Integrating Speculation Detection and Deep Learning to Extract Lung Cancer Diagnosis from Clinical Notes
Abstract
:1. Introduction
2. Related Work
3. A Hybrid Approach to Extract Lung Cancer Diagnosis
- Some cancer concepts are associated with a family member, but not the patient.
- Speculation and negation are two linguistic phenomena that frequently appear in clinical texts.
- Different events can occur to the patient (begin clinical trial, diagnosis, start treatment, etc.). All these events will appear with a date and can appear in a sentence in which a mention to a cancer concept is present. So it is needed to distinguish the events and the dates. As an example, let’s take the sentence: “Patient diagnosed with lung cancer, treated with surgery in March 2017”; in this case the date refers to the surgery and not to the diagnosis. Thus apart from cancer and date annotations, an approach to find events and to related dates to events is required.
- A medical record can contain hundreds of clinical notes for the same patient; this fact represents a challenge to extract the correct diagnosis date automatically. This problem is enhanced when a hospital is treating thousands of cancer patients, and therefore the number of clinical notes grows rapidly.
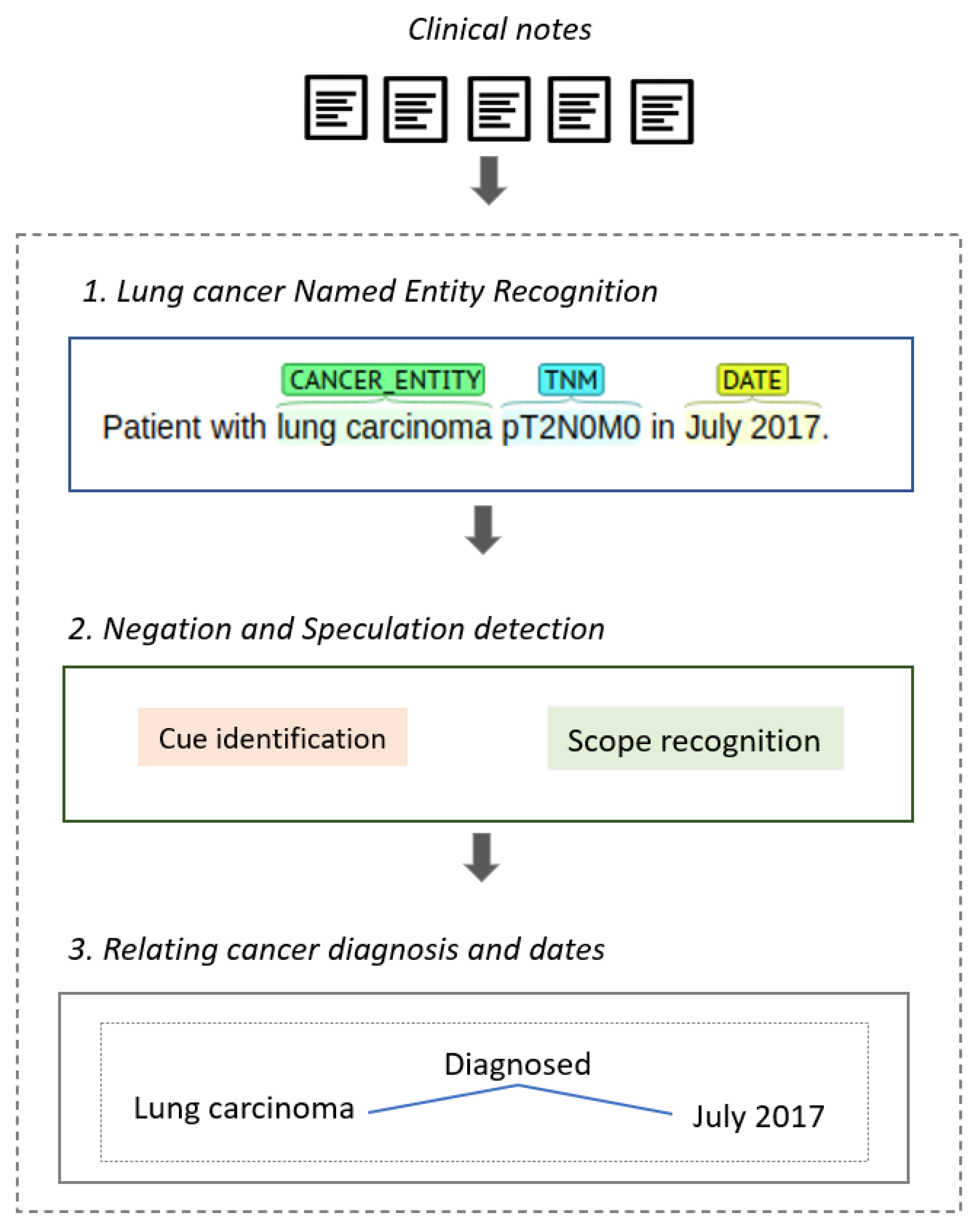
4. Lung Cancer Named Entity Recognition
4.1. Corpus Annotation
- Cancer entity: this entity captures both the cancer type (carcinoma, adenocarcinoma, cancer, etc.) together with the anatomical location (left lung, right lung, right lobe, etc.). For instance: “Patient diagnosed with right lung adenocarcinoma”. In the annotation process the more detailed description will be used. Thus in the sentence, “Patient diagnosed with small cell lung cancer”, the concept “Small cell lung cancer” will be annotated instead of “lung cancer”.
- Cancer stage: staging is the process of finding out how much cancer is in a person’s body and where it’s located. The cancer stage can be expressed on a scale that ranges from I to IV. Stage I indicates the initial stage and stage IV, the most advanced stage of cancer. The cancer staging can also be done using the TNM notation: Tumor (T), Nearby(N), and Metastasis(M). As a consequence, we have annotated both scales. On the one hand, entities such as stage II and stage IV are annotated as a stage entity while expressions such as cT3cN3cM1 and T3 N2 M0 are annotated as TNM entity.
- Dates: Represents dates and time expressions mentioned in clinical notes. Date entity is a crucial concept to obtain the natural history of the patient. Only explicit dates are annotated.
- Events: This entity is used to represent events such as being diagnosed, being treated, treatment start, begin clinical trial, etc. In the sentence “Patient diagnosed with lung cancer, begins treatment with chemotherapy on 5 December 2019, there are two events (shown in bold).
- Family members: represents concepts about family members of a patient. This entity commonly appears together with cancer concepts in the family antecedents, (e.g., ”Mother diagnosed with lung cancer in 2007)”. This entity is used to differentiate between a cancer concept associated to a patient, and one associated to a family member.
- Treatment: The kind of treatment ranging from chemotherapy, radiotherapy, and surgery will be included. These treatments will be annotated separately. This tag is only used to annotate generic mentions to treatments, without mentioning the specific drug, as in the case of sentences such as: “Patient with lung cancer, with chemotherapy in October 2018”.
- Drug: This entity is used to annotate particular names of drugs related to any kind of treatment, such as in the sentence: Lung cancer patient, treated with Carboplatin on October 2018.
4.2. A Deep Learning Approach to Extract Lung Cancer Entities
- Embedding layer: this layer makes it possible to represent words and documents using a dense vector representation. Word embeddings allows words with similar meanings to have a similar representation. The model that is proposed has been trained with the inclusion of different biomedical embedding:
- –
- SciELO Full-Text: This word embedding was created using full-text medical articles from Scielo, a scientific electronic library [41].
- –
- WikiHealth: This embedding was generated using a subset of Wikipedia articles comprised by the categories of Pharmacology, Medicine and Biology [41].
- –
- Lemma and Part of Speech (POS): We create in-house embeddings with lemmas and POS tags using as input the sentences in the annotated corpus.
- BiLSTM layer: as the input setting, this layer uses different embeddings and processes each vector representation of the text sequence in two ways:As an output, this layer produces a vector representation for each word, concatenating the left and right context values. These vectors contain scores for each label that is to be predicted. In the BiLSTM model, each input sentence is contextualized both on the left (L) and on the right (R). Both LSTM (left and right) are independent but contextualized with the same distribution function.
- CRF layer: This layer decodes the best label in all possible labels using the CRF (Conditional Random Fields) algorithm proposed by [49]. This algorithm considers the correlations between other labels and jointly decodes the best chain of labels for a given input sentence of text. Although the BiLSTM layer produces scores for each label, these scores are conditionally independent. For sequence labeling tasks, it is necessary to consider the correlations between labels. Therefore, the CRF layer aims to model dependencies between these labels to improve the predictions for each label. As an input setting, the CRF layer takes the output vectors from the BiLSTM layer and outputs the label sequence with the highest prediction score.
5. Negation and Speculation Detection
“Patient withpossiblelung cancer in July 2014, we recommend a chest test to confirm.”.
5.1. Developing a Cues Lexicon
- Cue Collection: The main goal of this step is to get an initial lexicon containing an initial set of cues in Spanish. Two public resources are used: BioScope corpus [52], and ConText [53]. The collected cues are translated to Spanish using Google translate, and later they are manually corrected. Bioscope and ConText are chosen as these resources are frequently used for negation and uncertainty detection in the English language.
- Semantic Enrichment: this step aims to enrich the initial lexicon by adding new cues that are semantically related. For each cue in the initial lexicon, similar words indicating negation or speculation are found. The semantic enrichment step is performed using the word embedding technique [38,54]. Since the word embedding technique generates high-dimensional vectors for each word, the t-distributed stochastic neighbor embedding (t-SNE) technique [55] was used to visualize the similarity between words using only two dimensions. Figure 5 shows in a two-dimensional diagram a set of words (printed in blue) semantically related to the cue “Probable” (printed in red), and other terms (printed in green) that appear within the context of this cue. The initial lexicon contains 390 cues, and after the semantic enrichment step, the cue’s lexicon contains 512 cues.
5.2. Cue Detection in Clinical Texts
- Prefix-cue: is found before the affected tokens. For example: in the sentence:“Paciente conprobablecarcinoma pulmonar”. (Patient with probable lung carcinoma), the cue is the word “probable” and the tokens affected are underlined. In these cases the tokens affected appear to the right of the cue.
- Postfix-cue: is found after the affected tokens. In the next sentence the cue is “negativa” and the tokens affected are underlined. In these cases the tokens affected appear to the left of the cue:“Biopsia líquida para cáncer: negativa”. (Liquid biopsy for cancer: negative).
“Nodolor, novómitos,nifiebre,notos”. (No pain, no vomiting, no fever, no cough), contains four contiguous cues. The affected tokens are underlined.
“Análisis de orina:Negativo, glóbulos rojos:negativo.” (Urinalysis: Negative, Red Blood Cells: Negative).
5.3. Sentence Analysis
- Sentence length: is the number of tokens in the sentence in which speculation or negation cues were detected. This property aims to analyze the behavior of negation and speculation according to the sentence’s length. The sentence length is used to define the short sentence heuristic. This heuristic indicates that a sentence is considered as a short sentence if its length belongs to the first quartile, and it has only one cue. Although short sentences are more frequently found in clinical text [51], long sentences can also be found [57]. Therefore, the short sentence heuristic is used to choose the proper rule in the scope recognition step according to sentence length.
- Presence of contiguous cues: this aims to analyze the behavior of negation and speculation in the presence of contiguous cues. This step is important as a condition needed to recognize the scope depending on the number of cues that the sentence contains.
5.4. Scope Recognition
- Rule 1: if the sentence contains a termination term, the scope is extracted using this term. A termination term is a word that indicates the end of the scope. Termination terms are previously created in a lexicon. In the next example the word “pero” (but) indicates the end of the scope (the scope is underlined).-“Probable carcinoma de pulmón, pero espero resultados de biopsia.”(“Probable lung carcinoma, but I will wait for biopsy results.”)
- Rule 2: if a cue is detected in a sentence containing contiguous cues , the scope for each will be given by the position of . For example in the sentence:-“No dolor, no inflamación, no dolores articulares, ni fiebre.” (No pain, no inflammation, no joint pain, no fever.), for each cue, the scope is given by the position of the next cue.
- Rule 3: if the sentence length corresponds to “a short sentence heuristic”, then the scope is given by the end of the sentence. The following example shows a short sentence with their cue and the scope (underlined):-“Posiblecancer pulmonar.” (“Possible lung cancer.”)
- Rule 4: if the sentence contains a token POS tagged with a conjunction or verb category. In this case, the scope is determined by the position of this token. The next sentence contains the token “proponemos”, which is tagged with the Verb category and indicates the end of the scope (the scope is underlined).-“Con la sospecha de cancer de pulmón, proponemos reunión del comité de tumores.”(Given the possibility of lung cancer, we propose a meeting with the tumor committee.)
- Rule 5: if the sentence does not match the previous rules, the algorithm generates a sentence parse tree. In this case, the scope is given by the sub-tree that contains the uncertainty or negation cue, as is shown in the next sentence. (The scope is underlined)-“Signos sugestivos de nódulos pulmonares, con fiebre alta desde ayer.”(Signs that suggest pulmonary nodules, with a high fever present since yesterday.)
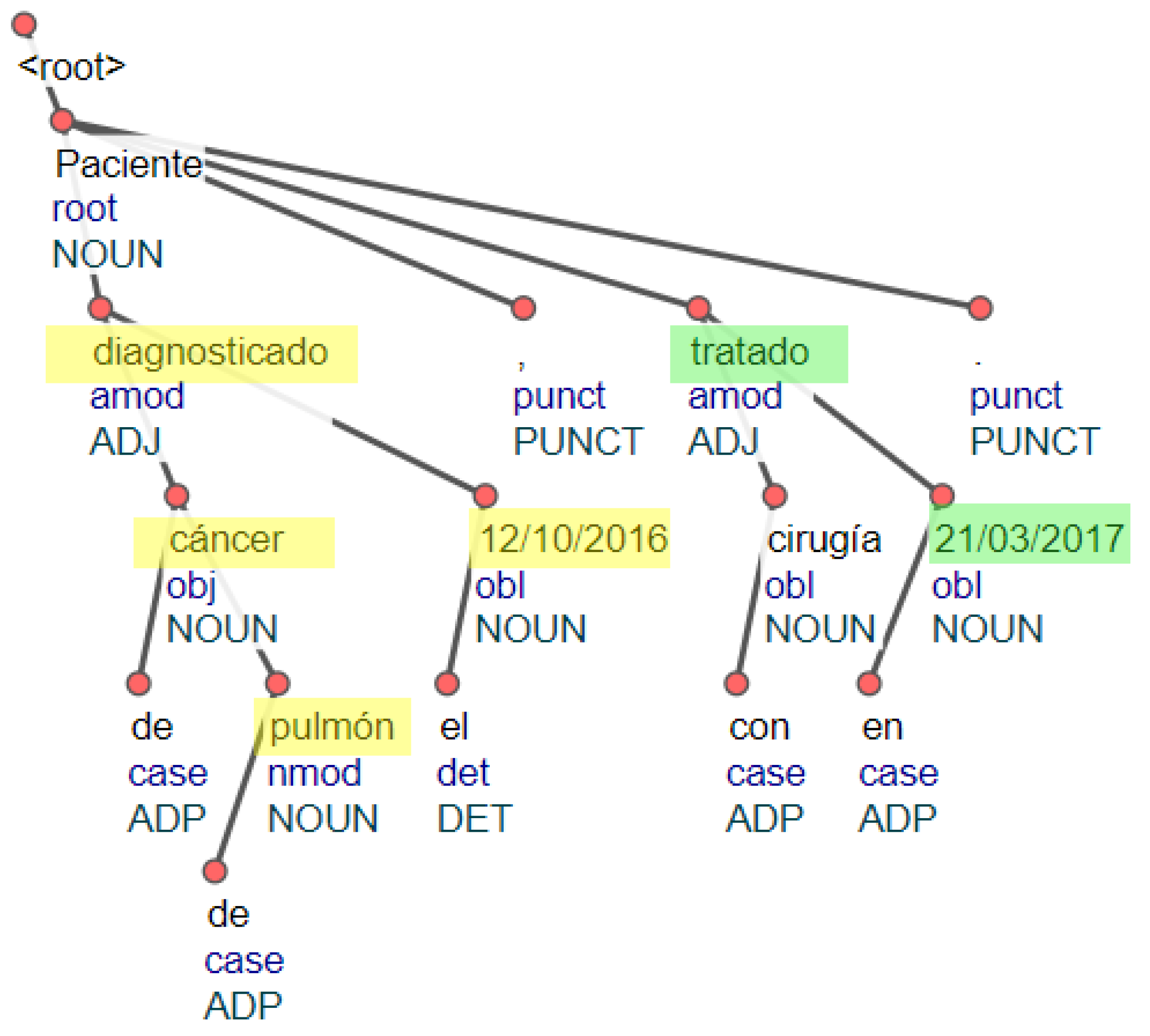
6. Relating Cancer Diagnosis and Dates
6.1. Linking Dates to Cancer Entities
- A cancer entity or an event entity is an ancestor of a date in the dependency path of the sentence.
- The cancer entity and a date entity appear contiguously or belong to the same predicate in the sentence.
- The sentence contains event entities: in this case, the event, the cancer entity, and the date are linked if some of the prior conditions are met.
6.2. Choosing the Proper Linkage
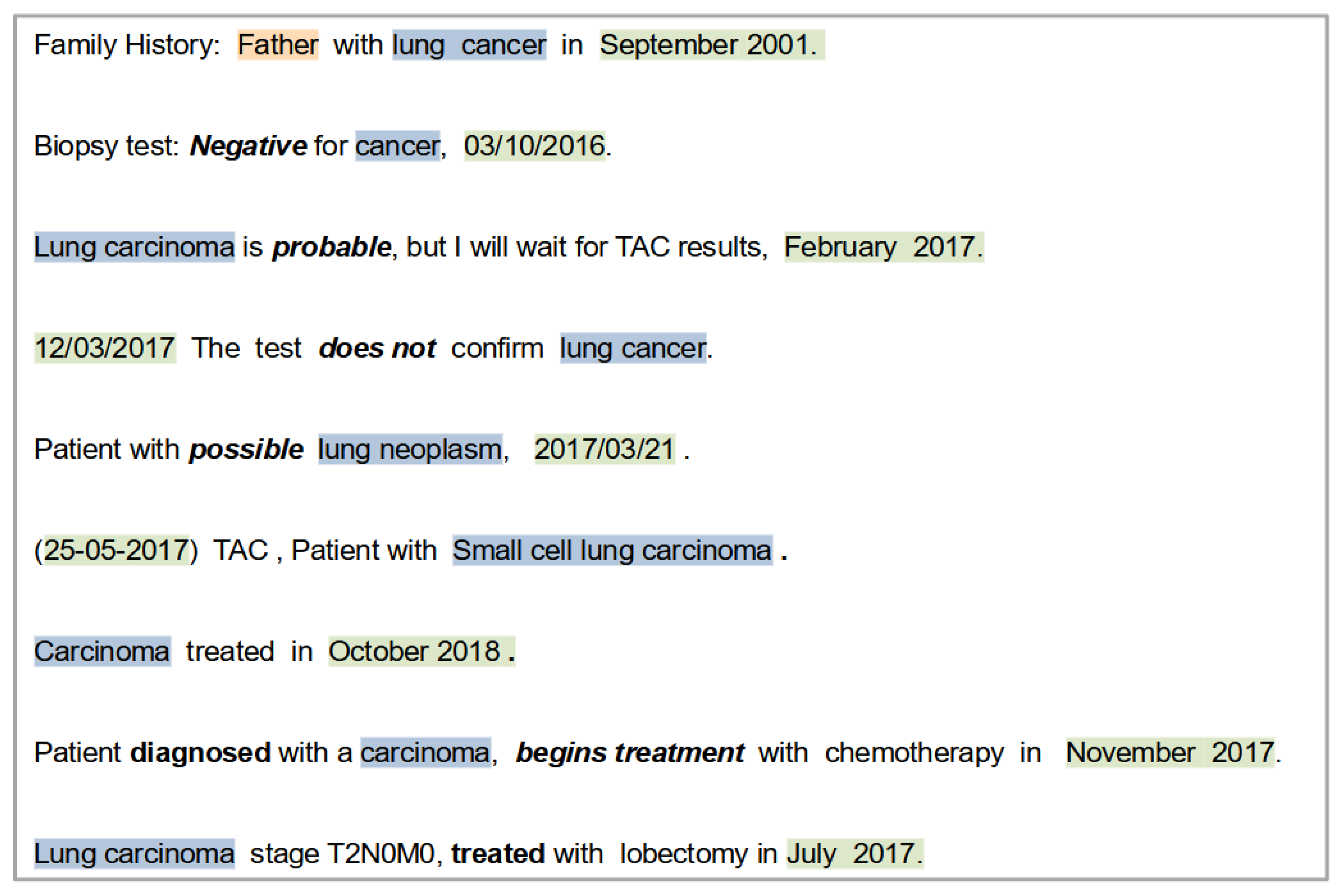
- There are different concepts indicating cancer diagnosis entity. Some of them are very generic (“Cancer”), while others are specific terms (“Small cell lung carcinoma”).
- There are different dates associated to cancer diagnosis entities.
- Not every record contains an event entity.
- Choosing the cancer diagnosis: A ranked list of UMLS identifiers is used. This list contains UMLS codes and their respective cancer diagnosis sorted according to those that more specifically describe the diagnosis. In this list, the concept “Squamous cell lung carcinoma” is more relevant than the concept “Lung cancer” because the former describes more specifically the patient’s diagnosis.
- Choosing the diagnosis date: two heuristics are applied to disambiguate the date for the chosen diagnosis:
- –
- Annotations containing events other than “diagnosed event” are eliminated.
- –
- Clinical notes are first ordered chronologically and then classified according to their type: (Anamnesis, Clinical Judgment, Medical Evolution, treatment, etc.). According to this classification, the date is assigned taking into account the earliest annotation coming from a document classified as Clinical Judgement or alike.
7. Validation and Results
7.1. Validation Methodology
- To evaluate the deep learning model (Figure 3) the corpus described in Section 4.1 was used, this corpus contains 14,750 annotated sentences. The corpus was shuffled and randomly split into three sets: training (80%), development (10%), and test (10%). This procedure was independently repeated ten times, while verifying that each set contained all the labels in the corpus. The test set was used to calculate the performance metrics, as shown in the Equations (1) and (2). The F-score (Equation (3)) is calculated as a weighted average of the Precision and Recall measurements.The BiLSTM-CRF model was developed using TensorFlow (https://www.tensorflow.org/?hl=es-419) and Keras (https://keras.io/) using the following parameters: learning rate as 0.001, dropout as 0.5, the number of epochs is 30, the BiLSTM hidden size is 300, and the batch size is 512.
- To validate the negation and speculation detection step, the NUBES corpus proposed by [12] was used. This public corpus is annotated with speculation and negation in clinical notes written in Spanish. We analyze the performance for each sub-task: cue detection and scope recognition.
- –
- A speculation or negation cue is correctly detected when given a sentence, the rule-based approach is able to recognize the cues indicated by the test corpus.
- –
- A scope is correctly detected when given a sentence and a detected cue, the approach is able to recognize the scope indicated in the corpus.
Equations (4) and (5) are used to calculate Precision and Recall for the cue detection sub-task. On the other hand, Equations (6) and (7) are used to calculate Precision and Recall for the scope detection sub-task. The F-score is calculated as a weighted average of the Precision and Recall. - To validate the diagnosis date extraction, a database containing data from “Hospital Universitario Puerta de Hierro Madrid” was used. It contains around 300,000 clinical notes corresponding to 1000 patients that were diagnosed with lung cancer in the last ten years. Information of the diagnosis date for each patient is available. A diagnosis date is correctly extracted when it corresponds to the diagnosis date given by the hospital dataset. Equations (8) and (9) are used to calculate the Precision and Recall respectively.
7.2. Named Entity Recognition Results
- The BiLSTM-CRF base model proposed by [32] is used.
- General domain embeddings training by Fast text (https://fasttext.cc/docs/en/pretrained-vectors.html) on Wikipedia are added to the BiLSTM-CRF model.
- Spanish medical embeddings proposed by [41] have been added to the BiLSTM-CRF model.
- Spanish medical embeddings and char embeddings are added to the model.
- A combination of medical embeddings, char embeddings, lemmas, and Part of Speech (POS) tagging features.
7.3. Negation and Speculation Results
7.3.1. Cue Detection Results
- Only the first and second Regex are used for cue detection (See Section 5.2). These Regex were adapted to Spanish from the popular rule-based Negex [56] proposal. The Negex adaptation to Spanish is used as baseline.
- The third Regex is added to measure the contiguous cues behavior.
- The fourth Regex was added, this experiment includes all Regex proposed in Section 5.2.
7.3.2. Scope Recognition Results
- The first rule is used to recognize the scope. As mentioned in Section 5.4, this rule searches for a termination term in the sentence which indicates the end of the scope. This approach is commonly used by previous rule-based approaches [53,60,61].
- The second rule is added to take into account contiguous cues for recognizing the scope.
- The short sentence heuristic is used by adding the third rule.
- Adding the fourth rule to include POS tagging features.
- The fifth rule is added to include the parse tree analysis for extracting the scope.
7.4. Relating Cancer Diagnosis and Date Results
- The diagnosis date is chosen from all extracted named entities which belong to the patient.
- The diagnosis date is chosen after filtering entities affected by negation.
- The diagnosis date is chosen after filtering entities affected by negation and speculation.
8. Discussion
- The proposed approach uses a lexicon that has been adapted to Spanish from two different resources specialized in negation and speculation in the biomedical domain. Additionally, the lexicon was semantically extended with a word embeddings technique that helped obtain new cues with similar meanings. Moreover, this lexicon was manually reviewed and evaluated. The resulting lexicon offers high precision in the cue detection task.
- Some weaknesses reported in [12], such as post-scope recognition are addressed in this proposal by using Regex 2 (see Section 5.2). When a cue is detected using this Regex, the scope is searched to the left of the cue.
- The rules proposed for scope recognition are adapted according to the analysis that has been performed on how speculation and negation are expressed in clinical notes written in Spanish (see Section 5.3).
- “Causas de sangrado: una complicación de la cirugíaouna patología asociada al cancer de pulmón.” (Causes of bleeding: a surgery-related complication or a lung cancer-related pathology)
- “Si acude a alguna consultaoservicio sanitario debe ser acompañado. ” (Patients must be accompanied when going to general appointments or when using the health service.))
- Sentences containing a sequence of contiguous cues where some part of the text is not affected by any cue. In the next sentence the scope is underlined and the text “sangrado pulmonar” is out the scope.“Novómitos, sangrado pulmonar desde ayer,notos, nodolor.”(No vomiting, lung bleeding since yesterday, no cough, no pain.)
- When the scope of a cue is in both directions, to the left and to the right of the cue. In the next sentence, the cue is the word “Versus”, and the scope is underlined.Cancer de pulmónVersusInfección en lóbulo derecho.(Lung cancer Versus right lobe infection).
9. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Spasić, I.; Livsey, J.; Keane, J.A.; Nenadić, G. Text mining of cancer-related information: Review of current status and future directions. Int. J. Med. Inform. 2014, 83, 605–623. [Google Scholar] [CrossRef] [PubMed]
- Demner-Fushman, D.; Chapman, W.W.; McDonald, C.J. What can natural language processing do for clinical decision support? J. Biomed. Inform. 2009, 42, 760–772. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.T.; Goryachev, S.; Weiss, S.; Sordo, M.; Murphy, S.N.; Lazarus, R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: Evaluation of a natural language processing system. BMC Med. Inform. Decis. Mak. 2006, 30, 327–348. [Google Scholar] [CrossRef]
- Wang, L.; Luo, L.; Wang, Y.; Wampfler, J.; Yang, P.; Liu, H. Natural language processing for populating lung cancer clinical research data. BMC Med. Inform. Decis. Mak. 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Zhang, Q.; Ren, Y.; Qiu, T.; Ma, J.; Sun, Q. Extracting comprehensive clinical information for breast cancer using deep learning methods. Int. J. Med. Inform. 2019, 132, 103985. [Google Scholar] [CrossRef]
- Sheikhalishahi, S.; Miotto, R.; Dudley, J.T.; Lavelli, A.; Rinaldi, F.; Osmani, V. Natural language processing of clinical notes on chronic diseases: Systematic review. J. Med. Internet Res. 2019, 21, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Najafabadipour, M.; Tuñas, J.M.; Rodríguez-González, A.; Menasalvas, E. Lung Cancer Concept Annotation from Spanish Clinical Narratives. In Data Integration in the Life Sciences; Auer, S., Vidal, M.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 153–163. [Google Scholar]
- Savova, G.K.; Tseytlin, E.; Finan, S.P.; Castine, M.; Timothy, A.; Medvedeva, O.P.; Harris, D.A.; Hochheiser, H.S.; Lin, C.; Girish, R. DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records. Cancer Res. 2017, 77, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Solarte-Pabon, O.; Torrente, M.; Rodriguez-Gonzalez, A.; Provencio, M.; Menasalvas, E.; Tunas, J.M. Lung cancer diagnosis extraction from clinical notes written in spanish. In Proceedings of the IEEE Symposium on Computer-Based Medical Systems, Rochester, MN, USA, 28–30 July 2020; pp. 492–497. [Google Scholar] [CrossRef]
- Alam, R.; Cheraghi-Sohi, S.; Panagiot, M.; Esmail, A.; Campbell, S.; Panagopoulou, E. Managing diagnostic uncertainty in primary care: A systematic critical review. BMC Fam. Pract. 2017, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.; Perez, N.; Cuadros, M.; Rigau, G. NUBES: A Corpus of Negation and Uncertainty in Spanish Clinical Texts. arXiv 2020, arXiv:2004.01092. [Google Scholar]
- Cruz Díaz, N.P.; Maña López, M.J. Negation and Speculation Detection; John Benjamins Publishing Company: Amsterdam, The Netherlands, 2019. [Google Scholar] [CrossRef]
- Agarwal, S.; Yu, H. Detecting hedge cues and their scope in biomedical text with conditional random fields. J. Biomed. Inform. 2010, 43, 953–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.; Chen, D.; He, H.; Liu, S.; Moon, S.; Peterson, K.J.; Shen, F.; Wang, L.; Wang, Y.; Wen, A.; et al. Clinical concept extraction: A methodology review. J. Biomed. Inform. 2020, 109, 103526. [Google Scholar] [CrossRef]
- Tulkens, S.; Šuster, S.; Daelemans, W. Unsupervised concept extraction from clinical text through semantic composition. J. Biomed. Inform. 2019, 91, 103120. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.W.; Yetisgen, M.; Harris, W.P.; Sharon, W.K. Natural Language Processing in Oncology Review. JAMA Oncol. 2016, 2, 797–804. [Google Scholar] [CrossRef]
- Warner, J.L.; Levy, M.A.; Neuss, M.N.; Warner, J.L.; Levy, M.A.; Neuss, M.N. ReCAP: Feasibility and Accuracy of Extracting Cancer Stage Information from Narrative Electronic Health Record Data. J. Oncol. Pract. 2016, 12, 157–158. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Lawley, M.J.; Hansen, D.P.; Bowman, R.V.; Clarke, B.E.; Duhig, E.E.; Colquist, S. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J. Am. Med. Inform. Assoc. 2010, 17, 440–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshmukh, P.R.; Phalnikar, R. TNM Cancer Stage Detection from Unstructured Pathology Reports of Breast Cancer Patients. In Proceedings of the International Conference on Computational Science and Applications, Pune, India, 7–9 August 2019; Bhalla, S., Kwan, P., Bedekar, M., Phalnikar, R., Sirsikar, S., Eds.; pp. 411–418. [Google Scholar]
- AAlAbdulsalam, A.K.; Garvin, J.H.; Redd, A.; Carter, M.E.; Sweeny, C.; Meystre, S.M. Automated Extraction and Classification of Cancer Stage Mentions from Unstructured Text Fields in a Central Cancer Registry. AMIA Jt. Summits Transl. Sci. Proc. 2018, 2017, 16–25. [Google Scholar]
- Evans, T.L.; Gabriel, P.E.; Shulman, L.N. Cancer Staging in Electronic Health Records: Strategies to Improve Documentation of These Critical Data. J. Oncol. Pract. 2016, 12, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Khor, R.C.; Nguyen, A.; O’Dwyer, J.; Kothari, G.; Sia, J.; Chang, D.; Ng, S.P.; Duchesne, G.M.; Foroudi, F. Extracting tumour prognostic factors from a diverse electronic record dataset in genito-urinary oncology. Int. J. Med. Inform. 2019, 121, 53–57. [Google Scholar] [CrossRef]
- Wang, Z.; Shah, A.D.; Tate, A.R.; Denaxas, S.; Shawe-Taylor, J.; Hemingway, H. Extracting diagnoses and investigation results from unstructured text in electronic health records by semi-supervised machine learning. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Zheng, S.; Jabbour, S.K.; O’Reilly, S.E.; Lu, J.J.; Dong, L.; Ding, L.; Xiao, Y.; Yue, N.; Wang, F.; Zou, W. Automated Information Extraction on Treatment and Prognosis for Non–Small Cell Lung Cancer Radiotherapy Patients: Clinical Study. JMIR Med. Inform. 2018, 6, e8. [Google Scholar] [CrossRef]
- Bitterman, D.; Miller, T.; Harris, D.; Lin, C.; Finan, S.; Warner, J.; Mak, R.; Savova, G. Extracting Relations between Radiotherapy Treatment Details. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, Online Conference, 19 November 2020; pp. 194–200. [Google Scholar] [CrossRef]
- Zeng, Z.; Espino, S.; Roy, A.; Li, X.; Khan, S.A.; Clare, S.E.; Jiang, X.; Neapolitan, R.; Luo, Y. Using natural language processing and machine learning to identify breast cancer local recurrence. BMC Bioinform. 2018, 19. [Google Scholar] [CrossRef]
- Isaksson, L.J.; Pepa, M.; Zaffaroni, M.; Marvaso, G.; Alterio, D.; Volpe, S.; Corrao, G.; Augugliaro, M.; Starzyńska, A.; Leonardi, M.C.; et al. Machine Learning-Based Models for Prediction of Toxicity Outcomes in Radiotherapy. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Forsyth, A.W.; Barzilay, R.; Hughes, K.S.; Lui, D.; Lorenz, K.A.; Enzinger, A.; Tulsky, J.A.; Lindvall, C. Machine Learning Methods to Extract Documentation of Breast Cancer Symptoms From Electronic Health Records. J. Pain Symptom Manag. 2018, 55, 1492–1499. [Google Scholar] [CrossRef] [Green Version]
- Hochreiter, S.; Schmidhuber, J. LSTM can solve hard long time lag problems. In Proceedings of the 9th International Conference on Neural Information Processing Systems, Denver, CO, USA, 2–5 December 1996; pp. 473–479. [Google Scholar]
- Goldberg, Y. Neural Network Methods for Natural Language Processing. Synth. Lect. Hum. Lang. Technol. 2017, 10, 1–311. [Google Scholar] [CrossRef]
- Lample, G.; Ballesteros, M.; Subramanian, S.; Kawakami, K.; Dyer, C. Neural architectures for named entity recognition. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL HLT 2016—Proceedings of the Conference, San Diego, CA, USA, 12–17 June 2016; pp. 260–270. [Google Scholar] [CrossRef]
- Lopez, M.M.; Kalita, J. Deep Learning applied to NLP. arXiv 2017, arXiv:1703.03091. [Google Scholar]
- Carta, S.; Ferreira, A.; Podda, A.S.; Reforgiato Recupero, D.; Sanna, A. Multi-DQN: An ensemble of Deep Q-learning agents for stock market forecasting. Expert Syst. Appl. 2021, 164, 113820. [Google Scholar] [CrossRef]
- Vázquez, J.J.; Arjona, J.; Linares, M.; Casanovas-Garcia, J. A Comparison of Deep Learning Methods for Urban Traffic Forecasting using Floating Car Data. Transp. Res. Procedia 2020, 47, 195–202. [Google Scholar] [CrossRef]
- Nguyen, G.; Dlugolinsky, S.; Tran, V.; Lopez Garcia, A. Deep learning for proactive network monitoring and security protection. IEEE Access 2020, 8, 19696–19716. [Google Scholar] [CrossRef]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed Representations of Words and Phrases and their Compositionality. In Advances in Neural Information Processing Systems 26; Burges, C.J.C., Bottou, L., Welling, M., Ghahramani, Z., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Nice, France, 2013; pp. 3111–3119. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. GloVe: Global Vectors for Word Representation. In Proceedings of the Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Bojanowski, P.; Grave, E.; Joulin, A.; Mikolov, T. Enriching Word Vectors with Subword Information. Trans. Assoc. Comput. Linguist. 2017, 5, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, S.; Afzal, N.; Rastegar-Mojarad, M.; Wang, L.; Shen, F.; Kingsbury, P.; Liu, H. A comparison of word embeddings for the biomedical natural language processing. J. Biomed. Inform. 2018, 87, 12–20. [Google Scholar] [CrossRef]
- Soares, F.; Villegas, M.; Gonzalez-Agirre, A.; Krallinger, M.; Armengol-Estapé, J. Medical word embeddings for Spanish: Development and evaluation. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, MN, USA, 7 June 2019; pp. 124–133. [Google Scholar] [CrossRef] [Green Version]
- Névéol, A.; Dalianis, H.; Velupillai, S.; Savova, G.; Zweigenbaum, P. Clinical Natural Language Processing in languages other than English: Opportunities and challenges. J. Biomed. Semant. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Najafabadipour, M.; Zanin, M.; Rodriguez-Gonzalez, A.; Gonzalo-Martin, C.; Garcia, B.N.; Calvo, V.; Bermudez, J.L.C.; Provencio, M.; Menasalvas, E. Recognition of time expressions in Spanish electronic health records. In Proceedings of the IEEE Symposium on Computer-Based Medical Systems, Cordoba, Spain, 5–7 June 2019; pp. 69–74. [Google Scholar] [CrossRef]
- Wang, L.; Wampfler, J.; Dispenzieri, A.; Xu, H.; Yang, P.; Liu, H. Achievability to Extract Specific Date Information for Cancer Research. In AMIA Annual Symposium Proceedings, AMIA Symposium; American Medical Informatics Association: Washington, DC, USA, 2019; Volume 2019, pp. 893–902. [Google Scholar]
- Garciá-Pablos, A.; Perez, N.; Cuadros, M. Vicomtech at cantemist 2020. CEUR Workshop Proc. 2020, 2664, 489–498. [Google Scholar]
- Carrasco, S.S.; Martínez, P. Using embeddings and bi-lstm+crf model to detect tumor morphology entities in Spanish clinical cases. CEUR Workshop Proc. 2020, 2664, 368–375. [Google Scholar]
- López-Úbeda, P.; Diáz-Galiano, M.C.; Martín-Valdivia, M.T.; Urenã-López, L.A. Extracting neoplasms morphology mentions in Spanish clinical cases throughword embeddings. CEUR Workshop Proc. 2020, 2664, 324–334. [Google Scholar]
- Miranda-Escalada, A.; Farré, E.; Krallinger, M. Named entity recognition, concept normalization and clinical coding: Overview of the cantemist track for cancer text mining in Spanish, corpus, guidelines, methods and results. CEUR Workshop Proc. 2020, 2664, 303–323. [Google Scholar]
- Lafferty, J.D.; McCallum, A.; Pereira, F.C.N. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In Proceedings of the Eighteenth International Conference on Machine Learning, ICML ’01, Williamstown, MA, USA, 28 June–1 July 2001; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2001; pp. 282–289. [Google Scholar]
- De Albornoz, J.C.; Plaza, L.; Diaz, A.; Ballesteros, M. UCM-I: A rule-based syntactic approach for resolving the scope of negation. In Proceedings of the *SEM 2012—1st Joint Conference on Lexical and Computational Semantics, Montréal, QC, Canada, 7–8 June 2012; Volume 1, pp. 282–287. [Google Scholar]
- Dalianis, H. Clinical Text Mining; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Vincze, V.; Szarvas, G.; Farkas, R.; Móra, G.; Csirik, J. The BioScope corpus: Biomedical texts annotated for uncertainty, negation and their scopes. BMC Bioinform. 2008, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harkema, H.; Dowling, J.N.; Thornblade, T.; Chapman, W.W. ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports. J. Biomed. Inform. 2009, 42, 839–851. [Google Scholar] [CrossRef] [Green Version]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Chapman, W.W.; Bridewell, W.; Hanbury, P.; Cooper, G.F.; Buchanan, B.G. A simple algorithm for identifying negated findings and diseases in discharge summaries. J. Biomed. Inform. 2001, 34, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Solarte-Pabón, O.; Menasalvas, E.; Rodriguez-González, A. Spa-neg: An approach for negation detection in clinical text written in Spanish. In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 6–8 May 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 323–327. [Google Scholar] [CrossRef]
- Elazhary, H. NegMiner: An automated tool for mining negations from electronic narrative medical documents. Int. J. Intell. Syst. Appl. 2017, 9, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Straka, M.; Hajič, J.; Straková, J. UDPipe: Trainable pipeline for processing CoNLL-U files performing tokenization, morphological analysis, POS tagging and parsing. In Proceedings of the 10th International Conference on Language Resources and Evaluation, LREC 2016, Portoroz, Slovenia, 23–28 May 2016; pp. 4290–4297. [Google Scholar]
- Stricker, V.; Iacobacci, I.; Cotik, V. Negated Findings Detection in Radiology Reports in Spanish: An Adaptation of NegEx to Spanish. In Proceedings of the Workshop on Replicability and Reproducibility in Natural Language Processing: Adaptive Methods, Resources and Software at IJCAI 2015, Buenos Aires, Argentina, 25–27 July 2015; pp. 1–7. [Google Scholar]
- Costumero, R.; Lopez, F.; Gonzalo-Martín, C.; Millan, M.; Menasalvas, E. An approach to detect negation on medical documents in Spanish. In Proceedings of the Brain Informatics and Health, Warsaw, Poland, 11–14 August 2014; pp. 366–375. [Google Scholar]




| Entity | Number of Annotations |
|---|---|
| Cancer entity | 4128 |
| Stage | 3274 |
| TNM | 1152 |
| Date | 4312 |
| Family member | 883 |
| Events | 2703 |
| Treatment | 1302 |
| Drug | 463 |
| Cancer Diagnosis Concept | Date | Event |
|---|---|---|
| Lung cancer | 25 March 2014 | |
| Cancer | March 2015 | Begins treatment |
| Adenocarcinoma | 17 May 2017 | |
| Lung carcinoma | 14 January 2014 | Begins clinical trial |
| Small cell lung carcinoma | July 2014 | Diagnosed |
| Lung neoplasm | 12 July 2016 | |
| Cancer | 17 May 2014 | |
| Adenoca | April 2018 | |
| Carcinoma | 18 September 2017 | Begins surgery |
| Model | P | R | F1 |
|---|---|---|---|
| BiLSTM-CRF | 0.83 | 0.78 | 0.80 |
| BiLSTM-CRF + General domain embeddings | 0.80 | 0.73 | 0.76 |
| BiLSTM-CRF + Medical embeddings | 0.87 | 0.84 | 0.85 |
| BiLSTM-CRF + Medical Embeddings + Char embeddings | 0.87 | 0.85 | 0.86 |
| BiLSTM-CRF + Medical Embeddings + Char embeddings + Lemmas + POS | 0.91 | 0.89 | 0.90 |
| Negation | Speculation | |||||
|---|---|---|---|---|---|---|
| Regex | P | R | F1 | P | R | F1 |
| Regex 1,2 | 0.84 | 0.81 | 0.82 | 0.91 | 0.88 | 0.89 |
| Regex 1,2,3 | 0.93 | 0.91 | 0.92 | 0.92 | 0.89 | 0.90 |
| Regex 1,2,3,4 | 0.96 | 0.94 | 0.95 | 0.92 | 0.89 | 0.90 |
| Negation | Speculation | |||||
|---|---|---|---|---|---|---|
| Rules | P | R | F1 | P | R | F1 |
| Rule 1 | 0.72 | 0.70 | 0.71 | 0.72 | 0.69 | 0.70 |
| Rules 1,2 | 0.78 | 0.75 | 0.76 | 0.72 | 0.69 | 0.70 |
| Rules 1, 2, 3 | 0.81 | 0.79 | 0.80 | 0.73 | 0.71 | 0.72 |
| Rules 1, 2, 3, 4 | 0.86 | 0.82 | 0.84 | 0.82 | 0.79 | 0.80 |
| Rules 1, 2, 3, 4, 5 | 0.91 | 0.87 | 0.89 | 0.87 | 0.84 | 0.85 |
| Experiment | P | R | F1 |
|---|---|---|---|
| Using all Named Entities | 0.67 | 0.62 | 0.64 |
| Filtering negated entities | 0.72 | 0.71 | 0.71 |
| Filtering negated and speculated entities | 0.92 | 0.87 | 0.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solarte Pabón, O.; Torrente, M.; Provencio, M.; Rodríguez-Gonzalez, A.; Menasalvas, E. Integrating Speculation Detection and Deep Learning to Extract Lung Cancer Diagnosis from Clinical Notes. Appl. Sci. 2021, 11, 865. https://doi.org/10.3390/app11020865
Solarte Pabón O, Torrente M, Provencio M, Rodríguez-Gonzalez A, Menasalvas E. Integrating Speculation Detection and Deep Learning to Extract Lung Cancer Diagnosis from Clinical Notes. Applied Sciences. 2021; 11(2):865. https://doi.org/10.3390/app11020865
Chicago/Turabian StyleSolarte Pabón, Oswaldo, Maria Torrente, Mariano Provencio, Alejandro Rodríguez-Gonzalez, and Ernestina Menasalvas. 2021. "Integrating Speculation Detection and Deep Learning to Extract Lung Cancer Diagnosis from Clinical Notes" Applied Sciences 11, no. 2: 865. https://doi.org/10.3390/app11020865
APA StyleSolarte Pabón, O., Torrente, M., Provencio, M., Rodríguez-Gonzalez, A., & Menasalvas, E. (2021). Integrating Speculation Detection and Deep Learning to Extract Lung Cancer Diagnosis from Clinical Notes. Applied Sciences, 11(2), 865. https://doi.org/10.3390/app11020865







