Feasibility of Ex Vivo Margin Assessment with Hyperspectral Imaging during Breast-Conserving Surgery: From Imaging Tissue Slices to Imaging Lumpectomy Specimen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Hyperspectral Imaging
2.1.1. Imaging Setup
2.1.2. Data Preprocessing: Calibration to Diffuse Reflectance
2.1.3. Data Preprocessing: Match Hyperspectral Data Obtained with Both Cameras
2.2. Data Acquisition and Histopathology Correlation
2.2.1. Tissue Slices Dataset
2.2.2. Lumpectomy Dataset
2.3. Hyperspectral Data Analysis
2.3.1. Characteristics of the Measured Surface
2.3.2. Influence of Tissue Thickness Underneath the Measured Surface on Measured Spectrum
2.3.3. Tissue Classification Using a Machine Learning Approach
3. Results
3.1. Data Description
3.2. Characteristics of the Measured Surface
3.3. Influence of Tissue Thickness Underneath the Measured Surface on Measured Spectrum
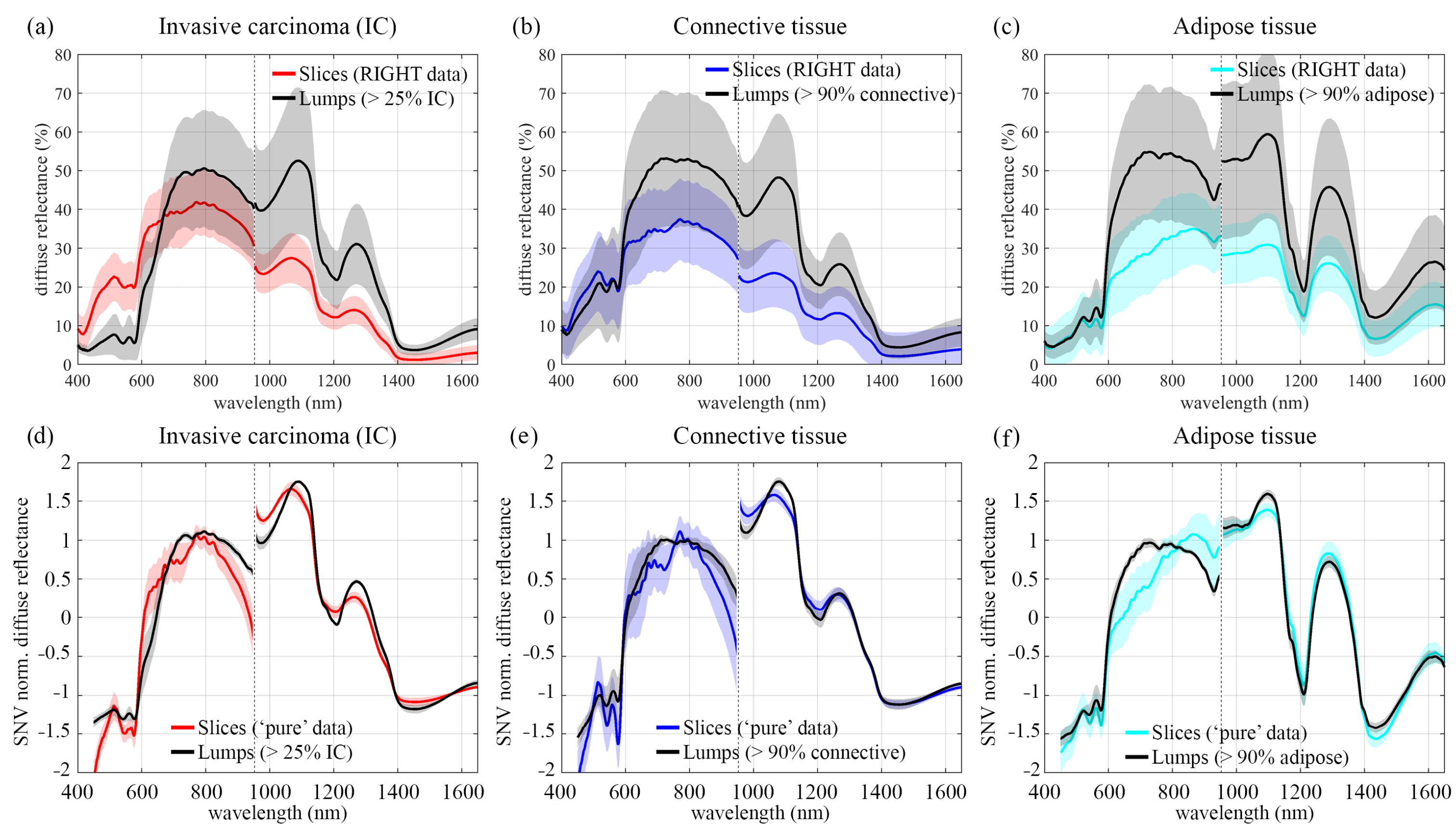
3.3.1. Spectral Comparison before Wavelength Reduction
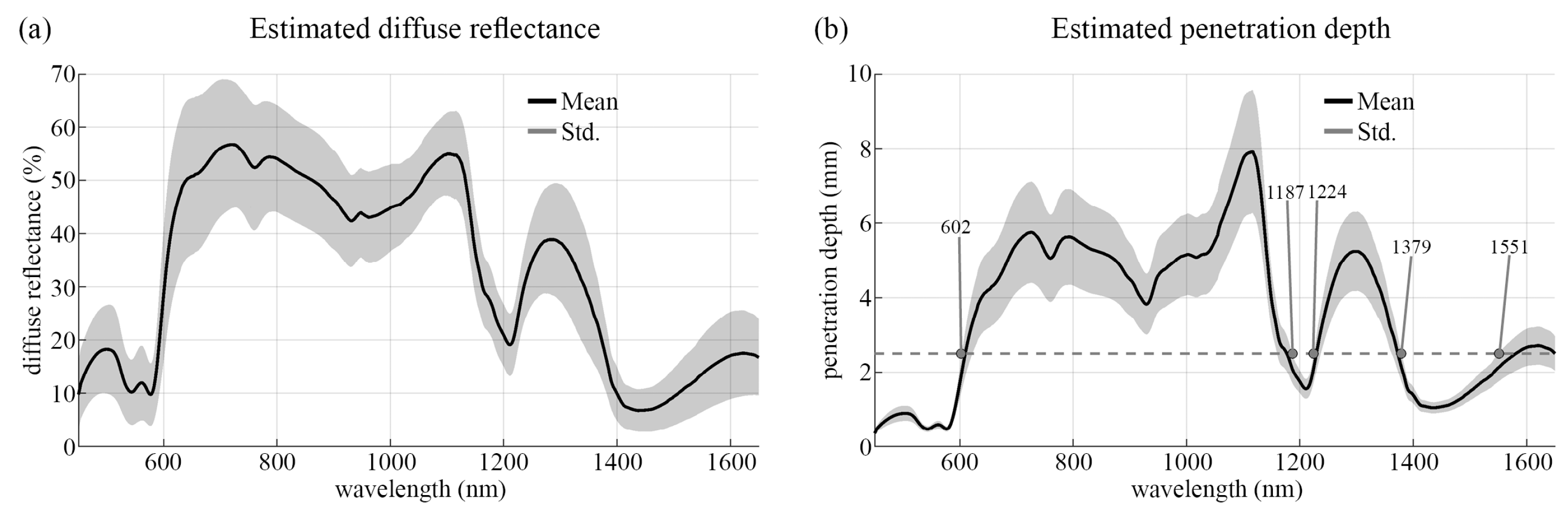
3.3.2. Estimated Penetration Depth
3.3.3. Spectral Comparison after Wavelength Reduction
3.4. Classification Results after Wavelength Reduction Using LDA
3.4.1. Tissue Slices
3.4.2. Lumpectomy Specimen
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Landercasper, J. Variability in Reexcision Following Breast Conservation Surgery. Breast Dis. Year Book Q. 2012, 4, 385–387. [Google Scholar] [CrossRef]
- Gentilini, O.; Gahir, J. An urgent need to reduce re-operation rates after breast conserving surgery. Gland. Surg. 2012, 1, 159. [Google Scholar] [CrossRef]
- Jeevan, R.; Cromwell, D.; Trivella, M.; Lawrence, G.; Kearins, O.; Pereira, J.; Sheppard, C.; Caddy, C.; Van Der Meulen, J. Reoperation rates after breast conserving surgery for breast cancer among women in England: Retrospective study of hospital episode statistics. BMJ 2012, 345, e4505. [Google Scholar] [CrossRef] [Green Version]
- Lagios, M.; Silverstein, M. Implications of New Lumpectomy Margin Guidelines for Breast-Conserving Surgery: Changes in Reexcision Rates and Predicted Rates of Residual Tumor. Breast Dis. Year Book Q. 2016, 4, 295–296. [Google Scholar] [CrossRef]
- Merrill, A.L.; Tang, R.; Plichta, J.K.; Rai, U.; Coopey, S.B.; McEvoy, M.P.; Hughes, K.S.; Specht, M.C.; Gadd, M.A.; Smith, B.L. Should new “no ink on tumor” lumpectomy margin guidelines be applied to ductal carcinoma in situ (DCIS)? A retrospective review using shaved cavity margins. Ann. Surg. Oncol. 2016, 23, 3453–3458. [Google Scholar] [CrossRef]
- Stewart, B.W.; Kleihues, P. World Cancer Report; IARC Press: Lyon, France, 2003. [Google Scholar]
- Havel, L.; Naik, H.; Ramirez, L.; Morrow, M.; Landercasper, J. Impact of the SSO-ASTRO margin guideline on rates of re-excision after lumpectomy for breast cancer: A meta-analysis. Ann. Surg. Oncol. 2019, 26, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Schulman, A.M.; Mirrielees, J.A.; Leverson, G.; Landercasper, J.; Greenberg, C.; Wilke, L.G. Reexcision surgery for breast cancer: An analysis of the American Society of Breast Surgeons (ASBrS) Mastery SM database following the SSO-ASTRO “no ink on tumor” guidelines. Ann. Surg. Oncol. 2017, 24, 52–58. [Google Scholar] [CrossRef]
- Vos, E.; Gaal, J.; Verhoef, C.; Brouwer, K.; Van Deurzen, C.; Koppert, L. Focally positive margins in breast conserving surgery: Predictors, residual disease, and local recurrence. Eur. J. Surg. Oncol. 2017, 43, 1846–1854. [Google Scholar] [CrossRef]
- Lu, G.; Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 2014, 19, 010901. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Little, J.V.; Wang, X.; Zhang, H.; Patel, M.R.; Griffith, C.C.; El-Deiry, M.W.; Chen, A.Y.; Fei, B. Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging. Clin. Cancer Res. 2017, 23, 5426–5436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halicek, M.; Dormer, J.D.; Little, J.V.; Chen, A.Y.; Myers, L.; Sumer, B.D.; Fei, B. Hyperspectral imaging of head and neck squamous cell carcinoma for cancer margin detection in surgical specimens from 102 patients using deep learning. Cancers 2019, 11, 1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltussen, E.J.; Kok, E.N.; de Koning, S.G.B.; Sanders, J.; Aalbers, A.G.; Kok, N.F.; Beets, G.L.; Flohil, C.C.; Bruin, S.C.; Kuhlmann, K.F.; et al. Hyperspectral imaging for tissue classification, a way toward smart laparoscopic colorectal surgery. J. Biomed. Opt. 2019, 24, 016002. [Google Scholar] [CrossRef] [PubMed]
- Kho, E.; de Boer, L.L.; Post, A.L.; Van de Vijver, K.K.; Jóźwiak, K.; Sterenborg, H.J.; Ruers, T.J. Imaging depth variations in hyperspectral imaging: Development of a method to detect tumor up to the required tumor-free margin width. J. Biophotonics 2019, 12, e201900086. [Google Scholar] [CrossRef] [PubMed]
- Kho, E.; Dashtbozorg, B.; De Boer, L.L.; Van de Vijver, K.K.; Sterenborg, H.J.; Ruers, T.J. Broadband hyperspectral imaging for breast tumor detection using spectral and spatial information. Biomed. Opt. Express 2019, 10, 4496–4515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabelo, H.; Halicek, M.; Ortega, S.; Szolna, A.; Morera, J.; Sarmiento, R.; Callico, G.M.; Fei, B. Surgical aid visualization system for glioblastoma tumor identification based on deep learning and in vivo hyperspectral images of human patients. In Medical Imaging 2019: Image-Guided Procedures, Robotic Interventions, and Modeling; International Society for Optics and Photonics: Washington, DC, USA, 2019; Volume 10951, p. 1095110. [Google Scholar] [CrossRef]
- Kho, E.; de Boer, L.L.; Van de Vijver, K.K.; van Duijnhoven, F.; Peeters, M.J.T.V.; Sterenborg, H.J.; Ruers, T.J. Hyperspectral imaging for resection margin assessment during cancer surgery. Clin. Cancer Res. 2019, 25, 3572–3580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Boer, L.L.; Kho, E.; Nijkamp, J.; Van de Vijver, K.K.; Sterenborg, H.J.; ter Beek, L.C.; Ruers, T.J. Method for coregistration of optical measurements of breast tissue with histopathology: The importance of accounting for tissue deformations. J. Biomed. Opt. 2019, 24, 075002. [Google Scholar] [CrossRef]
- Nederland, I.K. Landelijke Richtlijn Mammacarcinoom Versie 2.0 (Dutch Guideline Breast Cancer Version 2.0). 2016. Available online: https://www.oncoline.nl/uploaded/docs/mammacarcinoom/Dutch%20Breast%20Cancer%20Guideline%202012.pdf (accessed on 23 August 2021).
- Barnes, R.; Dhanoa, M.S.; Lister, S.J. Standard normal variate transformation and de-trending of near-infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Van der Meer, F. The effectiveness of spectral similarity measures for the analysis of hyperspectral imagery. Int. J. Appl. Earth Obs. Geoinf. 2006, 8, 3–17. [Google Scholar] [CrossRef]
- Randeberg, L.L.; Larsen, E.L.; Svaasand, L.O. Hyperspectral imaging of blood perfusion and chromophore distribution in skin. In Photonic Therapeutics and Diagnostics V; International Society for Optics and Photonics: Washington, DC, USA, 2009; Volume 7161, p. 71610C. [Google Scholar] [CrossRef]
- Zhang, H.; Salo, D.C.; Kim, D.M.; Komarov, S.; Tai, Y.C.; Berezin, M.Y. Penetration depth of photons in biological tissues from hyperspectral imaging in shortwave infrared in transmission and reflection geometries. J. Biomed. Opt. 2016, 21, 126006. [Google Scholar] [CrossRef] [PubMed]
- Flock, S.T.; Patterson, M.S.; Wilson, B.C.; Wyman, D.R. Monte Carlo modeling of light propagation in highly scattering tissues. I. Model predictions and comparison with diffusion theory. IEEE Trans. Biomed. Eng. 1989, 36, 1162–1168. [Google Scholar] [CrossRef] [PubMed]
- Nachabé, R.; Hendriks, B.H.W.; Lucassen, G.W.; van der Voort, M.; Evers, D.J.; Rutgers, E.J.; Peeters, M.-J.V.; Van der Hage, J.A.; Oldenburg, H.S.; Ruers, T.J.; et al. Diagnosis of breast cancer using diffuse optical spectroscopy from 500 to 1600 nm: Comparison of classification methods. J. Biomed. Opt. 2011, 16, 087010. [Google Scholar] [CrossRef] [PubMed]
- Bjorgan, A.; Milanic, M.; Randeberg, L.L. Estimation of skin optical parameters for real-time hyperspectral imaging applications. J. Biomed. Opt. 2014, 19, 066003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bydlon, T.M.; Barry, W.T.; Kennedy, S.A.; Brown, J.Q.; Gallagher, J.E.; Wilke, L.G.; Geradts, J.; Ramanujam, N. Advancing optical imaging for breast margin assessment: An analysis of excisional time, cautery, and patent blue dye on underlying sources of contrast. PLoS ONE 2012, 7, e51418. [Google Scholar] [CrossRef] [PubMed]
- Spliethoff, J.W.; Tanis, E.; Evers, D.J.; Hendriks, B.H.; Prevoo, W.; Ruers, T.J. Monitoring of tumor radio frequency ablation using derivative spectroscopy. J. Biomed. Opt. 2014, 19, 097004. [Google Scholar] [CrossRef]
- Adank, M.W.; Fleischer, J.C.; Dankelman, J.; Hendriks, B.H. Real-time oncological guidance using diffuse reflectance spectroscopy in electrosurgery: The effect of coagulation on tissue discrimination. J. Biomed. Opt. 2018, 23, 115004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouw, W.M.; Loog, M. An introduction to domain adaptation and transfer learning. arXiv 2018, arXiv:1812.11806. [Google Scholar]
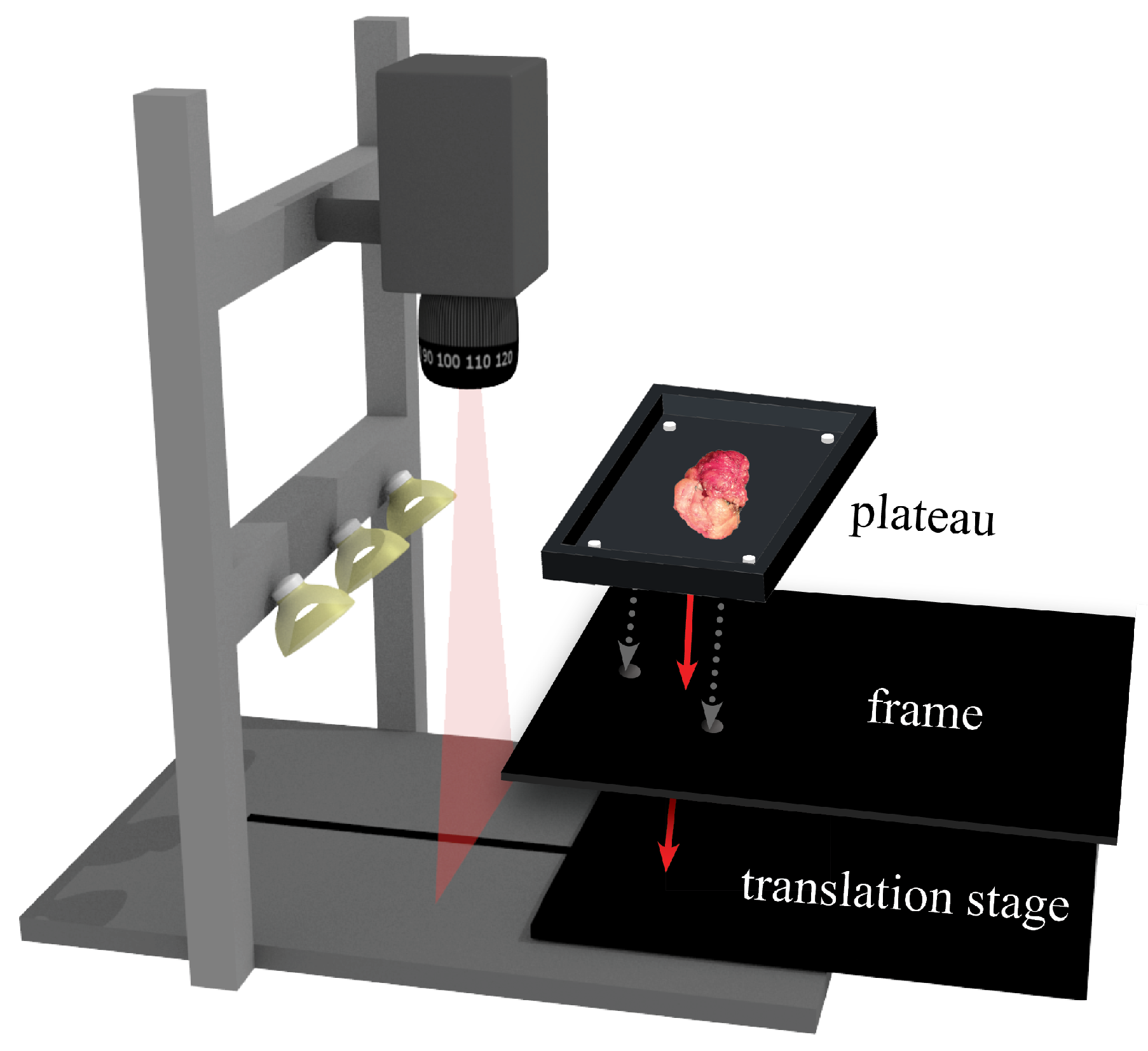
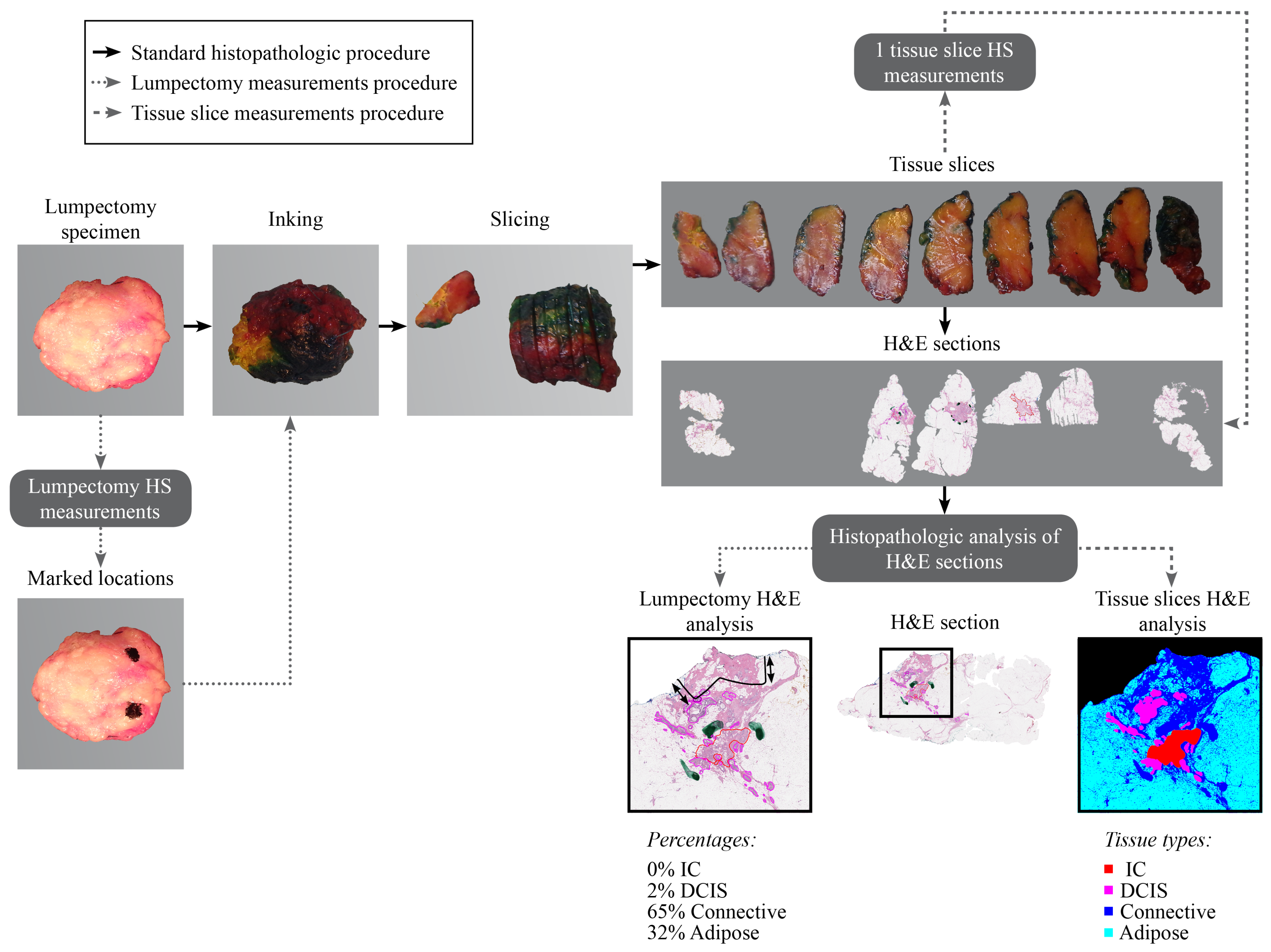


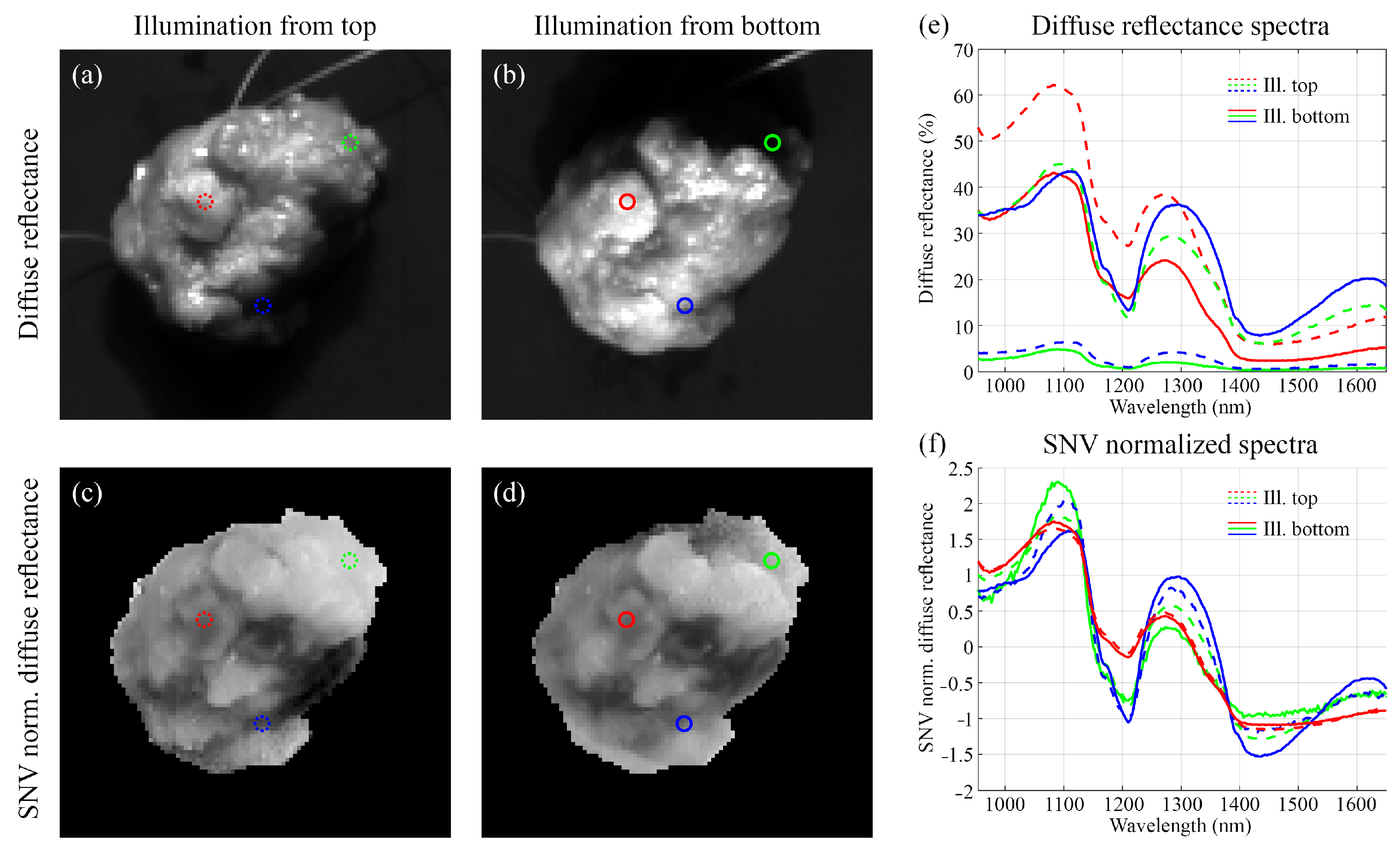
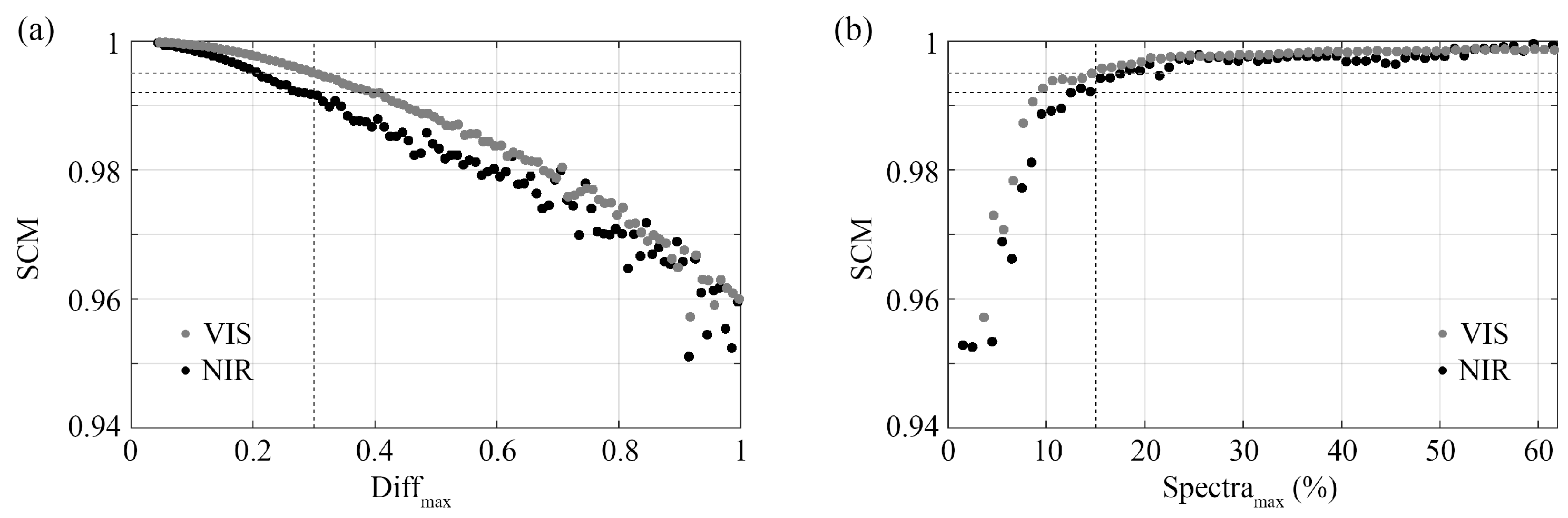



| Tissue Slices Dataset | Lumpectomy Dataset | |
|---|---|---|
| Data acquisition | ||
| Inclusion | Mastectomy and lumpectomy specimen | Lumpectomy specimen |
| Time in clinical workflow | After inking and slicing the specimen | Directly after resection |
| Measured surface | ||
| cut with | Metal knife (at the pathology department) | Ablation knife (during surgery) |
| flatness | As flat as possible | Spherical shaped |
| Histopathology | H & E section parallel | H & E section perpendicular |
| to measured side | to measured surface | |
| Spatial correlation | High | Low |
| Depth information | Low | High |
| Tissue Slices Dataset | Lumpectomy Specimen Dataset | |
|---|---|---|
| Patient and specimen characteristics | ||
| Number of patient | 42 | 52 |
| Patient age (mean ± std) | 55.8 ± 10.9 | 57.7 ± 10.4 |
| Patient ACR (mean ± std) | 2.81 ± 0.74 | 2.46 ± 0.80 |
| Specimen size (mean range) | surface: 15 cm (4–26) | volume: 51 cm (6–240) |
| thickness: 4.6 mm (2.5–8) | minimum thickness: 10 mm | |
| Specimens surface flatness | As flat as possible | Spherical shaped |
| Obtained data | ||
| Tissue type measurements | spectra/# patients | locations/# patients |
| Invasive carcinoma | 6054/20 | 5/5 |
| Ductal carcinoma in situ | 320/8 | 3/3 |
| Connective | 1180/15 | 44 /33 |
| Adipose | 60,622/42 | 58 /46 |
| Purity measurement | ‘pure’ and ‘mixed’ data | ‘mixed’ data |
| Margin assessment | No | Yes |
| Tissue Type | Percentage in H & E Section | Scattering | Absorption | |||||
|---|---|---|---|---|---|---|---|---|
| (%) | (%) | Blood (%) | Fat (%) | H2O (%) | ||||
| IC | 29 | 2.9 ± 0.9 | 100 ± 14 | 0.80 ± 0.76 | 45 ± 6 | 8.1 ± 0.7 | 31 ± 12 | 69 ± 12 |
| IC | 71 | 6.1 ± 1.2 | 100 ± 5 | 0.40 ± 0.37 | 55 ± 3 | 6.4 ± 1.0 | 36 ± 7 | 64 ± 7 |
| IC | 78.5 | 4.6 ± 1.3 | 100 ± 9 | 0.70 ± 0.64 | 24 ± 6 | 5.6 ± 1.6 | 35 ± 11 | 65 ± 11 |
| Connective | 98 | 24.9 ± 6.9 | 100 ± 8 | 0.82 ± 0.78 | 93 ± 3 | 2.7 ± 1.0 | 34 ± 15 | 66 ± 15 |
| Connective | 92 | 25.6 ± 14.8 | 100 ± 73 | 2.20 ± 2.33 | 87 ± 8 | 4.8 ± 3.6 | 37 ± 24 | 63 ± 24 |
| Connective | 93 | 5.4 ± 1.2 | 100 ± 4 | 0.20 ± 0.55 | 33 ± 6 | 3.6 ± 1.2 | 17 ± 16 | 83 ± 16 |
| Connective | 93 | 10.1 ± 1.8 | 100 ± 4 | 0.50 ± 0.46 | 75 ± 3 | 1.5 ± 0.3 | 43 ± 8 | 57 ± 8 |
| Connective | 91 | 11.0 ± 2.1 | 100 ± 4 | 0.35 ± 0.50 | 70 ± 3 | 4.6 ± 2.9 | 5 ± 1 | 95 ± 1 |
| Adipose | 95 | 15.0 ± 3.0 | 100 ± 6 | 0.59 ± 0.53 | 78 ± 3 | 4.6 ± 0.9 | 90 ± 3 | 10 ± 3 |
| Adipose | 96 | 6.1 ± 1.1 | 100 ± 6 | 0.54 ± 0.52 | 64 ± 4 | 3.7 ± 0.6 | 89 ± 2 | 11 ± 2 |
| Adipose | 94 | 10.2 ± 3.7 | 100 ± 19 | 1.34 ± 1.09 | 78 ± 6 | 4.4 ± 1.6 | 91 ± 5 | 9 ± 5 |
| Adipose | 92 | 10.9 ± 3.8 | 100 ± 23 | 1.49 ± 1.19 | 75 ± 6 | 6.7 ± 2.4 | 84 ± 8 | 16 ± 8 |
| Adipose | 92 | 11.1 ± 2.6 | 100 ± 10 | 1.00 ± 0.68 | 70 ± 4 | 3.4 ± 0.8 | 92 ± 2 | 9 ± 2 |
| Adipose | 92 | 5.7 ± 0.8 | 100 ± 8 | 0.45 ± 0.37 | 73 ± 3 | 3.2 ± 2.6 | 91 ± 1 | 9 ± 1 |
| Adipose | 92 | 20.3 ± 13.4 | 100 ± 70 | 2.10 ± 2.28 | 84 ± 10 | 8.6 ± 5.8 | 87 ± 10 | 13 ± 10 |
| Adipose | 93 | 7.2 ± 1.1 | 100 ± 5 | 0.54 ± 0.47 | 71 ± 3 | 2.6 ± 0.4 | 92 ± 1 | 8 ± 1 |
| Before Wavelength Reduction [15] | After Wavelength Reduction | ||
|---|---|---|---|
| Number of | VIS | VIS + NIR | |
| Spectra | NIR (450–1650 nm) | (450–602 & 1187–1224 & 1379–1551 nm) | |
| Tumor | 3312 | 99% | 79% |
| IC | 3200 | 99% | 80% |
| DCIS | 112 | 95% | 77% |
| Healthy | 21,227 | 99% | 90% |
| Connective | 468 | 94% | 68% |
| Adipose | 20,759 | 100% | 96% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kho, E.; Dashtbozorg, B.; Sanders, J.; Vrancken Peeters, M.-J.T.F.D.; van Duijnhoven, F.; Sterenborg, H.J.C.M.; Ruers, T.J.M. Feasibility of Ex Vivo Margin Assessment with Hyperspectral Imaging during Breast-Conserving Surgery: From Imaging Tissue Slices to Imaging Lumpectomy Specimen. Appl. Sci. 2021, 11, 8881. https://doi.org/10.3390/app11198881
Kho E, Dashtbozorg B, Sanders J, Vrancken Peeters M-JTFD, van Duijnhoven F, Sterenborg HJCM, Ruers TJM. Feasibility of Ex Vivo Margin Assessment with Hyperspectral Imaging during Breast-Conserving Surgery: From Imaging Tissue Slices to Imaging Lumpectomy Specimen. Applied Sciences. 2021; 11(19):8881. https://doi.org/10.3390/app11198881
Chicago/Turabian StyleKho, Esther, Behdad Dashtbozorg, Joyce Sanders, Marie-Jeanne T. F. D. Vrancken Peeters, Frederieke van Duijnhoven, Henricus J. C. M. Sterenborg, and Theo J. M. Ruers. 2021. "Feasibility of Ex Vivo Margin Assessment with Hyperspectral Imaging during Breast-Conserving Surgery: From Imaging Tissue Slices to Imaging Lumpectomy Specimen" Applied Sciences 11, no. 19: 8881. https://doi.org/10.3390/app11198881
APA StyleKho, E., Dashtbozorg, B., Sanders, J., Vrancken Peeters, M.-J. T. F. D., van Duijnhoven, F., Sterenborg, H. J. C. M., & Ruers, T. J. M. (2021). Feasibility of Ex Vivo Margin Assessment with Hyperspectral Imaging during Breast-Conserving Surgery: From Imaging Tissue Slices to Imaging Lumpectomy Specimen. Applied Sciences, 11(19), 8881. https://doi.org/10.3390/app11198881








