an NLP service launched under Amazon Web Services (AWS). Likewise, other research works have also addressed these topics, which motivates this review. A high-level overview of machine learning, neural networks, and evaluation metrics is presented below before we review clinical NER- and RE-related tasks.
2.1. Machine Learning
Machine learning (ML) is a type of data-driven Artificial Intelligence (AI) that provides the ability to learn about a system without explicit programming. ML algorithms are applied in many scientific domains and the most common applications include recommendation systems, data mining, and pattern recognition. ML is classified into one of the four subdomains:
- Supervised Learning: With these algorithms, the training data are given ground-truth labels, which can be used for learning the underlying patterns in the dataset. Classification and regression algorithms are most commonly used, including Naive Bayes [8], Support Vector Machines (SVM) [9], and Decision Trees [10].
- Unsupervised Learning: In this case, the training dataset is not given labels and, thus, many of the solutions attempt to find patterns without any prior guidance. Commonly used algorithms in this category are association rules and clustering methods, such as K-Means [11] or DBSCAN [12].
- Semi-Supervised Learning: Here, only some of the training data is labeled, putting these solutions in a space somewhere between fully supervised and unsupervised learning. Text classification [13] is one of the most common applications for semi-supervised learning.
- Reinforcement Learning: Using a reward system, a reinforcement learning agent optimizes future returns based on prior results. This iterative, continuous learning process mirrors how humans learn from their experiences when interacting with an environment. Deep Adversarial Networks [14] and Q-Learning [15] are well known reinforcement learning algorithms.
2.2. Neural Networks
The traditional machine learning algorithms often perform well with structured data but can struggle with unstructured or semi-structured data, i.e., human information processing mechanisms such as vision and speech [16]. Neural networks, specifically deep learning algorithms, have shown promising results with NLP and image analysis tasks. In neural networks, the input is processed through different layers of the network, where each layer transforms the features of the dataset following some mathematical function. The concept of neural networks follows the mechanism that the human brain uses to solve a problem. Once the data is processed through different layers within a neural network, the output layer performs the classification. In general, this approach does not require as much human intervention as the nested layers using different hierarchies try to find the hidden patterns on their own.
2.3. Common Evaluation Metrics
The F1-score is a popular evaluation metric for the two NLP functions reviewed in this paper. Comparisons can be classified as exact or relaxed match [17]. Relaxed match only considers the correct type and ignores the boundaries as long as there is an overlap with ground truth boundaries. In the case of an exact match, it is expected that the entity identified correctly should also detect boundary and type correctly at the same time [17]. The following keys are used to calculate the F-score, precision, and recall.
- True Positive (TP): A perfect match between the entity obtained by NER system and the ground truth.
- False Positive (FP): Entity detected by the NER system but not present in the ground truth.
- False Negative (FN): Entity not detected by the NER system but present in the ground truth.
- True Negative (TN): No match between the entity obtained by NER system and the ground truth.
Precision provides the number of correct results detected correctly whereas recall provides the total entities correctly detected; they are calculated as follows:
2.4. Named Entity Recognition
Named Entity Recognition (NER) is the task of identifying named entities such as specific location, treatment plan, medicines/drug, and critical health information from the clinical text. NER was first introduced in 1995 [18] where the three categories (Entity, Name, and Number) were defined. The original design idea for NER was to parse the text, to identify proper nouns from the text, and to categorize them.
NER is an extremely popular machine learning method and is also considered a base technique for many of the NLP tasks. Prior to 2011, all work on NERs was domain-specific and was designed to performing specific tasks based on ontologies. Collobert et al. [19] introduced a neural network-based NER, which for the first time, made it domain independent. This approach is now quite common, and there are many variations proposed over the last decade that leverage Recurrent Neural Networks (RNN) and word embeddings among others.
2.5. Relationship Extraction
A relationship can be extracted between any combination of named entities. An RE task is basically a classification of the semantic relationship between entities from textual data. RE between entities in any text is a vital task that facilitates its automated natural language understanding. The abundance and heterogeneity of unstructured data in any domain are hard to be fathomed by humans alone. Hence, the conversion of unstructured text into structured data by annotating its semantics needs to be automated. RE tasks are thus very useful in automating the process of identification of different relations from clinical data. Some important applications of clinical RE include gene–disease, drug–effect, disease–mutation, and disease–symptom relationships. In general, the pair-wise association between entities is considered, but in many cases, more than two entities are also involved. The process of checking whether a relationship exists between entities is a classification problem that can also be extended to multi-class classification or multi-label classification. In [20], a relation is defined as a tuple , where are the entities with a predefined relationship r within the document D. Similarly, all of the different relationships in a document can be defined.
Similar to NER, RE has been applied to many domains, including the healthcare domain. One of the oldest works on RE was published in 1999, which extracted informative patterns and relations from the World Wide Web [21]. In the following year, relationship extraction from the large plain text was conducted, where a system named Snowball introduced novel strategies for pattern generation [22]. Kernel-based methods such as dependency tree kernel-based technique [23], shortest path dependency kernel-based technique [24], and subsequence kernel-based techniques [25] were proposed. The integration of probabilistic models and data mining were also proven to be good techniques for extracting relations and patterns from text [26]. Although there are innumerable RE methods in place, the models and algorithms are very domain- and data-specific. The absence of generalized algorithms to perform RE makes it challenging to define and perform a new RE task; the state-of-the-art models vary between different datasets and from one domain to another. In general, RE is most commonly viewed as a supervised learning technique performing classification [27]. In such cases, a machine learning (ML) algorithm, either traditional ML or deep learning-based methods, is used. RE can also be achieved by using unsupervised learning and rule-based methods. In the following sections, we discuss the various RE tasks and techniques applied to the clinical and biomedical domains.
2.6. Motivation
The significant growth in Electronic Health Records (EHR) over the last decade has resulted in a rich availability of clinical text, which is unfortunately stored in an unstructured format. For example, in the radiation oncology domain, when analyzed using ML techniques, a lot of valuable information such as physician clinical assessments, which includes pre-existing conditions, clinical and social history, and clinical disease status embedded in free text and entered in clinical notes, can help physicians provide better treatment. Hence, there is a need to explore robust techniques to extract such information from the clinical text. NER and RE are the key components in information extraction. In this paper, our goal is to highlight the present status of NER and RE by evaluating the models and their performance and by discussing the challenges and factors affecting the NER and RE models that need to be considered while designing a clinical decision support system.
3. Methodology
We used Google Scholar to search for articles related to NER and RE and specifically papers used in the context of clinical text. We also checked for publications where the above mentioned techniques are used in the radiation oncology domain. We discovered that there is very limited work on NER and RE in the radiation oncology domain; however, we did notice that there are a plethora of publications in using NER and RE in the clinical text in general.
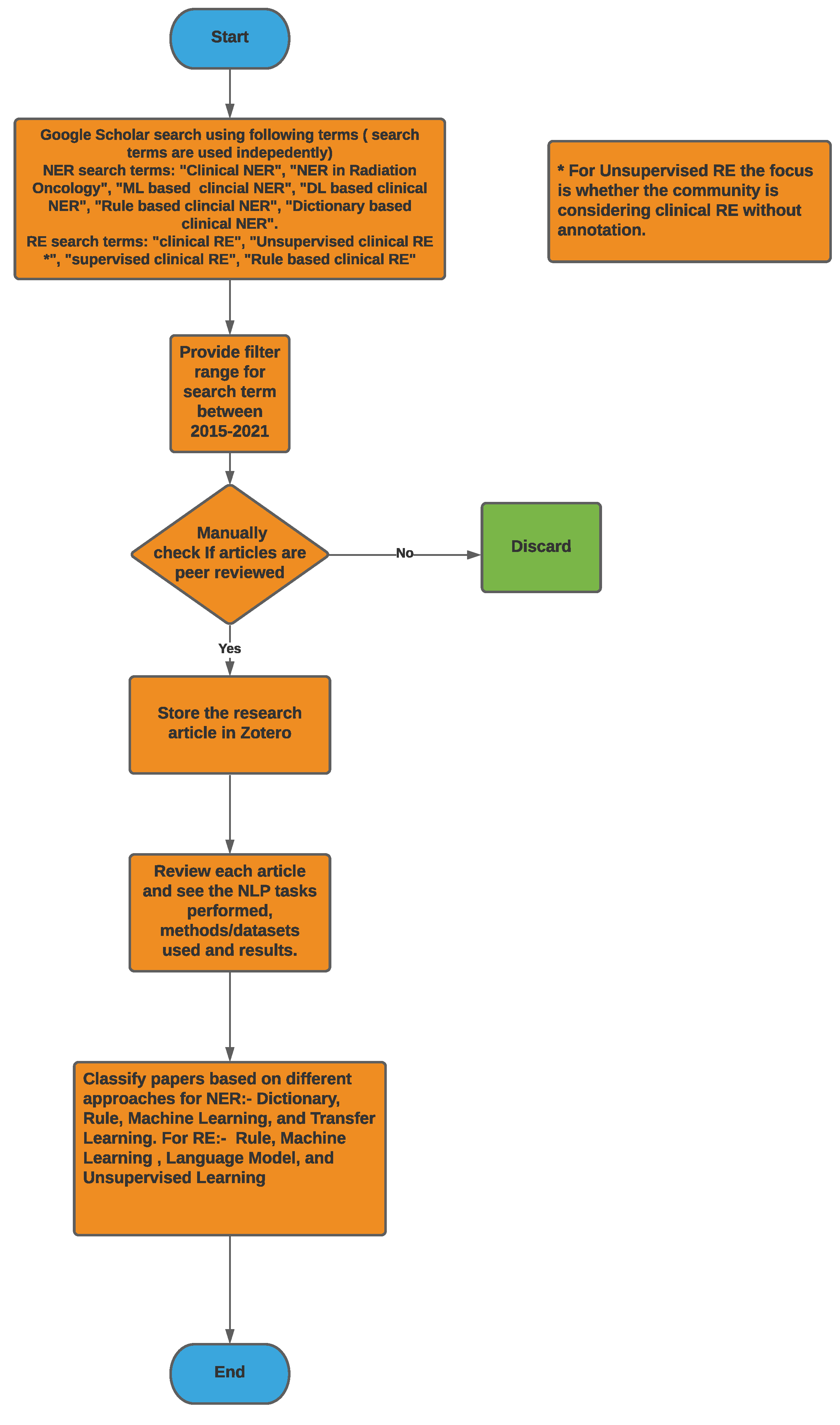
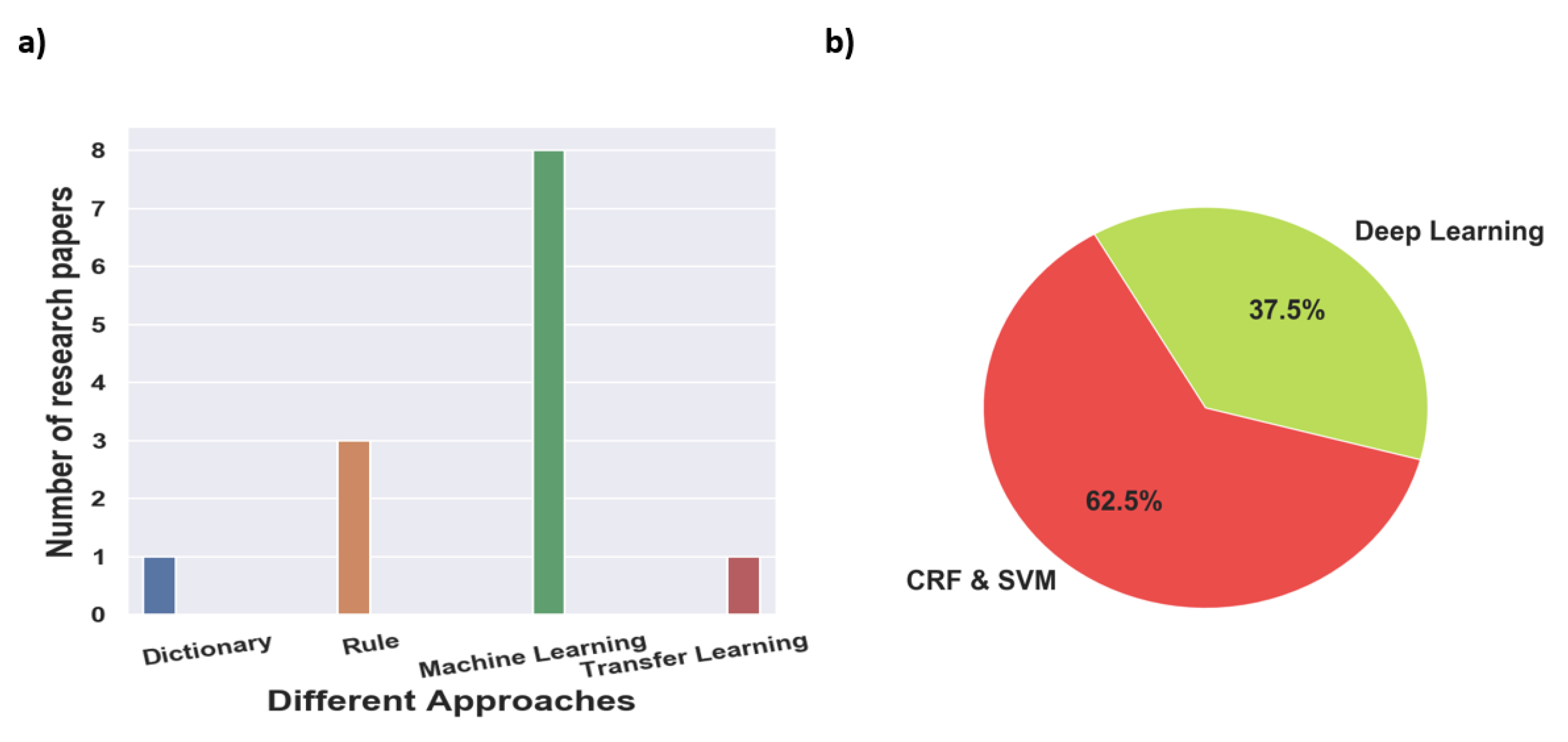
Figure 2 provides a high-level overview of the steps carried out to select research articles for the survey. For clinical NER, search terms such as ‘Clinical Named Entity Recognition’, ‘NER in Radiation Oncology’, ‘Deep learning Clinical NER’, and ‘Machine Learning based clinical NER’ were used. From the resulting articles, we categorized them based on the language used for NER i.e., English, Chinese, and Italian, among others. Next, we classified the articles based on the type of approach used for NER; we found that a majority of them used ML-based approaches, and only a few articles within the machine learning class used deep learning-based methods. Overall, for clinical NER, we selected around 23 papers, out of which 19 articles used machine learning-based approaches and 3 articles used rule-based methods while 1 article used a dictionary-based approach. Since 2018, most of the clinical NER models used only ML models, we discuss such methods in greater detail. Figure 3a shows a representation of various clinical NER models that were identified; we came across ∼8 papers that use ML approaches to develop NER models for clinical text. Figure 3b represents the distribution of ML-based clinical NER models.
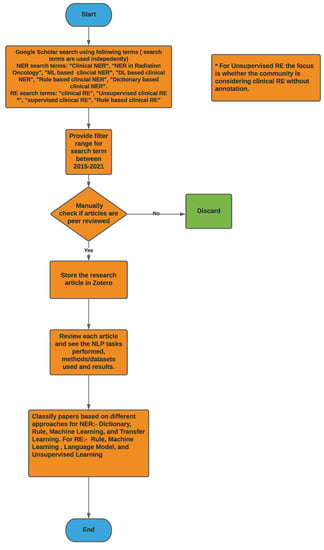
Figure 2.
Methodology flowchart used here for both NER and RE to select articles.
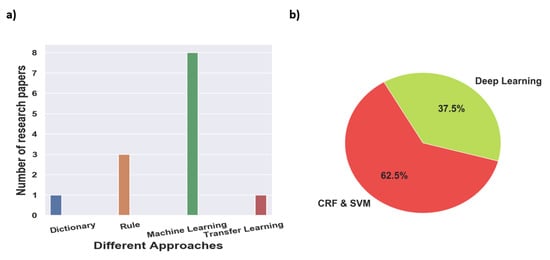
Figure 3.
(a) Representation of the various clinical NER models based on different approaches for this survey paper and (b) percentage of NLP models identified based on different machine learning approaches.
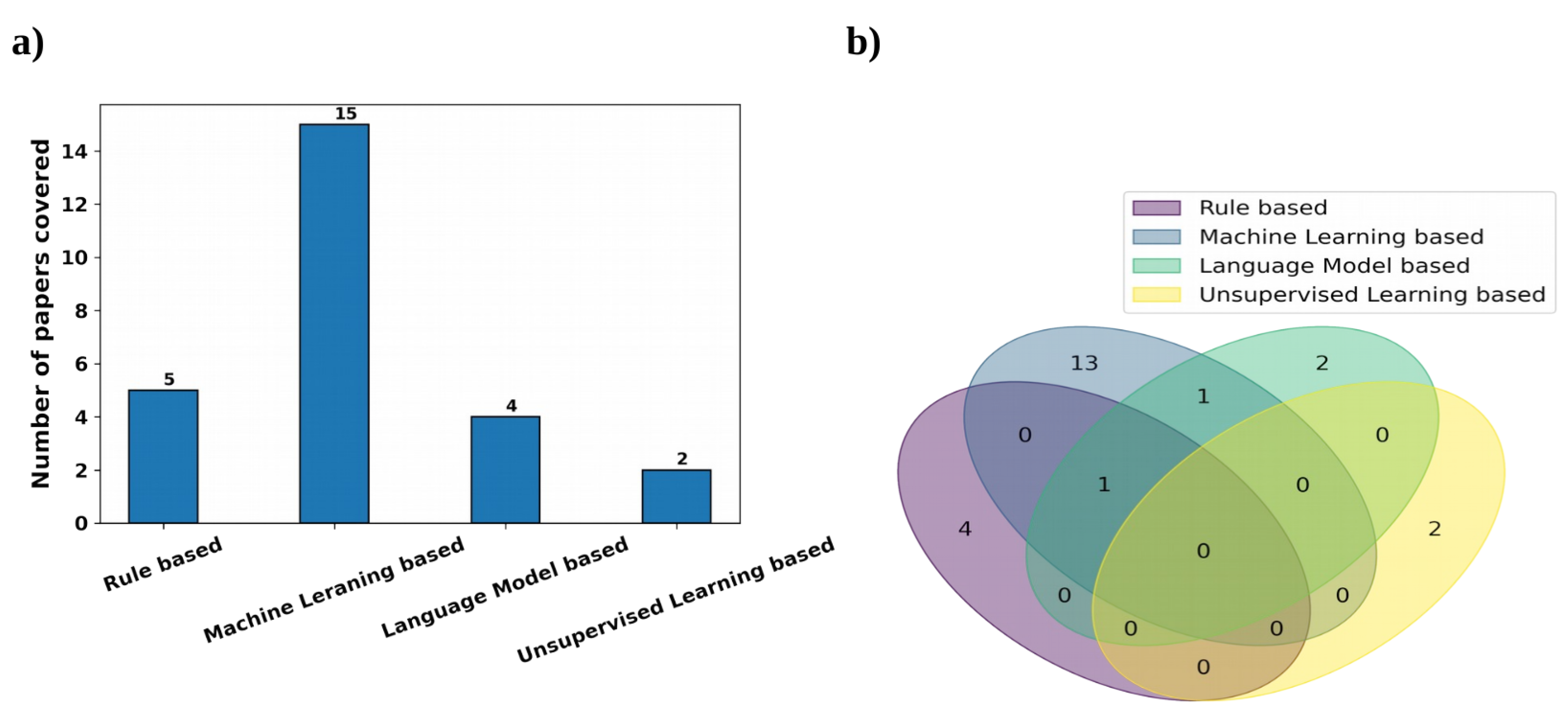
For clinical RE, we used the search term, ‘Clinical Relationship Extraction’ and obtained a number of research papers on clinical information extraction. After going through them, we found out that most people consider this to be a classification problem using machine learning models. Hence to filter out more of these papers, we again used the search term, ‘Supervised Clinical Relationship Extraction’. Next, we used our judgement to use the search term, ‘Unsupervised Clinical Relationship Extraction’ to see if the community focuses on clinical RE without data-annotation. The last search term for clinical RE is ‘Rule based Clinical Relationship Extraction’ as we found out from the first search that rule-based methods are also used to some extent besides ML-based methods. From the top results of this search process, we manually identified the relevant papers based on their closeness to clinical RE and considering the diversity of the presented methods. We also kept the search results mostly limited to papers after 2016; however, this filter could not effectively find clinical RE-based articles using rule-based and unsupervised learning-based approaches. Not much work was conducted on clinical RE using unsupervised learning-based approaches because, in the clinical domain, most datasets are annotated and the supervised approaches are able to outperform these approaches in most cases, which are discussed later; we could only find two papers in this area. Rule-based methods have been used for clinical RE to some extent, but most of the noteworthy work was conducted before 2015–2016. After that, the application of supervised learning-based approaches for clinical RE started escalating distinctly and the focus on other approaches diminished. Hence, we manually identified two papers using rule-based approaches after 2016; both were published in 2021. We also manually chose three earlier papers using rule-based methods as they were popular in the past. We manually chose the 16 top, relevant, diverse papers using supervised learning-based approaches after 2016. We also considered another noteworthy supervised learning-based method for clinical RE before 2016. Out of these 17 articles, 15 papers used traditional ML and deep learning-based approaches and 4 papers used language models, with 2 papers using both language models and ML. Overall, we were able to choose 23 papers for clinical RE that used rule-based, deep learning-based, or language model-based methods. Figure 4 shows the distribution of the clinical RE research articles considered here on the basis of the methods therein using a bar chart and a Venn diagram.
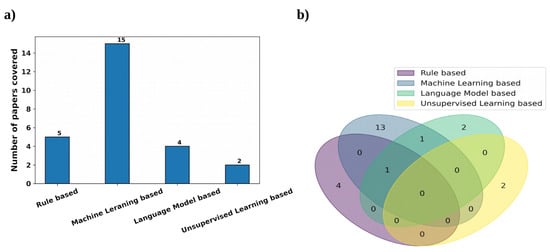
Figure 4.
Representation of the clinical RE research papers used, based on the variety of the methods by using (a) a bar chart and (b) a Venn diagram.
We used software tools such as Zotero to collect all of the papers and to perform the literature survey. The next step was to categorize all of the articles and to prepare an outline for this survey. We evaluated the architectures used, how the results were reported, and the data used in the experiments. In total, we came across 51 articles (28 for clinical NER, and 23 for RE), and 46 of them were used for this survey paper; only peer reviewed articles were considered. It is worth mentioning that a couple of survey papers [17,28] also provide an in-depth view of each topic separately; however, we did not find any such survey that discusses these two related topics together specifically with respect to the clinical domain such as radiation oncology. To the best of our knowledge, this paper surveys clinical NER and RE for the first time and discusses various approaches along with their outcomes and limitations. The paper is organized as follows: Section 5 discusses the tasks associated with clinical NER, followed by a brief overview of various approaches and their results. Similarly for RE, we review the various approaches and their performance in Section 6. Finally, in Section 7, we provide our inference about the latest trends, state-of-the-art techniques, and what we believe the community (both for clinical NER and RE) needs to focus on in the future.
4. NLP Competitions and Datasets for Clinical Text
In this section, we review the different NLP competitions and datasets that are more geared towards clinical text.
4.1. Competitions
Competitions and datasets are considered assets in NLP tasks. Although most of these challenges are for data from the general domains, clinical domain-related challenges have come up in the past. Clinical-NER based competitions were mostly focused on the de-identification of Personal Health Information (PHI). In 2014, there was a i2b2 UTHealth challenge that had longitudinal data [29], and the goal of the competition was to perform de-identification on clinical narratives, with a second track focused on determining risk factors for heart disease over time. Stubbs et al. [30] provides a comprehensive review of a workshop that includes how data were released and how the submissions were evaluated. The 2016-CEGS N-GRID shared tasks that the workshop used in gathering psychiatric data [31] for addressing text de-identification, symptom severity detection, and the proposal of new research questions. Stubbs et al. [31] explained how the data were generated; discussed the challenges with psychiatric data as it contains higher occurrence of PHI; and the outcomes, which showcase the best performing systems and how the submitted models were evaluated. There was also another competition on clinical NER for de-identification on Japanese text (2012 NTCIR-10) [32]. Coffman et al. [33] organized a competition, which was also a final deliverable for the Applied NLP course taught at UC Berkeley. The objective of the competition was to develop an algorithm that predicts/assigns an ICD-9 (International Classification of Diseases, 9th revision) code to clinical free text [33]. MADE1.0 [34] is a competition for detecting Adverse Drug Events (ADEs) from EHR. The goal of the NLP task is to detect medication names and other attributes such as frequency and duration. Around 11 teams participated in at least one of the three tasks. There was a total of 41 submissions, among which Wunnava et al. [35] ranked first for the NER task, with a micro-averaged F1-score of 0.892. SemEval-2014 [36] Task 7 was another competition on analyzing clinical text; it had two subtasks, namely, identification and normalization of disease and disorders in a clinical text from the ShARe [37] corpus. Around 21 teams participated in the identification task, and the best F1-score reported was 81.3, while for the normalization task, 18 teams participated, reporting a best accuracy of ∼74.1.
National NLP Clinical Challenges, also called n2c2, is a very popular competition for different clinical NLP tasks. Between 2004 and 2014, the competition was called Informatics for Integrating Biology and the Bedside (i2b2) but was then changed to n2c2 in 2018. They introduced the following clinical RE tasks over the years, with datasets generated by the NIH-funded National Centers for Biomedical Computing (NCBC).
- 2010 i2b2/VA Challenge on Concepts, Assertions, and Relations in Clinical Text [38]: In this competition, 16 teams participated in the relationship extraction task that showed that rule-based methods can be augmented with machine learning-based methods. SVM-based supervised learning algorithm performed the best with an F1-score of 0.737 [39].
- 2011 Evaluating the state-of-the-art in co-reference resolution for electronic medical records [40]: In this competition, 20 teams participated and rule-based and machine learning-based approaches performed best, with an augmentation of the external knowledge sources (coreference clues) from the document structure. The best results on the co-reference resolution on the ODIE corpus with the ground truth concept mentions and the ODIE clinical records were provided by Glinos et al. [41], with an F1-score of 0.827. The best results on both the i2b2 and the i2b2/UPMC data were provided by Xu et al. [42], with F1-scores of 0.915 and 0.913, respectively.
- Evaluating temporal relations in clinical text, 2012 i2b2 Challenge [43]: 18 teams participated in this challenge, where for the temporal relations task, the participants first determined the event pairs and temporal relations exhibiting temporal expressions and then identified the temporal relation between them. This competition also showed that hybrid approaches based on machine learning and heuristics performed the best for the relationship classification. Rule-based pair selection with CRF and SVM by Vanderbilt University provided the best results here (F1-score: 0.69).
- 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records [44]: a total of 21 teams participated in the relationship classification task on adverse drug events (ADEs) and medication. Team UTHealth/Dalian (UTH) [45] designed a BiLSTM–CRF-based joint relation extraction system that performed the best (F1-score: 0.9630).
4.2. Datasets
The datasets are important in understanding the different entities and relations extracted in the clinical domain. This subsection gives an overview of the different datasets used for clinical NER and RE tasks for a better understanding of the challenges in the domain.
We came across a few publicly available datasets for clinical NER; however, these datasets are restricted to specific NLP tasks in clinical domain. Below is a list of datasets that were used in NER challenges or used as training for NER models, which are discussed in Section 5.3 for training, testing, and validation:
- Mayo Clinic EMR: It has around 273 clinical notes, which includes 61 consult, 4 educational visits and general medical examinations, and a couple of exam notes. A few models, such as Savova et al. [46], generated a clinical corpus from Mayo Clinic EMR [47].
- MADE1.0 Data set: This dataset consists of 1092 medical notes from 21 randomly selected cancer patients’ EHR notes at the University of Massachusetts Memorial Hospital.
- FoodBase Corpus: It consists of 1000 recipes annotated with food concepts. The recipes were collected from a popular recipe sharing social network. This is the first annotated corpora with food entities and was used by Popovski et al. [48] to compare food-based NER methods and to extract food entities from dietary records for individuals that were written in an unstructured text format.
- Swedish and Spanish Clinical Corpora [49]: This dataset consists of annotated corpora clinical texts extracted from EHRs; the Spanish dataset consists of annotated entities for disease and drugs, while the Swedish dataset has entities annotated for body parts, disorder, and findings. This dataset is mostly used for training and validation for NER on Swedish and Spanish clinical text.
- i2b2 2010 dataset [38]: This dataset includes discharge data summaries from Partners Healthcare, Beth Israel Deaconess Medical center, and University of Pittsburgh (also contributed progress reports). It consists of 394 training, 477 test, and 877 unannotated reports. All of the information are de-identified and released for challenge. These datasets are used for training and validation in many of the NER models used for clinical text.
- MIMIC-III Clinical Database [50]: This is a large and freely available dataset consisting of de-identified clinical data of more than 40,000 patients who stayed at the Beth Israel Deaconess Medical Center between 2001 and 2012. This dataset also consists of free-text notes, besides also providing a demo dataset with information for 100 patients.
- Shared Annotated Resources (shARe) Corpus [37]: This dataset consists of a corpus annotated with disease/disorder in clinical text.
- CanTeMiST [51]: It comprises 6933 clinical documents that does not contain any PHI. The dataset is annotated for the synonyms of tumor morphology and was used for clinical NER on a Spanish text by Vunkili et al. [51].
Specific relations annotated in the datasets from the various clinical RE challenges mentioned in Section 4.1 are as follows:
- 1.
- 2010 i2b2/VA Challenge on Concepts, Assertions, and Relations in Clinical Text [38]: A wide variety of relations were identified as follows:
- Medical problem–treatment relations:
- -
- TrIP indicates that treatment improves medical problems, such as hypertension being controlled by hydrochlorothiazide.
- -
- TrWP indicates that treatment worsens medical conditions, such as the tumor growing despite the available chemotherapeutic regimen.
- -
- TrCP indicates that treatment causes medical problems, such as Bactrium possibly being a cause of abnormalities.
- -
- TrAP indicates that treatment is administered for medical problems, e.g., periodic Lasix treatment preventing congestive heart failure.
- -
- TrNAP indicates that treatment is not administered because of medical problems e.g., Relafen being contraindicated because of ulcers.
- -
- Others that do not fit into medical problem–treatment relations.
- Medical problem–test relations:
- -
- TeRP indicates that the test reveals medical problems, such as an MRI revealing a C5-6 disc herniation.
- -
- TeCP indicates that the test was conducted to investigate a medical problem, such as a VQ scan being performed to investigate a pulmonary embolus.
- -
- Others that do not fit into medical–test relations.
- Medical problem–medical problem relations:
- -
- PIP indicates any kind of medical problem such as a C5–6 disc herniation with cord compression.
- -
- Other relations with respect to medical problems that do not fit into the PIP relationship.
- 2.
- 2011 Evaluating the state-of-the-art in coreference resolution for electronic medical records [40]: The data for this challenge was similar to the 2010 i2b2/VA challenge as the dataset contained two separate corpora, i.e., the i2b2/VA corpus and the Ontology Development and Information Extraction (ODIE) corpus, which contained de-identified clinical reports, pathology reports, etc.
- 3.
- Evaluating temporal relations in clinical text, 2012 i2b2 Challenge [43]: The temporal relations or links in the dataset indicate how two events or two time expressions or an event and a time expression is related to each other. The possible links annotated in the dataset were BEFORE, AFTER, SIMULTANEOUS, OVERLAP, BEGUN_BY, ENDED_BY, DURING, and BEFORE_OVERLAP.Ex: OVERLAP -> She denies any fever or chills.Ex: ENDED_BY -> His nasogastric tube was discontinued on 05-26-98.
- 4.
- 2018 n2c2 shared a task on adverse drug events and medication extraction in electronic health records [44]: The different relations identified between two entities in this case are either of the following types: Strength–Drug, Form–Drug, Dosage–Drug, Frequency–Drug, Route–Drug, Duration–Drug, Reason–Drug, and ADE–Drug.
5. Discussion on Clinical Named Entity Recognition
The goal of using NER on clinical text is to extract entities or subjects of interest from the clinical text. The clinical text, in general, has many medical terms such as the disease name, location, and medical procedures, and hence, the named entities can help in finding useful patterns. The nature of the clinical text, in general, is dictated by notes from physicians based on their interaction with the patients. In most cases, it is in free text format, which can be split into multiple paragraphs, and is mostly narrative in nature. For example, the clinical text written by physicians in the consultation notes from the radiation oncology domain has the following information:
- Physical Exam: This section can have both structured and unstructured information such as toxicity and review of systems, where we try to store information such as dizziness, cough, and rectal bleeding.
- Past Medical History: This has all of the allergy information, medications, prior military service, prior surgery information, and prior diseases for patients and is mostly stored as unstructured free text.
- Oncologic History: This includes all of the prior oncologic information in unstructured format and varies based on the types of cancer.
- Diagnostic Test: Various tests may be performed on patients and vary based on cancer types. They are mostly in structured format; however, some tests may be specific to patients that can be documented and stored in unstructured free text format such as Bone Scan and CT Pelvis.
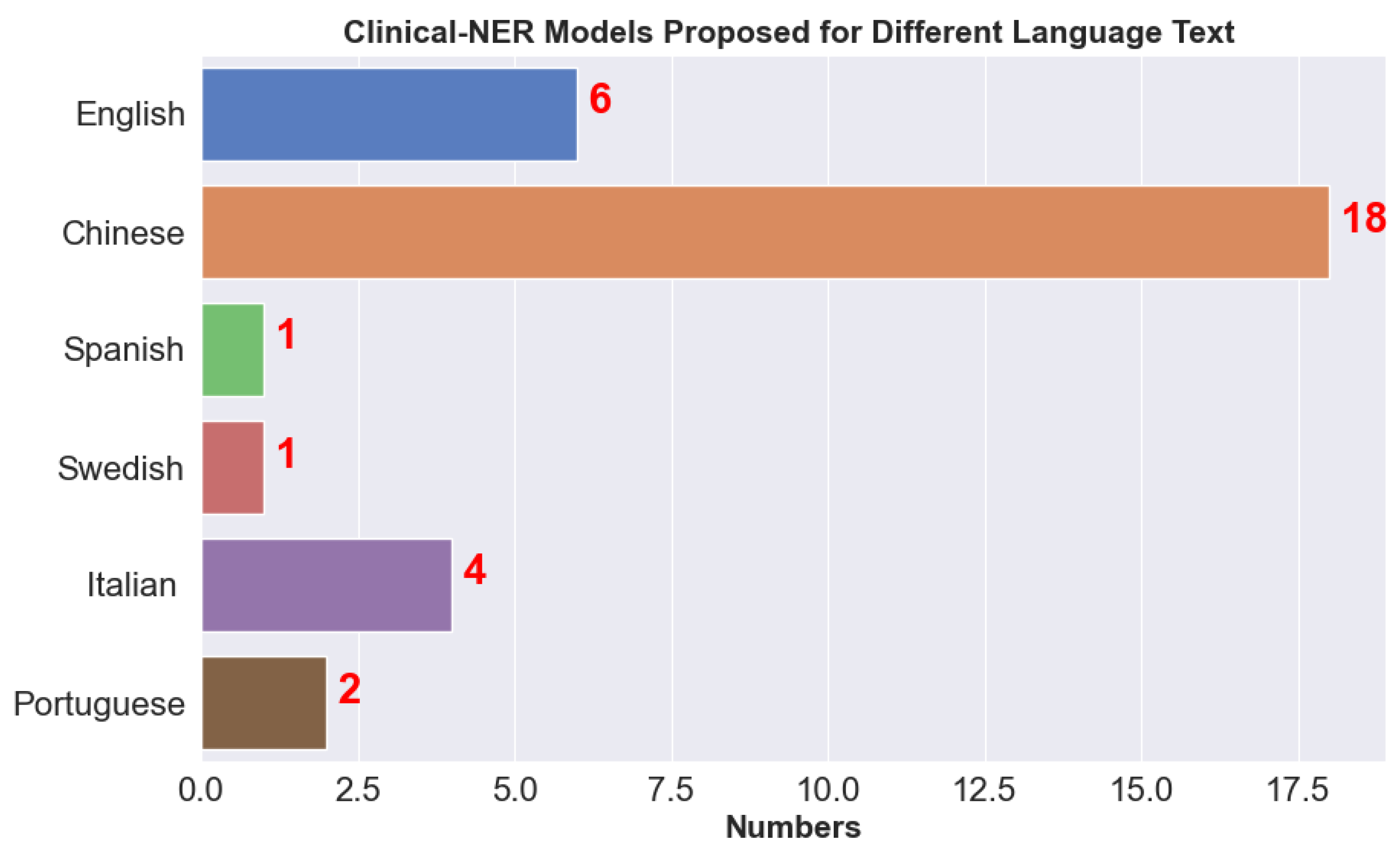
Clinical NER is very common these days due to the massive growth in EHRs and is considered the first step in processing clinical text. The output of clinical NER is further used for other tasks such as decision-making in precision treatment. Due to the unstructured nature of the clinical text, there are challenges in designing effective clinical NER systems, as discussed below. We observed that many clinical NER models are developed for different languages such as Chinese and Italian; Figure 5 shows the number of clinical NER models that we came across for different languages. Due to the strict privacy rules in the EU (European Union) and HIPAA compliance in the US, it is difficult to disseminate medical information. We found very few articles that use clinical NER models for de-identification where medical document is parsed and any Protected/Personal Health Information (PHI) is removed; for example, recently Catelli et al. [52] developed a clinical NER model for Italian COVID-19 clinical text.
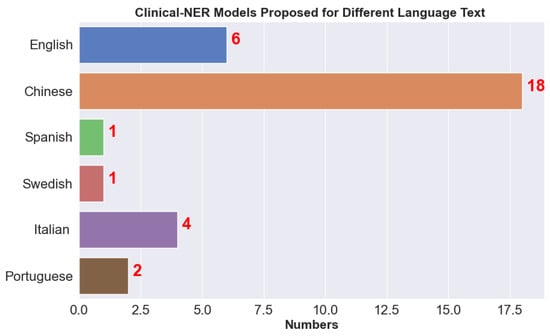
Figure 5.
Clinical NER models available for text in different languages.
5.1. Challenges in Clinical NER
- Nested Entities and Ambiguity: Most clinical terms are often confusing as there is no common ontology. Physicians often use abbreviations or acronyms, which makes it very difficult to standardize clinical text. In the radiation oncology domain, a common challenge is that physicians dictate their clinical assessment based on the style they were trained in and it varies significantly for different types of cancers, which makes it very difficult to develop a standard NER model for processing radiation oncology notes that cater to all of the different types.
- Meaning of Context: The clinical terms used can have different meanings, which vary based on the context. Although this problem mostly applies to non-clinical notes, for clinical NER, this becomes more challenging as the model should understand the complete clinical context along with the entity. A common issue is negative medical findings, where text is written in such a manner that it reports findings in a negative context; however, the NER considers that as a positive.
To address the nested entities and ambiguity, there are efforts to standardize the nomenclature of clinical entities [53,54]. However, this is still in an initial phase, and to be successful, it needs to be widely adopted.
5.2. Clinical NER Methods
One of the important challenges in designing a clinical NER is how to extract meaningful information without much human effort. Prior to NER, the NLP techniques used required a lot of human effort to process the text. There are various NER models proposed over the last decade to extract information from the clinical text that can be broadly classified into four types of approaches:
- Dictionary-Based Approach: In this approach, a predefined set of named entities are defined that are later used as a lookup while parsing the clinical text for entities. For example, Savova et al. [46] used a dictionary-based approach to detect NERs from clinical text using their NLP toolkit.
- Rule-Based Approach: Here, the rules/entities are predefined by domain experts. Most of the rules are handcrafted and are used to detect entities in a specific text. The limitation of this approach is generalizability or extensibility, as most of them are applicable to the domain they were defined in. This approach certainly requires a lot of effort where experts spend time defining the entities, and then, it is used as a lookup while parsing the clinical notes.
- Machine Learning-Based Approach: The purpose of this approach is to completely automate the NER process. Commonly used ML algorithms such as Random Forest (RE), Support Vector Machines (SVMs), and Neural Networks (NN) are used to learn the pattern (entities and boundaries) using the training set. Once the training is over, the model can classify the clinical text into predefined classes. This approach is garnering much attention due to recent advancements in ML and the easy availability of computational resources. The majority of the articles collected for this survey used this approach.
- Conditional Random Field (CRF)-Based Approach: The CRF approaches fall under the ML category and mostly solve a label sequencing problem, where for a given input sequence , CRF tries to find the best label sequence . At first, the entities are annotated with tags; in general, the BIO (Beginning, Inside, and Outside of Entity) schema is used for annotation, where each word is assigned to a label. The input for CRF models is mostly designed by humans and represented as a bag-of-words style vector. Wu et al. [4] introduced seven tags and three CRF baselines using different features. All of the commonly used CRF-based implementations in clinical NER can be found in the CRF++ package. In Table A1 and Table A2, we observe that there are many models using CRFs for NER with good accuracy.
- Deep Learning-Based Methods: This is similar to the CRF label sequencing problem using the BIO schema, where the input is a raw sequence of words. An added layer performs the word embedding by converting words into densely valued vectors. In the training phase, it learns the dependencies and features to determine entities. Deep learning methods are very popular for clinical NER as they achieve state-of-the-art results and can also detect hidden features automatically. The first neural network architecture for NER was proposed by Collobert et al. [19], with a convolution layer, several standard layers, and a non-linear layer. This architecture achieved state-of-the-art performance in clinical NER. Details on the CNN model for clinical NER can be found in [17]. New studies have recently shown that RNNs (Recurrent Neural Networks) perform much better than CNNs and are capable of capturing long-term dependencies for sequence data. Lample et al. [55] introduced Long Short-Term Memory (LSTM), a popular implementation of RNN architecture, for this problem. Wu et al. [4] evaluated the performance of CNNs, RNNs, and CRFs with different features and concluded that the RNN implementation outperformed the other two.
- Hybrid Approaches: here, any of the above approaches are combined and then used to determine entities.
5.3. Clinical NER Models
- Savova et al. [46] proposed a dictionary look up algorithm, where each named entity is mapped to a terminology. The dictionary was constructed using the terms from UMLS, SNOMED CT, and RxNORM. This implementation also involves a parser in which the output is used further to search for noun phrases. The limitation of this implementation is that it fails to resolve ambiguities while working with results from multiple terms in the same text. They datasets for NER are derived from Mayo clinic EMR. For exact and overlapping matches F1-score reported were 0.715 and 0.824 respectively.
- Skeppstedt et al. [56] used CRF model and a rule-based approach to detect NER on Swedish health records and identified four entities: Drug, Finding, Disorder, and Body structure. They also compared it on English clinical text. They reported precision and recall for all of their findings: 0.88 and 0.82 for body structure, 0.80 and 0.82 for disorders, 0.72 and 0.65 for findings, and 0.95 and 0.83 for pharmaceutical drugs.
- Chen et al. [57] developed a rule-based NER system that was designed to detect patients for clinical trial. They used the n2c2-1 challenge dataset for training and achieved an F1-score of 0.90.
- Eftimov et al. [48] developed a rule-based approach to detect extraction of food, nutrient, and dietary recommendations from text. They discussed four methods FoodIE, NCBO, NCBO (OntoFood), and NCBO(FoodON). Based on their comparison, they identified that FoodIE performs well. Their model was trained on the FoodBase Corpus and was able to identify entities from dietary recommendation.
- Xu et al. [58] developed a joint model based on which CRF performs word segmentation and NER. Generally, both systems are developed independently, but the joint model used to detect Chinese discharge summaries performed well. There was no score reported in this publication; they only reported that the joint model performance is better when they compared it with the two individual tasks.
- Magge et al. [59] developed an NLP pipeline, which processed clinical notes and performed NER using bi-directional LSTM coupled with CRF in the output layer. They used 1092 notes from 21 cancer patients, from which 800 notes were used for NER training. They reported NER precision, recall, F1-score for the entities individually and reported a macro-averaged F1-score of 0.81.
- Nayel et al. [60] proposed a novel ensemble approach using the strength of one approach to overcome the weakness of other approaches. In their proposed two-stage approach, the first step is to identify base classifiers using SVM, while in the second phase, they combined the outputs of base classifiers based on voting. They used the i2b2 dataset and reported an F1-score of 0.77.
- Wu et al. [4] performed a comparison study between two well-known deep learning architectures, CNN and RNN, with three other implementations: CRFs and two state-of-the-art NER systems from the i2b2 2010 competition to extract components from clinical text. The comparison created a new state-of-the art performance for the RNN model and achieved an F1-score of 85.94%.
- Wang et al. [61] proposed a model to study symptoms from Chinese clinical text. They performed an extensive set of experiments and compared CRF with HMM and MEMM for detecting symptoms. They also used label sequencing and the CRF approach outperformed the other methods.
- Yadav et al. [17] provided a comprehensive survey of deep neural architectures for NER and compared it with other approaches including supervised and semi-supervised learning algorithms. Their experiments showed good performance when they include neural networks, and they claim that integrating neural networks with earlier work on NER can help obtain better results.
- Vunikili et al. [51] used Bidirectional Encoder Representations from Transformers (BERT) [62] and Spanish BERT (BETO) [63] for transfer learning. This model is used to extract tumor information from clinical reports written in Spanish. They reported an F1-score of 73.4%.Jiang et al. [64] developed ML-based approaches to extract entities such as discharge summaries, medical problems, tests, and treatment from the clinical text. They used a dataset comprising 349 annotated notes for training and evaluated their model on 477 annotated notes to extract entities. They reported an F1-score of 0.83 for concept extraction.
- Yang et al. [65] proposed a deep learning model to extract family history, and they compared LSTM, BERT, and ensemble models using a majority voting.
5.4. Clinical NER Evaluation Metrics
The outputs from clinical NER systems are usually compared with human annotations. In general, a comparison can be either exact or relaxed matches [17]. A relaxed match only considers the correct type and ignores the boundaries as long as there is an overlap with ground truth boundaries. We observed from our cohort of selected articles on clinical NER that all of them reported exact matches, which is the F1-score and variations such as macro F1-score. In the case of an exact match, it is expected that the entity identified correctly should also detect boundary and type correctly at the same time [17]. We also observed that a few NER models report performance in macro- and micro-average. In macro-averaging, the F1-scores of all entities are calculated independently and then averaged. In micro-averaging, the sums of the false positive, false negative, and true positive across all entities are taken. Other commonly used metrics in ML such as sensitivity, specificity, ROC (Receiver Operator Characteristic), and AUC (Area Under the Curve) were not used in the clinical NER articles reviewed here. There are however many studies such as [66] that point to the limitations of using F1-score as an evaluation metric in NLP; one of the major issues is that the F1-score metric is biased towards the majority class. The class imbalance problem has been recently garnering attention for both binary and multi-class classification. Accuracy and precision scores are relevant if we focus on majority classes; for a minority, those metrics evaluations do not have any significant incluence. Branco et al. [67] provided a comprehensive list of metrics both for binary class and multi-class classification such as classes average accuracy, and Matthews Correlation Coefficient. Along with the list of metrics provided, they claim that the metrics available are not suitable for all cases. We also found very few papers that tested for statistical significance between experimental methods.
6. Discussion on Clinical Relationship Extraction
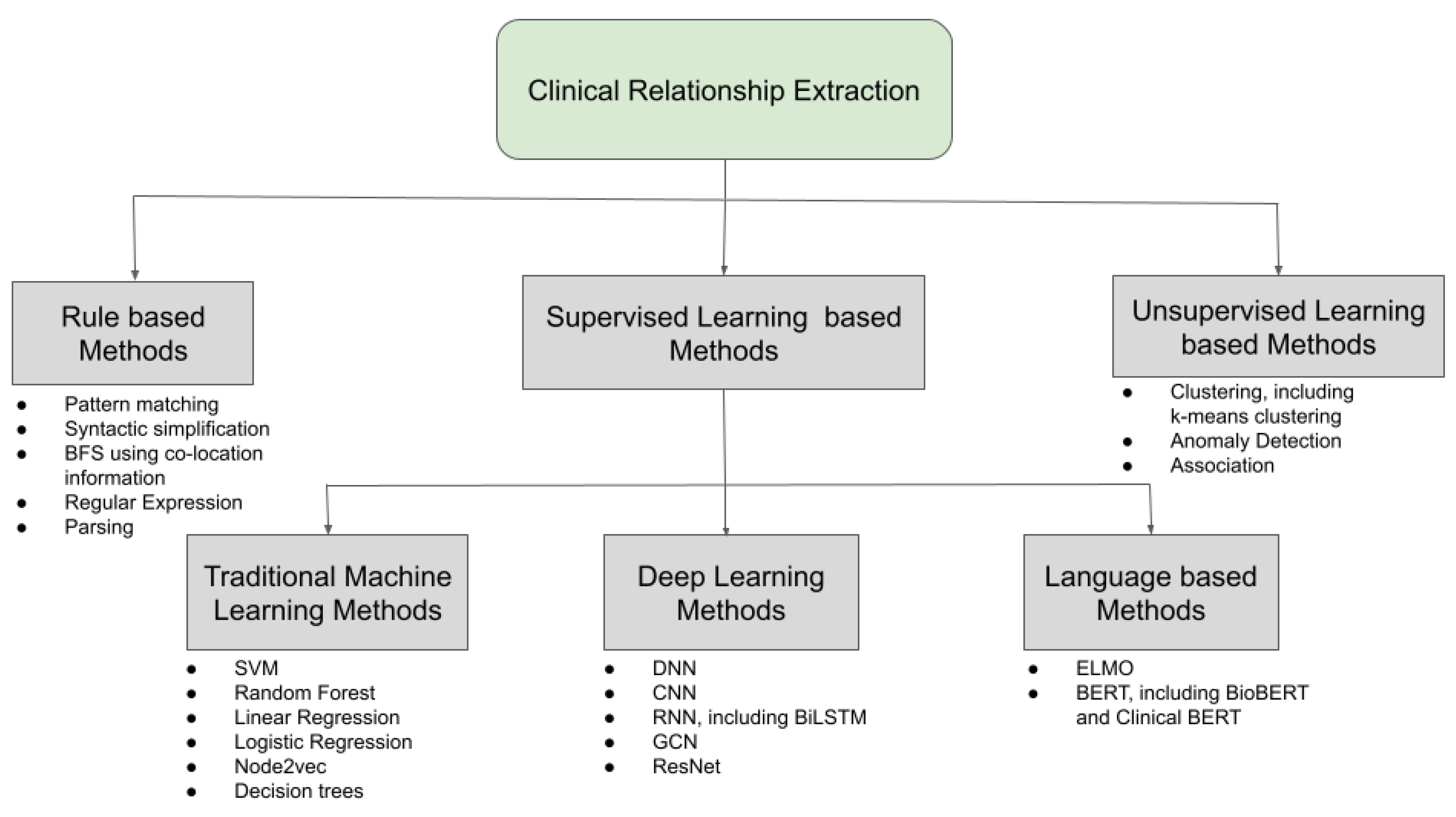
RE is a specialized task of collecting meaningful structured information from unstructured text. In clinical and biomedical domains, RE has been applied to drug–gene relationships [68], disease–gene relationships [69], semantic classes for radiology report text identification [70,71], relation extraction for biological pathway construction [72], relationship between lexical contexts and category of medical concepts [73], and disease–mutation relationship from biomedical literature [74]. Temporal relationship extraction from clinical texts is another important RE task [75]. In all these of different tasks, the NLP-based methods that are used to extract the relations between different entities are very much specific to the particular dataset, i.e.,the particular combination of feature representation and learning algorithm is very distinct from any other case. Due to this fact, the methods that are used to extract relations such as an ML problem are not very generalizable. However, RE from clinical texts has also been performed by using a domain invariant convolutional neural network (CNN) [76]. Most RE tasks are based on finding the relationship between entities inside the same sentence but there are some instances of RE tasks across sentences as well [77,78,79]. Since, in most cases, RE is treated as a classification problem, both multi-label classification [80] and multi-class classification [76,81] were proposed to extract clinical relations. An RE task consists of syntactic processing modules, which deals with the process of text representation and feature generation such as tokenization, word embeddings, etc., and semantic processing modules, which deals with meaningful information collection such as relationship identification and classification, in this case. In clinical texts, a variety of feature generation techniques are used to extract relations from various data, which can range from contextualized word embeddings, part-of-speech (POS) tagging, etc. The next or the final step is to select a learning algorithm such as supervised, unsupervised, or even rule-based methods on the features in order to identify the relations. The various feature representation and learning methods used in the clinical and biomedical text are discussed next. A pictorial representation of the different learning methods used to learn the different relations from clinical texts is shown in Figure 6.
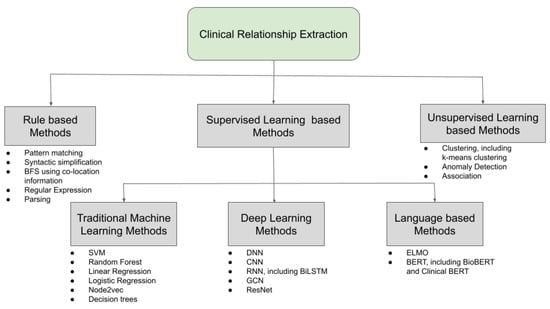
Figure 6.
Different learning methods used for clinical RE.
6.1. Feature Generation
Feature generation is an important step for RE, where features are extracted from the unstructured text and then represented only with numbers. This step is particularly very important for the supervised and the unsupervised learning methods because these methods require inputs in the form of numbers only. The performance of these ML models depends not only on the actual algorithm but also on how the input features were represented. The first step before representing the features is preprocessing and tokenizing the text. In many deep learning-based approaches, the whole instance is considered the input, which is basically a featureless representation. The various features that can be considered for RE tasks are the word, the words distance from both the entities, chunk tag of the word, POS tag of the word, type of the word, n-grams, etc. Sahu et al. (2016) [76] introduced a domain-invariant RE technique using CNN, where the inputs were represented with the word, its distance from the first entity, its distance from the second entity, a Part-of-Speech (POS) tag, chunk, and the type of the word. Singhal et al. (2016) [79] used Nearness to Target Disease Score, Target Disease Frequency Score, Other Disease Frequency Score, Same Sentence Disease-Mutation Co-occurrence Score, Within Text Sentiment Score, and Text Sentiment Subjectivity Score as input features to the decision trees to extract the disease–mutation relationship. Hasan et al. (2020) [82] used word embedding, POS embedding, IOB embedding, relative distance, concept embedding, and dependency tree to represent the input features. Alimova et al. (2020) [83] compared the performance of BERT with that of random forest based on a multitude of features such as word distance; character distance; sentence distance; punctuation distance; position distance; bag of words; bag of entities; entity types; entities embedding; concept embedding; sentence embedding; the similarity between entities; and some knowledge features such as UMLS, MeSH, etc. Mahendran et al. (2021) [70] also divided the sentences into five segments based on the location of the context to represent the input features of the segment-CNN model. Textual input features used for various ML algorithms are mostly a combination of the features mentioned above.
6.2. Rule-Based Methods
Though rule-based methods are not the most popular method nowadays to extract relations from clinical texts, they are still being used and have been used in the past in good numbers. These methods require defining some rules in the beginning based on the nature of the input dataset. These methods of extracting information by using well-defined rules and patterns are often not very computationally efficient such as the machine learning models with respect to their performance, and hence, these methods are not very popular these days. Segura-Bedmar et al. (2011) [84] developed a linguistic hybrid rule-based method to extract drug–disease interactions via the combined use of shallow parsing, syntactic simplification, and pattern matching. A pharmacist defined the domain-specific lexical patterns of the drug–disease interactions that were matched with the generated sentences. This method did not perform well with an average precision and a very low recall. Xu et al. (2011) [85] combined rule-based methods with ML to engineer features for structured RE from clinical discharge summaries as provided by the i2b2 2010 challenge. The RE task received a micro-averaged F1-score of 0.7326. Li et al. (2015) [86] matched the drug names to their attributes in a prescription list, and then the matching was confirmed by means of the co-location information and RxNorm dictionary. It helped in identifying the medication discrepancies with very high performances. Veena et al. (2021) [87] used NLP-based regular expressions to extract the words from the text document of different medical data using scraping and POS tagging. Then, the relations between different medical terms were extracted using a path similarity analysis. Mahendran et al. (2021) [70] used the co-location information between the drug and the non-drug entity types by using a breadth-first-search (BFS) algorithm to find the adverse drug effects. The left-only rule-based approach (macro-average F1-score: 0.83) eclipsed the performance of other rule-based models. Overall, the rule-based approaches for clinical RE can perform well, depending on how the rules are defined. Some clinical RE tasks using rule-based methods are tabulated in Table A3.
6.3. Supervised Learning Methods
As mentioned before, supervised learning algorithms have been extensively considered for RE. This method uses a classifier to determine the presence or absence of a relationship between two entities. Computers cannot understand the unstructured text, and hence, this kind of learning method requires features about the text as an input. As a result of this, there is an absolute necessity to annotate the clinical texts by domain experts. Annotating or labeling examples is a time-consuming procedure as it takes a lot of effort to manually annotate the data. This is an important limitation of these methods although they have high accuracy. These methods used in clinical RE however suffer from the difficulty of adding new relations. Supervised learning algorithms can also be extended to include distantly supervised RE or weakly supervised learning or semi-supervised learning.
6.3.1. Traditional Machine Learning and Deep Learning-Based Methods
Supervised learning is defined as an ML task to learn a function that maps the input to the output of each input–output data point [88]. This requires the annotated data to be divided into training and testing samples. The model learns the function based on the values of the inputs and the outputs of the training examples. Analyzing the inputs and the outputs, the model comes up with an inferred function. Then, the efficiency of the inferred function is analyzed by testing the function on the testing set. Supervised ML algorithms can be classified into two categories: (i) traditional supervised learning algorithms and (ii) Artificial Neural Network (ANN) based algorithms. The traditional methods are heavily dependant on the well-defined features, and hence, their performance relies on the efficacy of the feature extraction process. Moreover, these shallow algorithms are found to be overshadowed by ANNs where data is large and of high dimension. Still, the shallow traditional ML algorithms perform better in the case of low-dimensional data or data with a limited number of training samples. ANNs can be very deep, depending on the number of hidden layers between the input and the output, leading to deep learning-based methods. The differences between the traditional shallow methods and ANNs are surveyed by Janiesch et al. (2021) [89]. Examples of traditional algorithms include but are not limited to Support Vector Machines (SVM), Linear Regression, Logistic Regression, Naive Bayes, Linear Discriminant Analysis, Decision Trees, K-Nearest Neighbor (KNN), Node2vec, etc., whereas Dense Neural Networks (DNN), Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), Graph Neural Networks (GNN), autoencoders, etc. are some of the Deep Learning algorithms. These algorithms have been extensively used in clinical domain for a variety of tasks [90,91,92,93,94].
Swampillai et al. (2011) [78] first used an SVM-based approach on adapted features to extract relations between entities spread across different sentences. Their work showed that the structured features used for intra-sentential RE can be adopted for inter-sentential RE as they both performed comparably. Later on, inter-sentential RE tasks were defined on clinical notes too. In the 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text [38], SVM-based supervised learning algorithm performed the best with an F1-score of 0.737 [39]. The domain-invariant CNN on multiple features for clinical RE used by Sahu et al. (2016) [76] showed a decent performance with various filter length combinations; filter combination of {4, 6} performed the best (precision: 0.7634, recall: 0.6735, and F1-score: 0.7116). Singhal et al. (2016) [74] used a C4.5 decision tree because of its superior performance on the features extracted from various biomedical literature for disease-mutation RE. It demonstrated improved performance when compared with the previous state-of-the-art models with F1-scores of 0.880 and 0.845 for prostate and breast cancer mutations, respectively. The performance of a sparse deep autoencoder-based model introduced by Lv et al. (2016) [95] outperformed the performance of a deep autoencoder on most of the clinical relation types. Lin et al. (2017) [80] presented a multi-label structured SVM for Disorder Recognition in the 2013 Conference and Labs of the Evaluation Forum (CLEF) textual dataset. This model achieved an F1-score of 0.7343, i.e., 0.1428 higher than their baseline BIOHD1234 scheme. Mondal et al. (2017) [73] compared the performance of a rule-based approach with a feature-oriented SVM-based supervised learning approach for clinical RE, where the supervised learning model reported higher F1-scores. Magge et al. (2018) [59] used a bidirectional LSTM-CRF for the clinical NER and a random forest-based binary classifier for the clinical RE. The various features used for RE as an input to the random forest classifier such as entity types, number of words in the entity, and an average of the entity word embeddings resulted in a micro-averaged F1-score of 0.88 (precision: 0.82; recall: 0.94). Kim et al. (2018) [72] used node2vec to learn the features from texts in networks in order to extract relations for biological pathways, which outshone the previous methods to detect relationships in the type 2 diabetes pathway. Munkhdalai et al. (2018) [96] compared the performance of an SVM model with a deep learning-based LSTM model to extract relations towards drug surveillance. SVM showed better performance (89.1% F1-score) on the test data compared with that of LSTM. Li et al. (2019) [97] introduced a novel approach for RE in clinical texts by using neural networks to model the shortest dependency path between the target entities along with the sentence sequence. This approach used on the 2010 i2b2 relation extraction dataset improved the performance to an F1-score of 74.34%. The multi-class SVM model on this dataset, introduced by Minard et al. (2019) [81] achieved an F1-score of 0.70, which is lower than the previous models. Christopoulou et al. (2020) [79] proposed an ensemble deep learning method to extract the adverse drug events and medications relations, which achieved a micro-averaged F1-score of 0.9472 and 0.8765 for RE and end-to-end RE, respectively. Hasan et al. (2020) [82] compared the performance of different deep learning methods such as CNN, GCN, GCN-CDT, ResNet, and BiLSTM on various combinations of features, as mentioned in the previous subsection for clinical RE. BiLSTM achieved the highest 9 class F1-score of 0.8808 in that dataset. Both CNN models used by Mahendran et al. (2021) [70], segment-CNN and the sentence-CNN, failed to surpass the performance of the rule-based model proposed for this dataset. Research has shown that the traditional ML methods have outperformed deep learning methods in many clinical RE tasks where the dataset has limited data instances, whereas in some cases where more data is present, deep learning methods given better performance. Additionally, the level of performance depends on the complexity of the data. Currently, it is not possible to generalize whether traditional ML methods or deep learning methods perform the best for clinical RE as the performance is very data-dependent. Some clinical RE tasks using traditional machine learning and deep learning-based methods are tabulated in Table A4.
6.3.2. Language Model-Based Methods
Language model-based approaches have shown improved performance in many NLP tasks as these language models use contextual information into account to represent the features. Then, a classifier is added on top of the language model output to perform the classification of relationships in the end. It is also a supervised learning model as the inputs are well defined for each instance. The language models popularly used in NLP tasks are ULMFit, ELMO, BERT, etc. Out of them, BERT [62], introduced by Google in 2019, has become extremely popular for various NLP tasks including RE. Its breakthrough has resulted in improved performance in many NLP tasks because of its strong ability to pretrain deep bidirectional representations of any unlabelled text by conditioning on its context on both sides in all the 12 transformer layers. For biomedical clinical texts, two BERT-based models were later introduced such as BioBERT [98], trained on biomedical PubMed corpus, and Clinical BERT [99], trained on a biomedical corpus, clinical notes, and only discharge summaries. These models have the same model architecture as that of BERT, but they were trained on a medical corpus.
BERT and the biological and clinical versions of BERT gained high popularity for RE tasks on clinical texts. Since these are language models, there is no need to generate and represent the features. The entire text, i.e., the complete sentence or the complete paragraph of each instance, is taken as input to the model. Lin et al. (2019) [77] established state-of-the-art results in temporal RE in clinical domain using pretrained domain-specific as well as fine-tuned BERT: 0.684F for in-domain texts and 0.565F for cross-domain texts. Alimova et al. (2020) [83] used BERT-based models, including BioBERT and Clinical BERT. The BERT models used there performed really well for some of the classes, but for other classes, the Random Forest Classifier using different input features performed better. Wei et al. (2020) [100] established that the Fine-Tuned BERT eclipsed the performance of other models for RE on clinical narratives. Overall, the language models have shown superior performances than other models on clinical RE tasks. BERT (cased and uncased), BioBERT and Clinical BERT were the language models used by Mahendran et al. (2021) [70]. All of the BERT models, with an impressive macro-averaged F1-score of 0.93, outshone the performance of all of the other rule-based or deep learning methods on this dataset. Therefore, in most cases, language models such as BERT have outshone other ML and deep learning methods for clinical RE due to their capability to learn from the context. Some clinical RE tasks using traditional language model-based methods are tabulated in Table A5.
6.4. Unsupervised Learning Methods
Unsupervised Learning is defined as an ML technique where users are not required to supervise the model, but it allows the model to run and learn by itself to excavate interesting patterns that were earlier undetected. These methods do not require annotated texts as they are capable of working on unlabelled data on their own. The level of processing needed for these kind of tasks is very high, but due to their simplicity, these algorithms are more suitable for simpler tasks and, hence, unsupervised learning algorithms can be unpredictable for RE. The different types of unsupervised learning techniques are Clustering, Anomaly Detection, and Association. In clinical RE, unsupervised learning algorithms have been used to identify the different types of relations in the text that needs to be later reviewed and annotated by domain specialists to evaluate the performance of the model. In real life, the text contains a lot of noise and unsupervised learning is not always effective in identifying the different relations with a high level of accuracy. However, this method is less time expensive and is preferred in some cases.
Unsupervised learning has been the least popular in RE on clinical texts because of the limitations of the unsupervised algorithms to identify relation patterns from complex textual data. Without proper clinical annotations by the clinicians, this learning task is far more ambiguous, which might result in the decreased accuracy of these models. Out of the very few works, Quan et al. (2014) [101] were the pioneers in proposing an unsupervised text mining method for RE on clinical data. The unsupervised clustering-based method that is a combination of dependency and phase structure parsing for RE performed moderately with respect to the previous models but their proposed semi-supervised model surpassed its performance to become the second-best model on this dataset. Alicante et al. (2016) [102] used unsupervised methods for entity and relation extraction from Italian clinical records. The performance of the unsupervised clustering algorithm in the space of entity pairs, being represented by an ad hoc feature vector, is found to be promising in labeling the clinical records by using the most significant features. Since the dataset was not annotated here, similarity measures such as Manhattan, Binary, and Cosine similarities are used to measure the goodness of the clustering models. Not many other unsupervised methods have been proposed for RE on clinical notes. Some clinical RE tasks using unsupervised learning-based methods are tabulated in Table A6.
7. Trends and Future Research Directions
Our main observation from this review is that the clinical-NER community is more focused on deep learning as it has shown promising results. The other approaches such as dictionary or rule-based methods have lost popularity in the last few years. We believe that the upcoming research on clinical NER will develop models using hybrid approaches where the ML-based and rule/dictionary-based approaches can be combined. One of the major challenges while evaluating different clinical NER models was how to measure their effectiveness. The F1-score measure has its own limitations, as mentioned earlier; simply comparing the F1-score does not give much insight into the models. We have seen recently that there are few attempts to address the limitations of F1-score and suggest alternative metrics such as [103]. However, currently, we did not see any attempts to standardize an evaluation metric for clinical NER. For the class imbalance problem discussed in this survey paper, we believe that the community should consider using metrics that address the multi-class imbalance problem. We did see multiple metrics available; however, the selection of correct metric is based on the user interest towards majority or minority classes. Alternatively, we recommend using multiple metrics to obtain a better idea of the balanced performance. We have seen many recent works published on performing clinical NER on text from different languages apart from English and Chinese text such as [52] in Italian text. There are attempts to use transfer learning from the text in different languages to improve the performance such as [52]; although this is still in an initial phase, we believe that, in the next few years, more work will follow this approach. As mentioned in the Clinical NER section, one of the major issues in clinical NER is that most of the models developed are only limited to specific clinics or centers, and specific domains. In order to address this and to make clinical NER models widely available for usage, the clinical terms should be standardized and widely adopted. We found a few attempts on the standardization of clinical terms such as [53]; however, there is not much work currently available that attempts to perform clinical NER on standardized clinical terms and is available for adoption. We believe that the community will move towards a standardization of clinical terms and that future models developed will aim to use those terms. We also noticed that the clinical NER tasks performed vary based on different domains; our survey found that none of them have used transfer learning approaches to train their models from different domains. We believe that, with the success of transfer learning in [52], the community will be looking to develop their deep learning models using transfer learning from different clinical NER tasks.
Most of the clinical NER tasks that we came across aimed to identify the entities from clinical text and then to use them for other NLP tasks. Given the sensitive nature of the clinical text, it is becoming difficult to publish models that are developed for clinical NER. The community is trying to overcome this by developing clinical NER models that identify sensitive terms/entities from clinical text, remove them, and make them available for publishing. Recently, other ML communities are using GANs (Generative Adversarial Networks) [104], which automatically discover patterns in the data and can develop synthetic data that looks similar to the actual data. This approach has many benefits such as handling privacy as no real data is compromised or used in a training phase, and it is capable of handling under sampling and oversampling for multiple classes. We believe that, in the future, clinical NER models will use GANs to develop more robust and scalable models. Likewise, this approach can be one of the potential approaches for clinical RE.
NER reconciliation is a process of collecting data from multiple sources, gathering and mapping them to a real-world object. In clinical NER, this problem can be more severe, as in the radiation oncology domain, different physicians can assign different names to the same structure. Most of the datasets discussed in this paper are annotated and follow the standard naming convention, but this process is not scalable if multiple data sources are used for integration. We performed an extensive search to find any literature on clinical NER reconciliation. To date, we did not find any attempts to perform clinical NER reconciliation. However, we found a few attempts for NER reconciliation in other domains such as Isaac et al. [105] and Van Holland et al. [106]; these approaches are geared towards vocabulary reconciliation. We believe that clinical NER reconciliation is an open research problem. As mentioned earlier, there are ongoing attempts to standardize the clinical terms, and if such a standardization is widely adopted by physicians, it can make the integration process a lot simpler.
After surveying the clinical RE papers, it was found that, lately, the community is most interested in investigating traditional ML-based approaches, deep learning-based approaches, and language models to perform clinical RE. Very little research using rule-based approaches are coming up but unsupervised learning-based methods for clinical RE have become somewhat dormant because of the uncertainty in the results generated by these methods. Rule-based methods were used in many research works before 2016. With the introduction of newer techniques and newer research over the years, the performance of the clinical RE tasks kept on improving. Later on, traditional ML-based methods and deep learning methods along with different feature representation techniques were adopted for this purpose. It was observed that the traditional methods outperformed the deep learning methods in many cases. In some cases, deep learning methods performed poorer than rule-based methods. This may be due to the limited data used in most of these works. Deep learning methods generally perform better than traditional methods in case of a large amount of data, but clinical data is often limited. This is a practical limitation of using deep learning methods for clinical RE. In this era of supervised learning on clinical texts, it was found that the language models such as BERT and its variations vastly perform the best in extracting relations from clinical texts. This shows that the language models are somewhat capable of understanding the intricacies of the language better. However, experimentation with newer and advanced supervised algorithms for relationship classification in the clinical domain should continue in the future as the performance of the algorithms often vary with the data.
In all of the articles we found on clinical RE, F1-score is the metric used for evaluating the performance of the methods. Although other statistical metrics can be used for this purpose, these works chose to only use the F1-score perhaps because of its popularity. When the dataset is not annotated and unsupervised learning-based algorithms have to be used [102], only then other statistical measures are used to quantify the goodness of those measures; for example, Manhattan, Binary, and Cosine similarities were used for comparing the performance of the various clustering models such as Model-Based, K-Means, and Hierarchical Clustering. However, these measures are only used for assessing the goodness of unsupervised learning-based clustering algorithms to provide high-level model performance estimates as they do not serve as a direct evaluation metric for NER/RE tasks. It was observed that most of the clinical RE tasks from a computational point of view are multi-class classification tasks. However, multi-label classification tasks are not used in large numbers for clinical RE because most datasets are annotated into multiple classes but not into multiple labels most of the time.
8. Conclusions
In this paper, we present the first review of the various interrelated NER and RE methods in the context of clinical text. Our literature survey highlights the increasing popularity of various traditional machine learning-based approaches and deep learning models over the past few years, which has somewhat led to a sharp decline in the usage of rule-based methods for both NER and RE or dictionary-based methods for NER only. Hence, hybrid approaches by combining machine learning-based and rule/dictionary-based approaches have the potential to be one of the dominant approaches for these tasks in the future. On top of that, various other machine learning approaches, deep learning approaches, and language model-based approaches for clinical NER and RE will most probably continue to come up in good numbers in the next few years. GANs, which can automatically discover patterns in the data, can potentially also be a good architecture for clinical NER and RE.
In the case of both NER and RE, the F1-score is the most frequentlly used evaluation metric. For unsupervised clinical RE, some work used different similarity measures such as Manhattan, Binary, and Cosine similarities to measure the goodness of the various unsupervised clustering approaches. A few clinical NER papers have mentioned the usage of t-tests on the models to find out their statistical significance. Other popular metrics used in ML-like sensitivity, specificity, ROC, and AUC can also be used in the future to evaluate the performance of the different approaches used for both NER and RE.
We also believe that the community will move towards a standardization of clinical terms and that the future models developed will aim to use these terms. Standardization will help us integrate data from multiple sources and will also help in NER reconciliation. Clinical NER tasks vary based on different domains; we observed that none of them use transfer learning approaches to train their models from different domains. Developing deep learning models using transfer learning from different clinical NER tasks can be a promising future research direction. In the case of clinical RE, relationships are mostly extracted between entities present in a sentence and the types of relationships are mostly multiclass but not multilabel in most cases. Therefore, from the computational angle, it may be worthwhile to carry out more research on RE across sentences besides also multilabel RE but these tasks require data preparation and annotation in some specific formats.
Author Contributions
The contributions of the authors are listed as follows: conceptualization, P.B., S.S. and P.G.; methodology, P.B. and S.S.; investigation, P.B. and S.S.; resources, W.C.S.IV, R.K., J.P. and P.G.; writing—original draft preparation, P.B. and S.S.; writing—review and editing, P.G., W.C.S.IV and R.K.; supervision, R.K., J.P. and P.G.; project administration, R.K., J.P. and P.G.; funding acquisition, R.K. and J.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been funded by the US Veterans Health Administration-National Radiation Oncology Program (VHA-NROP). The results, discussions, and conclusions reported in this paper are completely those of the authors and are independent from the funding sources.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| IDC | International Data Corporation |
| TM | Text Mining |
| NLP | Natural Language Processing |
| NE | Named Entity |
| NER | Named Entity Recognition |
| RE | Relationship Extraction |
| ML | Machine Learning |
| AI | Artificial Intelligence |
| EHR | Electronic Health Record |
| WEKA | Waikato Environment for Knowledge Analysis |
| CLAMP | Clinical Language Annotation, Modeling, and Processing |
| AWS | Amazon Web Services |
| EU | European Union |
| HIPAA | Health Insurance Portability and Accountability Act |
| n2c2 | National NLP Clinical Challenges |
| i2b2 | Informatics for Integrating Biology and the Bedside |
| NIH | National Institutes of Health |
| NCBC | National Centers for Biomedical Computing |
| EMR | Electronic Medical Record |
| SVM | Support Vector Machine |
| RF | Random Forest |
| NN | Neural Network |
| CRF | Conditional Random Field |
| ME | Maximum Entropy |
| BERT | Bidirectional Encoder Representations from Transformers |
| BETO | SPanish BERT |
| BIO | Beginning, Inside, Outside of Entity |
| POS | Parts of Speech |
| BFS | Breadth First Search |
| CNN | Convolutional Neural Network |
| RNN | Recurrent Neural Network |
| LSTM | Long Short Term Memory |
| GCN | Graph Convolutional Network |
| CDT | Concept Dependency Tree |
| GNN | Graph Neural Network |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
| ROC | Receiver Operator Characteristic |
| AUC | Area Under the Curve |
| ODIE | Ontology Development and Information Extraction |
| ADE | Adverse Drug Events |
| PHI | Personal Health Information |
Appendix A

Table A1.
Summary of previous works in clinical NER.
Table A1.
Summary of previous works in clinical NER.
| Publication | Task | Methods | Performance |
|---|---|---|---|
| Savova et al. [46] | Extraction of entities from EMR using NLP tools | Dictionary look-up algorithm | Conducted multiple performance evaluation on different NLP tasks; for NER, the F1-scores reported were 0.71 (exact match) and 0.82 (overlapping matches). |
| Skeppstedt et al. [56] | Detecting disorders, findings, and body structures from Swedish clinical text | Rule-based and CRF approach | Precision and recall for detecting body structure are 0.88 and 0.82, respectively, while for disorder, they were reported as 0.72 and 0.65; for finding, they are 0.72 and 0.65; and for drug, they are 0.95 and 0.83 |
| Chen et al. [57] | Detecting patients who are qualified for clinical trial | Rule-Based approach using knowledge input defined by lexical, syntactic, or meta-level tasks | F1-score reported was 0.90 |
| Eftimov et al. [48] | Extraction of food entity, nutrient entity, and quantity/unit from dietary recommendations | Rule-based approach | TP for food, nutrient, and quantity was reported as 538, 557, and 86. FN for food, nutrient, and quantity was reported as 25, 17, 11. FP for food, nutrient, and quantity was reported as 5, 2, and none. |
| Xu et al. [58] | Combined Segmentation and NER on Chinese text | CRF using three features | 96% F1-score was recorded as the best performance; the authors also provided a comparison between individual, incremental, and joint models. |
| Magge et al. [59] | Identification of specific entities from clinical notes such as drug, dose, and route; a total of nine terms were used for identification | Machine; earning-based approach: bidirectional LSTM-CRF | F1-score average for all nine terms is 0.81; they used the standard gold annotated dataset available at the University of Massachusetts comprising about 1092 medical notes. Around 800 notes were used for training, 76 was for validation, and the rest was used for testing. |

Table A2.
Summary of previous work for clinical NER.
Table A2.
Summary of previous work for clinical NER.
| Publication | Task | Methods | Performance |
|---|---|---|---|
| Nayel et al. [60] | Detection of annotated data from clinical text | Designed an ensemble approach which combined the results of base classifiers and used SVM for learning base classifiers | The proposed ensemble learning model reported an F1-score of 77%. |
| Wu et al. [4] | Concept extraction from clinical text by using and comparing CNN and RNN | Deep learning-based approach | RNN model performed better when compared with CNN and achieved an F1-score of 86%. |
| Wang et al. [61] | Studying symptoms and parthenogenesis in Chinese EHR | ML-based approach used CRF, SVM, and Maximum Entropy (ME) | Among all three methods applied, CRF outperformed the others. |
| Yadav et al. [17] | Advancement and improvement in NER from deep learning models | ML-based approach but focus was more on using deep learning | Better performance reported using deep learning compared with other supervised and semi-supervised learning algorithms. |
| Vunikili et al. [51] | NER on Spanish Clinical Text to extract tumor morphology | Transfer learning using BERT and BETO | 73% F1-score was reported without any features. |
| Jiang et al. [64] | Extraction of clinical entities from 349 clinical annotated notes with different features | ML-based approach (SVM and CRF) | CRF outperformed SVM and their hybrid system achieved an F1-score of 0.84 for concept extraction and 0.93 for assertion classification. |
| Yang et al. [65] | Extraction of family history from clinical narratives | Deep learning-based models such as LSTM, BERT, and ensemble models using majority voting strategy | Micro-averaged F1-score of 0.7944 for concept extraction. |

Table A3.
Summary of the rule-based approaches for clinical RE.
Table A3.
Summary of the rule-based approaches for clinical RE.
| Publication | Task | Methods | Performance |
|---|---|---|---|
| Segura-Bedmar et al. (2011) [84] | Drug–disease interaction extraction from clinical texts | Linguistic hybrid rule-based method using shallow parsing, syntactic simplification, and pattern matching | Did not perform well with an average precision and a very low recall |
| Xu et al. (2011) [85] | Clinical RE on 2010 i2b2 dataset | Combination of Rule-based and ML methods | Model performed decently with a micro-average F1-score of 0.7326 |
| Li et al. (2015) [86] | Automated extraction of medication discrepancy | Matching of drug names with their attributes from a prescription list and confirming it by means of co-location information | Performed well in identifying the medical discrepancies |
| Veena et al. (2021) [87] | RE between different clinical words | Path similarity analysis on the terms extracted by scraping and POS tagging | Successfully converted the data into a classified form |
| Mahendran et al. (2021) [70] | Adverse drug event extraction on 2018 n2c2 dataset | BFS based on the co-location information between the drug and the non-drug entity types | Left-only rule-based approach (macro-average F1-score: 0.83) performed the best amongst other rule-based models |

Table A4.
Summary of the machine learning-based approaches for clinical RE.
Table A4.
Summary of the machine learning-based approaches for clinical RE.
| Publication | Task | Methods | Performance |
|---|---|---|---|
| Roberts et al. (2011) [39] | 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text [38] | SVM-based supervised learning algorithm | Best performance with an F1-score of 0.737 |
| Sahu et al. (2016) [76] | Clinical RE on 2010 i2b2 dataset | Domain invariant CNN on multiple features | Decent performance: filter combination of [4, 6] performed the best (F1-score: 0.7116) amongst CNNs |
| Singhal et al. (2016) [79] | Disease-mutation RE on biomedical texts | C4.5 decision trees on various features | State-of-the art performance thus far; F1-score of 0.880 and 0.845 on prostate and lung disease mutations |
| Lv et al. (2016) [95] | Clinical RE on 2010 i2b2 dataset | Deep autoencoder-based model and sparse deep autoencoder-based model | Sparse deep autoencoder-based model performed better with an F1-score above 80% |
| Lin et al. (2017) [80] | Disorder Recognition in the 2013 CLEF task-1 dataset | multi-label structured SVM | Improved Performance: F1-score: 0.7343, i.e., 0.1428 more than the baseline BIOHD1234 scheme. |
| Mondal et al. (2017) [73] | Clinical RE based on the categories of medical concepts | Feature-oriented SVM-based supervised learning | Better performance (F1-score: 0.86) than the rule-based approach (F1-score: 0.79) |
| Kim et al. (2018) [72] | Clinical RE for biological pathway | Node2vec to learn the features from texts in networks | Best performance for type 2 diabetes pathway |
| Munkhdalai et al. (2018) [96] | Clinical RE towards drug surveillance | SVM model and a deep learning-based LSTM model | SVM performed better (89.1% F1-score) than all of the LSTM models |
| Li et al. (2019) [97] | Clinical RE on 2010 i2b2 dataset | NNs to model the shortest dependency path between entities and sentences | Resulted in an improved performance with an F1-score of 74.34% |
| Minard et al. (2019) [81] | Clinical RE on 2010 i2b2 dataset | Multi-class SVM | Poor performance (F1-score: 0.70) compared with the previous models |
| Christopoulou et al. (2020) [79] | Extraction of the adverse drug events and medications relations | An ensemble deep learning method | Achieved a micro-averaged F1-score of 0.9472 and 0.8765 for RE and end-to-end RE, respectively |
| Hasan et al. (2020) [82] | Clinical RE on 2010 i2b2 dataset | Deep learning methods such as CNN, GCN, GCN-CDT, ResNet, and BiLSTM | BiLSTM performed the best with a nine-class F1-score of 0.8808 and a six-class F1-score of 0.8894 |
| Mahendran et al. (2021) [70] | Adverse drug event extraction on 2018 n2c2 dataset | Sentence-CNN and segment-CNN | The CNN models did not perform better (micro-average F1-score: 0.78 and macro-average F1-score: 0.77) than the other models mentioned |

Table A5.
Summary of the language model-based approaches for clinical RE.
Table A5.
Summary of the language model-based approaches for clinical RE.
| Publication | Task | Methods | Performance |
|---|---|---|---|
| Lin et al. (2019) [77] | Temporal RE in clinical domain | Pretrained domain-specific as well as fine-tuned BERT | State-of-the art performance; 0.684 F1-score for in-domain texts and 0.565 F1-score for cross-domain texts |
| Alimova et al. (2020) [83] | Drug–disease RE from biomedical and clinical texts | BERT, BioBERT and Clinical BERT and Random Forest | The BERT models performed much better on the MADE corpus |
| Wei et al. (2020) [100] | RE on two clinical corpus: 2018 n2c2 dataset and 2010 i2b2 dataset | Fine-tuned and feature-combined BERT along with some deep learning methods | MIMIC fine-tuned BERT performed the best: F1-score of 0.9409 and 0.7679 on the n2c2 and the i2b2 datasets, respectively |
| Mahendran et al. (2021) [70] | Adverse drug event extraction on 2018 n2c2 dataset | BERT (cased and uncased), BioBERT, and Clinical BERT along with other methods | All of the BERT models performed the best, with a micro-averaged F1-score of 0.94 and a macro-averaged F1-score of 0.93 |

Table A6.
Summary of the unsupervised learning approaches for clinical RE.
Table A6.
Summary of the unsupervised learning approaches for clinical RE.
| Publication | Task | Methods | Performance |
|---|---|---|---|
| Quan et al. (2014) [101] | Protein–protein interactions and gene–suicide association extraction | Clustering based on dependency and phased structure parsing | Performed moderately but the proposed semi-supervised model surpassed its performance |
| Alicante et al. (2016) [102] | Domain-relevant entities and RE from Italian clinical records | Model Based, K-Means, and Hierarchical Clustering for pattern discovery | Promising performance to introduce a semi-automatic relation labelling |
References
- Gantz, J.; Reinsel, D. The digital universe in 2020: Big data, bigger digital shadows, and biggest growth in the far east. IDC IView IDC Anal. Future 2012, 2007, 1–16. [Google Scholar]
- Tan, A.H. Text mining: The state of the art and the challenges. In Proceedings of the Pakdd 1999 Workshop on Knowledge Disocovery from Advanced Databases, Bejing, China, 26–28 April 1999; Volume 8, pp. 65–70. [Google Scholar]
- Kong, H.J. Managing unstructured big data in healthcare system. Healthc. Inform. Res. 2019, 25, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Jiang, M.; Xu, J.; Zhi, D.; Xu, H. Clinical Named Entity Recognition Using Deep Learning Models. AMIA Annu. Symp. Proc. 2017, 2017, 1812–1819. [Google Scholar]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA Data Mining Software: An Update. SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Soysal, E.; Wang, J.; Jiang, M.; Wu, Y.; Pakhomov, S.; Liu, H.; Xu, H. CLAMP—A toolkit for efficiently building customized clinical natural language processing pipelines. J. Am. Med. Inform. Assoc. 2017, 25, 331–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, P.; Celikkaya, B.; Khalilia, M.; Senthivel, S. Comprehend Medical: A Named Entity Recognition and Relationship Extraction Web Service. In Proceedings of the 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA), Boca Raton, FL, USA, 16–19 December 2019; pp. 1844–1851. [Google Scholar] [CrossRef] [Green Version]
- Rish, I. An empirical study of the naive Bayes classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, WA, USA, 4–6 August 2001. [Google Scholar]
- Vishwanathan, S.; Murty, M.N. SSVM: A simple SVM algorithm. In Proceedings of the 2002 International Joint Conference on Neural Networks, IJCNN’02 (Cat. No. 02CH37290), Honolulu, HI, USA, 12–17 May 2002; Volume 3, pp. 2393–2398. [Google Scholar]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef] [Green Version]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Society. Ser. C (Appl. Stat.) 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Schubert, E.; Sander, J.; Ester, M.; Kriegel, H.P.; Xu, X. DBSCAN revisited, revisited: Why and how you should (still) use DBSCAN. ACM Trans. Database Syst. (TODS) 2017, 42, 1–21. [Google Scholar] [CrossRef]
- Aggarwal, C.C.; Zhai, C. A survey of text classification algorithms. In Mining Text Data; Springer: Boston, MA, USA, 2012; pp. 163–222. [Google Scholar]
- Derr, T.; Karimi, H.; Liu, X.; Xu, J.; Tang, J. Deep Adversarial Network Alignment. arXiv 2019, arXiv:cs.SI/1902.10307. [Google Scholar]
- Watkins, C.J.; Dayan, P. Q-learning. Mach. Learn. 1992, 8, 279–292. [Google Scholar] [CrossRef]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef] [Green Version]
- Yadav, V.; Bethard, S. A survey on recent advances in named entity recognition from deep learning models. arXiv 2019, arXiv:1910.11470. [Google Scholar]
- Grishman, R.; Sundheim, B. Message understanding conference-6: A brief history. In Proceedings of the 1995 International Conference on Computational Linguistics (COLING), Copenhagen, Denmark, 5–9 August 1995. [Google Scholar]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural language processing (almost) from scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Bach, N.; Badaskar, S. A review of relation extraction. Lit. Rev. Lang. Stat. II 2007, 2, 1–15. [Google Scholar]
- Brin, S. Extracting Patterns and Relations from the World Wide Web. In The World Wide Web and Databases, Proceedings of the International Workshop WebDB’98, Valencia, Spain, 27–28 March 1998; Atzeni, P., Mendelzon, A., Mecca, G., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 172–183. [Google Scholar]
- Agichtein, E.; Gravano, L. Snowball: Extracting Relations from Large Plain-Text Collections. In Proceedings of the Fifth ACM Conference on Digital Libraries (DL’00), San Antonio, TX, USA, 2–7 June 2000; Association for Computing Machinery: New York, NY, USA, 2000; pp. 85–94. [Google Scholar] [CrossRef]
- Culotta, A.; Sorensen, J. Dependency Tree Kernels for Relation Extraction. In Proceedings of the 42nd Annual Meeting of the Association for Computational Linguistics (ACL-04), Barcelona, Spain, 21–26 July 2004; pp. 423–429. [Google Scholar] [CrossRef] [Green Version]
- Bunescu, R.C.; Mooney, R.J. A Shortest Path Dependency Kernel for Relation Extraction. In Proceedings of the HLT/EMNLP, Vancouver, BC, Canada, 6–8 October 2005; pp. 724–731. [Google Scholar]
- Bunescu, R.C.; Mooney, R.J. Subsequence Kernels for Relation Extraction. In Proceedings of the 18th International Conference on Neural Information Processing Systems (NIPS’05), Vancouver, BC, Canada, 5–8 December 2005; MIT Press: Cambridge, MA, USA, 2005; pp. 171–178. [Google Scholar]
- Culotta, A.; McCallum, A.; Betz, J. Integrating Probabilistic Extraction Models and Data Mining to Discover Relations and Patterns in Text. In Proceedings of the HLT-NAACL, New York, NY, USA, 4–9 June 2006. [Google Scholar]
- Allahyari, M.; Pouriyeh, S.; Assefi, M.; Safaei, S.; Trippe, E.D.; Gutierrez, J.B.; Kochut, K. A Brief Survey of Text Mining: Classification, Clustering and Extraction Techniques. arXiv 2017, arXiv:cs.CL/1707.02919. [Google Scholar]
- Hedderich, M.A.; Lange, L.; Adel, H.; Strötgen, J.; Klakow, D. A survey on recent approaches for natural language processing in low-resource scenarios. arXiv 2020, arXiv:2010.12309. [Google Scholar]
- Stubbs, A.; Uzuner, Ö. Annotating longitudinal clinical narratives for de-identification: The 2014 i2b2/UTHealth corpus. J. Biomed. Inform. 2015, 58, S20–S29. [Google Scholar] [CrossRef]
- Stubbs, A.; Kotfila, C.; Xu, H.; Uzuner, Ö. Identifying risk factors for heart disease over time: Overview of 2014 i2b2/UTHealth shared task Track 2. J. Biomed. Inform. 2015, 58, S67–S77. [Google Scholar] [CrossRef]
- Stubbs, A.; Filannino, M.; Uzuner, Ö. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID Shared Tasks Track 1. J. Biomed. Inform. 2017, 75, S4–S18. [Google Scholar] [CrossRef]
- Goto, I.; Chow, K.P.; Lu, B.; Sumita, E.; Tsou, B.K. Overview of the Patent Machine Translation Task at the NTCIR-10 Workshop. In Proceedings of the NTCIR, Tokyo, Japan, 18–21 June 2013. [Google Scholar]
- Coffman, A.; Wharton, N. Clinical Natural Language Processing: Auto-Assigning ICD-9 Codes. Overview of the Computational Medicine Center’s. 2007. Available online: https://courses.ischool.berkeley.edu/i256/f09/Final%20Projects%20write-ups/coffman_wharton_project_final.pdf (accessed on 2 September 2012).
- Jagannatha, A.; Liu, F.; Liu, W.; Yu, H. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug Saf. 2019, 42, 99–111. [Google Scholar] [CrossRef]
- Liu, F.; Jagannatha, A.; Yu, H. Towards Drug Safety Surveillance and Pharmacovigilance: Current Progress in Detecting Medication and Adverse Drug Events from Electronic Health Records. Drug Saf. 2019, 42, 95–97. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Chapman, W.; Man, S.; Savova, G. Semeval-2014 task 7: Analysis of clinical text. In Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014), Dublin, Ireland, 23–24 August 2014. [Google Scholar]
- Pradhan, S.; Elhadad, N.; South, B.R.; Martinez, D.; Christensen, L.; Vogel, A.; Suominen, H.; Chapman, W.W.; Savova, G. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J. Am. Med. Inform. Assoc. 2015, 22, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Uzuner, Ö.; South, B.R.; Shen, S.; DuVall, S.L. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inform. Assoc. 2011, 18, 552–556. [Google Scholar] [CrossRef] [Green Version]
- Roberts, K.; Rink, B.; Harabagiu, S. Extraction of medical concepts, assertions, and relations from discharge summaries for the fourth i2b2/VA shared task. In Proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data, Washington, DC, USA, 12–13 November 2010. [Google Scholar]
- Uzuner, O.; Bodnari, A.; Shen, S.; Forbush, T.; Pestian, J.; South, B.R. Evaluating the state of the art in coreference resolution for electronic medical records. J. Am. Med. Inform. Assoc. 2012, 19, 786–791. [Google Scholar] [CrossRef] [Green Version]
- Glinos, D. A search based method for clinical text coreference resolution. In Proceedings of the 2011 i2b2/VA/Cincinnati Workshop on Challenges in Natural Language Processing for Clinical Data, Washington, DC, USA, 21–22 October 2011. [Google Scholar]
- Xu, Y.; Liu, J.; Wu, J. EHUATUO: A mention-pair coreference system by exploiting document intrinsic latent structures and world knowledge in discharge summaries: 2011 i2b2 challenge. In Proceedings of the 2011 i2b2/VA/Cincinnati Workshop on Challenges in Natural Language Processing for Clinical Data, Washington, DC, USA, 21–22 October 2011. [Google Scholar]
- Sun, W.; Rumshisky, A.; Uzuner, O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J. Am. Med. Inform. Assoc. 2013, 20, 806–813. [Google Scholar] [CrossRef] [Green Version]
- Henry, S.; Buchan, K.; Filannino, M.; Stubbs, A.; Uzuner, O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J. Am. Med. Inform. Assoc. 2019, 27, 3–12. [Google Scholar] [CrossRef]
- Xu, J.; Lee, H.J.; Ji, Z.; Wang, J.; Wei, Q.; Xu, H. UTH_CCB System for Adverse Drug Reaction Extraction from Drug Labels at TAC-ADR 2017; TAC: Gaithersburg, MD, USA, 2017. [Google Scholar]
- Savova, G.K.; Masanz, J.J.; Ogren, P.V.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, J.E.; Ryu, E.; Johnson, K.J.; Koenig, B.A.; Maschke, K.J.; Morrisette, J.A.; Liebow, M.; Takahashi, P.Y.; Fredericksen, Z.S.; Sharma, R.G.; et al. The Mayo Clinic Biobank: A building block for individualized medicine. Mayo Clin. Proc. 2013, 88, 952–962. [Google Scholar] [CrossRef] [Green Version]
- Popovski, G.; Seljak, B.K.; Eftimov, T. A survey of named-entity recognition methods for food information extraction. IEEE Access 2020, 8, 31586–31594. [Google Scholar] [CrossRef]
- Weegar, R.; Pérez, A.; Casillas, A.; Oronoz, M. Recent advances in Swedish and Spanish medical entity recognition in clinical texts using deep neural approaches. BMC Med. Inform. Decis. Mak. 2019, 19, 274. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Li-Wei, H.L.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Vunikili, R.; SH, N.; Marica, G.; Farri, O. Clinical NER using Spanish BERT Embeddings. In Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), Malaga, Spain, 23 September 2020. [Google Scholar]
- Catelli, R.; Gargiulo, F.; Casola, V.; De Pietro, G.; Fujita, H.; Esposito, M. Crosslingual named entity recognition for clinical de-identification applied to a COVID-19 Italian data set. Appl. Soft Comput. 2020, 97, 106779. [Google Scholar] [CrossRef]
- Nalluri, J.; Kapoor, R.; Sleeman, W.; Soni, P.; Ghosh, P.; Khajamoinuddin, S.; Hagan, M.; Palta, J. Health Information and Gateway Exchange (HINGE): Big Data Curation Tool for Radiation Oncology. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, E132. [Google Scholar] [CrossRef]
- Kapoor, R.; Sleeman, W.C., IV; Nalluri, J.J.; Turner, P.; Bose, P.; Cherevko, A.; Srinivasan, S.; Syed, K.; Ghosh, P.; Hagan, M.; et al. Automated data abstraction for quality surveillance and outcome assessment in radiation oncology. J. Appl. Clin. Med. Phys. 2021, 22, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural architectures for named entity recognition. arXiv 2016, arXiv:1603.01360. [Google Scholar]
- Skeppstedt, M.; Kvist, M.; Dalianis, H. Rule-based Entity Recognition and Coverage of SNOMED CT in Swedish Clinical Text. In Proceedings of the LREC, Istanbul, Turkey, 23–25 May 2012; pp. 1250–1257. [Google Scholar]
- Chen, L.; Gu, Y.; Ji, X.; Lou, C.; Sun, Z.; Li, H.; Gao, Y.; Huang, Y. Clinical trial cohort selection based on multi-level rule-based natural language processing system. J. Am. Med. Inform. Assoc. 2019, 26, 1218–1226. Available online: https://academic.oup.com/jamia/article-pdf/26/11/1218/36089031/ocz109.pdf (accessed on 13 July 2019). [CrossRef]
- Xu, Y.; Wang, Y.; Liu, T.; Liu, J.; Fan, Y.; Qian, Y.; Tsujii, J.; Chang, E.I. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. J. Am. Med. Inform. Assoc. 2014, 21, e84–e92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magge, A.; Scotch, M.; Gonzalez-Hernandez, G. Clinical NER and relation extraction using bi-char-LSTMs and random forest classifiers. In Proceedings of the International Workshop on Medication and Adverse Drug Event Detection, Virtual, 4 May 2018; pp. 25–30. [Google Scholar]
- Nayel, H.; Shashirekha, H. Improving NER for clinical texts by ensemble approach using segment representations. In Proceedings of the 14th International Conference on Natural Language Processing (ICON-2017), Kolkata, India, 18–21 December 2017; pp. 197–204. [Google Scholar]
- Wang, Y.; Yu, Z.; Chen, L.; Chen, Y.; Liu, Y.; Hu, X.; Jiang, Y. Supervised methods for symptom name recognition in free-text clinical records of traditional Chinese medicine: An empirical study. J. Biomed. Inform. 2014, 47, 91–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Canete, J.; Chaperon, G.; Fuentes, R.; Pérez, J. Spanish pre-trained bert model and evaluation data. In Proceedings of the PML4DC, ICLR 2020, Addis Aboba, Ethiopia, 26 April 2020. [Google Scholar]
- Jiang, M.; Chen, Y.; Liu, M.; Rosenbloom, S.T.; Mani, S.; Denny, J.C.; Xu, H. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J. Am. Med. Inform. Assoc. 2011, 18, 601–606. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, H.; He, X.; Bian, J.; Wu, Y. Extracting Family History of Patients From Clinical Narratives: Exploring an End-to-End Solution With Deep Learning Models. JMIR Med. Inform. 2020, 8, e22982. [Google Scholar] [CrossRef]
- Hsu, T.C.; Feldt, L.S. The effect of limitations on the number of criterion score values on the significance level of the F-test. Am. Educ. Res. J. 1969, 6, 515–527. [Google Scholar]
- Branco, P.; Torgo, L.; Ribeiro, R.P. Relevance-based evaluation metrics for multi-class imbalanced domains. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Jeju, Korea, 23–26 May 2017; Springer: Cham, Switzerland, 2017; pp. 698–710. [Google Scholar]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2013, 42, D1091–D1097. Available online: https://academic.oup.com/nar/article-pdf/42/D1/D1091/3559045/gkt1068.pdf (accessed on 13 July 2019). [CrossRef] [PubMed] [Green Version]
- Hebbring, S.J. The challenges, advantages and future of phenome-wide association studies. Immunology 2014, 141, 157–165. Available online: https://onlinelibrary.wiley.com/doi/pdf/10.1111/imm.12195 (accessed on 13 July 2019). [CrossRef] [PubMed]
- Mahendran, D.; McInnes, B.T. Extracting Adverse Drug Events from Clinical Notes. arXiv 2021, arXiv:cs.CL/2104.10791. [Google Scholar]
- Sarker, A.; Gonzalez, G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J. Biomed. Inform. 2015, 53, 196–207. [Google Scholar] [CrossRef] [Green Version]
- Kim, M. Relation extraction for biological pathway construction using node2vec. BMC Bioinform. 2018, 19, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondal, A.; Das, D.; Bandyopadhyay, S. Relationship Extraction based on Category of Medical Concepts from Lexical Contexts. In Proceedings of the 14th International Conference on Natural Language Processing (ICON-2017), Kolkata, India, 18–21 December 2017; NLP Association of India: Kolkata, India, 2017; pp. 212–219. [Google Scholar]
- Singhal, A.; Simmons, M.; Lu, Z. Text mining for precision medicine: Automating disease-mutation relationship extraction from biomedical literature. J. Am. Med. Inform. Assoc. 2016, 23, 766–772. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.G.; Choi, H.J. Temporal Relationship Extraction for Natural Language Texts by Using Deep Bidirectional Language Model. In Proceedings of the 2020 IEEE International Conference on Big Data and Smart Computing (BigComp), Busan, Korea, 19–22 February 2020; pp. 555–557. [Google Scholar] [CrossRef]
- Sahu, S.K.; Anand, A.; Oruganty, K.; Gattu, M. Relation extraction from clinical texts using domain invariant convolutional neural network. arXiv 2016, arXiv:cs.CL/1606.09370. [Google Scholar]
- Lin, C.; Miller, T.; Dligach, D.; Bethard, S.; Savova, G. A BERT-based universal model for both within-and cross-sentence clinical temporal relation extraction. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, MN, USA, 7 June 2019; pp. 65–71. [Google Scholar]
- Swampillai, K.; Stevenson, M. Extracting relations within and across sentences. In Proceedings of the International Conference Recent Advances in Natural Language Processing 2011, Hissar, Bulgaria, 12–14 September 2011; pp. 25–32. [Google Scholar]
- Christopoulou, F.; Tran, T.T.; Sahu, S.K.; Miwa, M.; Ananiadou, S. Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. J. Am. Med. Inform. Assoc. 2019, 27, 39–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Ji, D.; Lu, Y. Disorder recognition in clinical texts using multi-label structured SVM. BMC Bioinform. 2017, 18, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minard, A.L.; Ligozat, A.L.; Grau, B. Multi-class SVM for relation extraction from clinical reports. In Proceedings of the Recent Advances in Natural Language Processing, Varna, Bulgaria, 2–4 September 2011. [Google Scholar]
- Hasan, F.; Roy, A.; Pan, S. Integrating Text Embedding with Traditional NLP Features for Clinical Relation Extraction. In Proceedings of the 2020 IEEE 32nd International Conference on Tools with Artificial Intelligence (ICTAI), Baltimore, MD, USA, 9–11 November 2020; pp. 418–425. [Google Scholar] [CrossRef]
- Alimova, I.; Tutubalina, E. Multiple features for clinical relation extraction: A machine learning approach. J. Biomed. Inform. 2020, 103, 103382. [Google Scholar] [CrossRef]
- Segura-Bedmar, I.; Martínez, P.; de Pablo-Sánchez, C. A linguistic rule-based approach to extract drug-drug interactions from pharmacological documents. BMC Bioinform. 2011, 12, S1. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Hong, K.; Tsujii, J.; Chang, E.I.C. Feature engineering combined with machine learning and rule-based methods for structured information extraction from narrative clinical discharge summaries. J. Am. Med. Inform. Assoc. 2012, 19, 824–832. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Spooner, S.A.; Kaiser, M.; Lingren, N.; Robbins, J.; Lingren, T.; Tang, H.; Solti, I.; Ni, Y. An end-to-end hybrid algorithm for automated medication discrepancy detection. BMC Med. Inform. Decis. Mak. 2015, 15, 37. [Google Scholar] [CrossRef] [Green Version]
- Veena, G.; Hemanth, R.; Hareesh, J. Relation Extraction in Clinical Text using NLP Based Regular Expressions. In Proceedings of the 2019 2nd International Conference on Intelligent Computing, Instrumentation and Control Technologies (ICICICT), Kannur, India, 5–6 July 2019; Volume 1, pp. 1278–1282. [Google Scholar] [CrossRef]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach; Prentice Hall: Hoboken, NJ, USA, 2002. [Google Scholar]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021. [Google Scholar] [CrossRef]
- Bose, P.; Sleeman, W.C.; Syed, K.; Hagan, M.; Palta, J.; Kapoor, R.; Ghosh, P. Deep Neural Network Models to Automate Incident Triage in the Radiation Oncology Incident Learning System. In Proceedings of the 12th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (BCB’21), Gainesville, FL, USA, 1–4 August 2021; Association for Computing Machinery: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Watson, D.S.; Krutzinna, J.; Bruce, I.N.; Griffiths, C.E.; McInnes, I.B.; Barnes, M.R.; Floridi, L. Clinical applications of machine learning algorithms: Beyond the black box. BMJ 2019, 364, l886. [Google Scholar] [CrossRef] [Green Version]
- Weng, S.F.; Reps, J.; Kai, J.; Garibaldi, J.M.; Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE 2017, 12, e0174944. [Google Scholar] [CrossRef] [Green Version]
- Sleeman, W.; Bose, P.; Ghosh, P.; Palta, J.; Kapoor, R. Using CNNs to Extract Standard Structure Names While Learning Radiomic Features. In Medical Physics; Wiley: Hoboken, NJ, USA, 2021; Volume 48. [Google Scholar]
- Bose, P.; Sleeman, W.; Srinivasan, S.; Palta, J.; Kapoor, R.; Ghosh, P. Integrated Structure Name Mapping with CNN. In Medical Physics; Wiley: Hoboken, NJ, USA, 2021; Volume 48. [Google Scholar]
- Lv, X.; Guan, Y.; Yang, J.; Wu, J. Clinical relation extraction with deep learning. Int. J. Hybrid Inf. Technol. 2016, 9, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Munkhdalai, T.; Liu, F.; Yu, H. Clinical Relation Extraction Toward Drug Safety Surveillance Using Electronic Health Record Narratives: Classical Learning Versus Deep Learning. JMIR Public Health Surveill. 2018, 4, e29. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Z.; Shen, C.; Xu, J.; Zhang, Y.; Xu, H. Integrating shortest dependency path and sentence sequence into a deep learning framework for relation extraction in clinical text. BMC Med. Inf. Decis. Mak. 2019, 19, 22. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef]
- Alsentzer, E.; Murphy, J.R.; Boag, W.; Weng, W.H.; Jin, D.; Naumann, T.; McDermott, M.B.A. Publicly Available Clinical BERT Embeddings. arXiv 2019, arXiv:cs.CL/1904.03323. [Google Scholar]
- Wei, Q.; Ji, Z.; Si, Y.; Du, J.; Wang, J.; Tiryaki, F.; Wu, S.; Tao, C.; Roberts, K.; Xu, H. Relation Extraction from Clinical Narratives Using Pre-trained Language Models. AMIA Annu. Symp. Proc. 2020, 2019, 1236–1245. [Google Scholar]
- Quan, C.; Wang, M.; Ren, F. An Unsupervised Text Mining Method for Relation Extraction from Biomedical Literature. PLoS ONE 2014, 9, e102039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alicante, A.; Corazza, A.; Isgrò, F.; Silvestri, S. Unsupervised entity and relation extraction from clinical records in Italian. Comput. Biol. Med. 2016, 72, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Hand, D.J.; Christen, P.; Kirielle, N. F*: An interpretable transformation of the F-measure. Mach. Learn. 2021, 110, 451–456. [Google Scholar] [CrossRef]
- Aggarwal, A.; Mittal, M.; Battineni, G. Generative adversarial network: An overview of theory and applications. Int. J. Inf. Manag. Data Insights 2021, 1, 100004. [Google Scholar]
- Isaac, A.; Schlobach, S.; Matthezing, H.; Zinn, C. Integrated access to cultural heritage resources through representation and alignment of controlled vocabularies. Libr. Rev. 2008, 57, 187–199. [Google Scholar] [CrossRef]
- Van Hooland, S.; Verborgh, R.; De Wilde, M.; Hercher, J.; Mannens, E.; Van de Walle, R. Evaluating the success of vocabulary reconciliation for cultural heritage collections. J. Am. Soc. Inf. Sci. Technol. 2013, 64, 464–479. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).