Assessment of Skin Deep Layer Biochemical Profile Using Spatially Offset Raman Spectroscopy
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Phantom Preparation
2.2. Human Tissue Samples
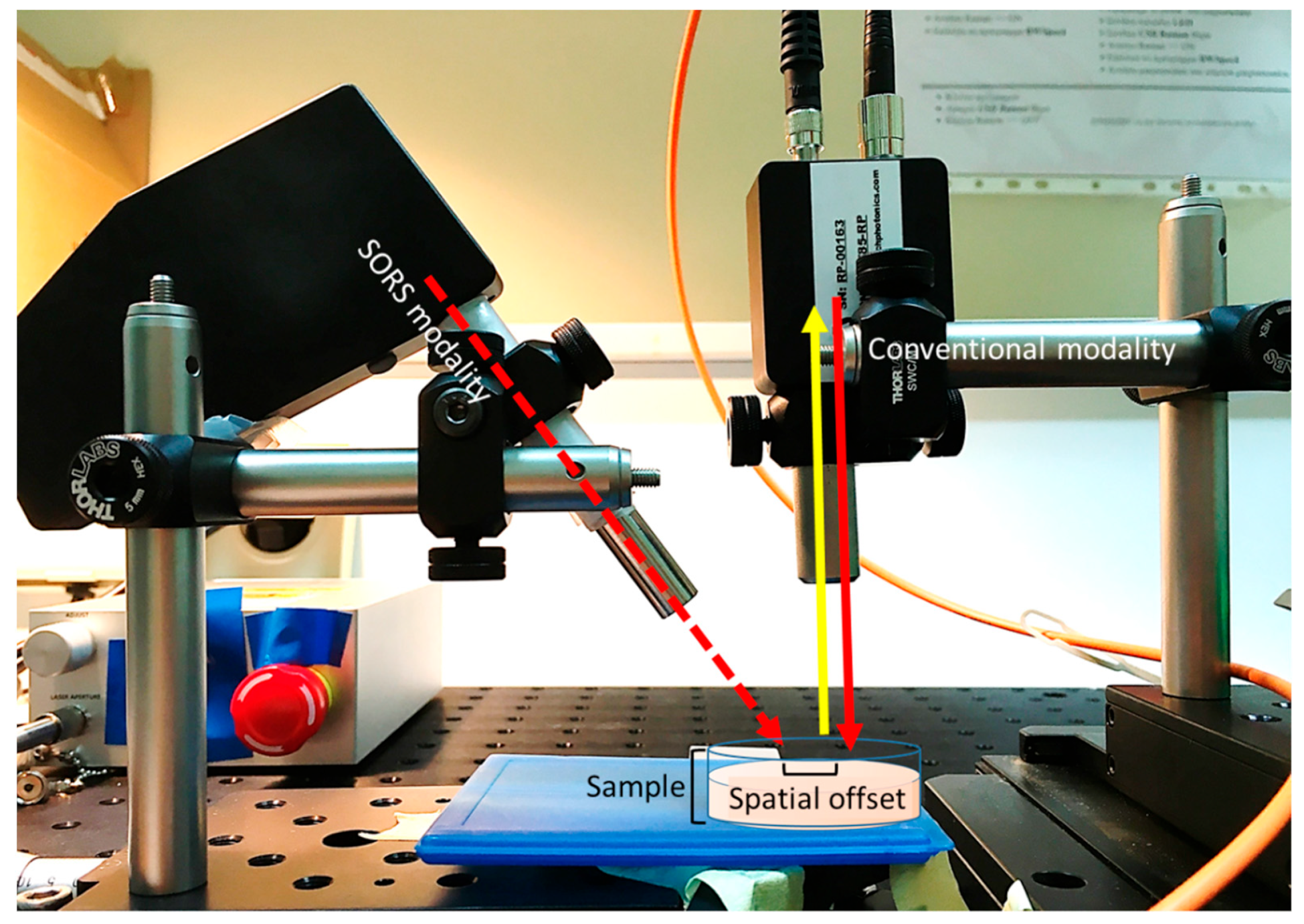
2.3. Raman Setup
2.4. Raman Measurements
2.5. Data Analysis
3. Results
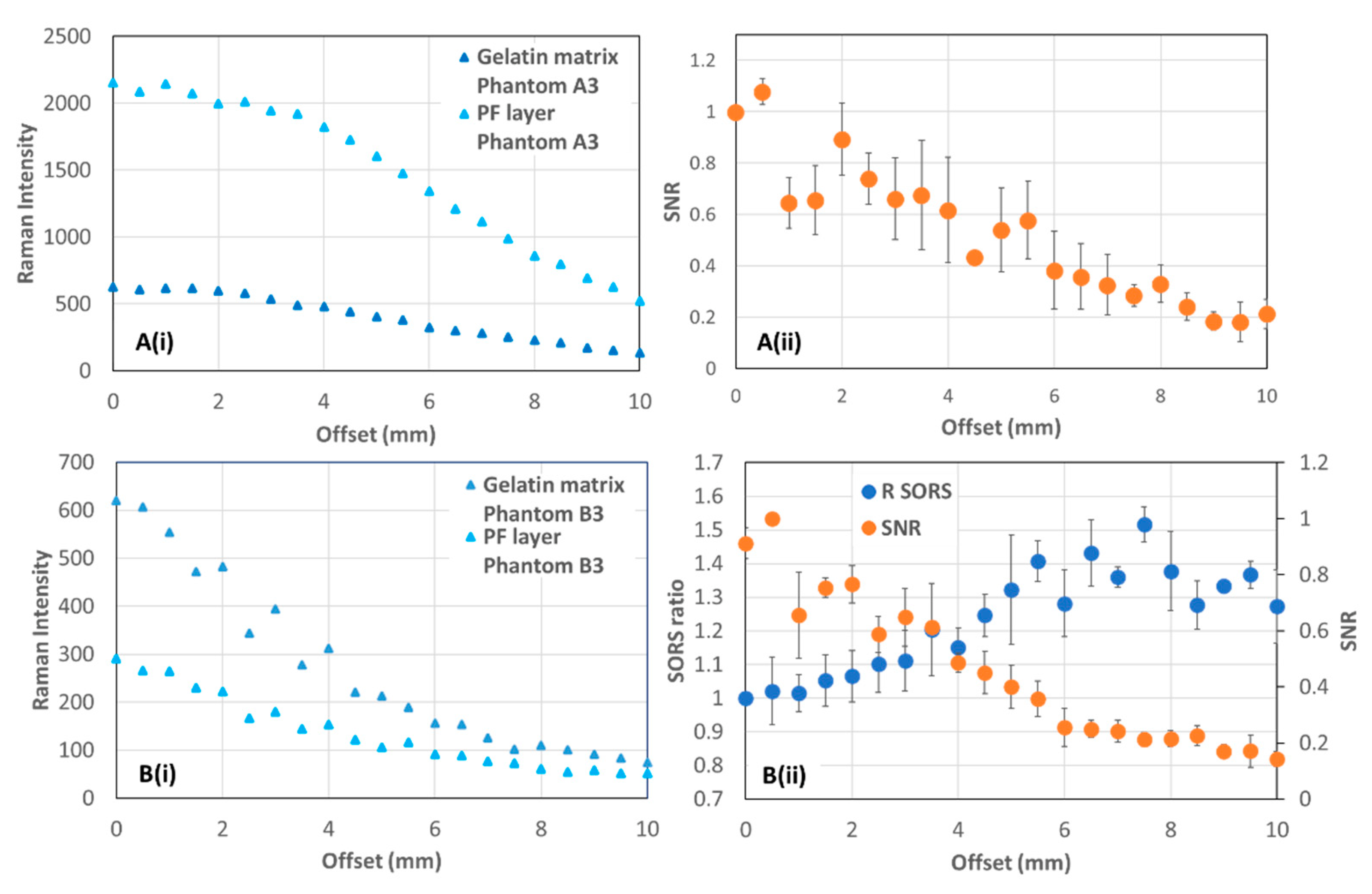
3.1. Tissue Phantoms
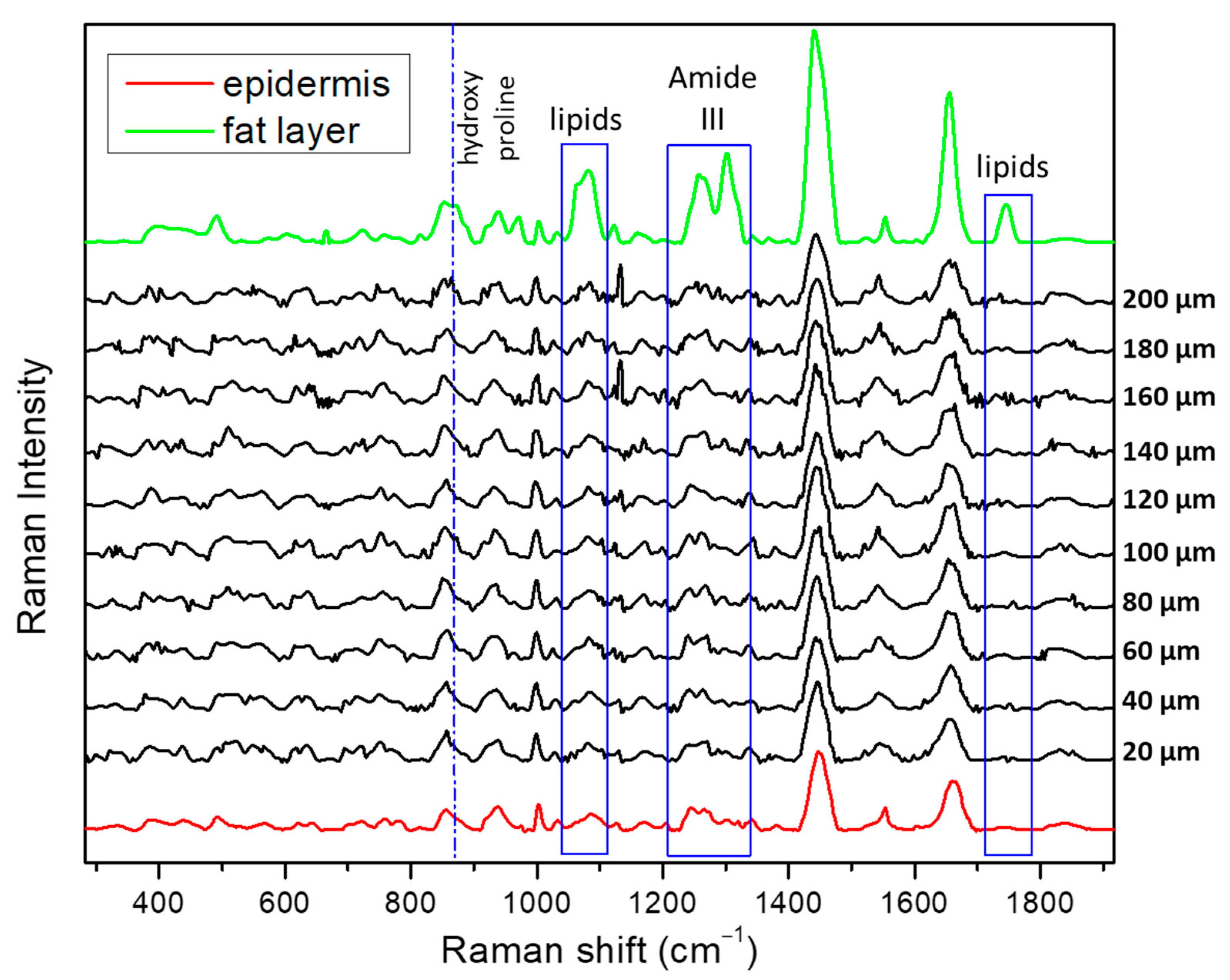
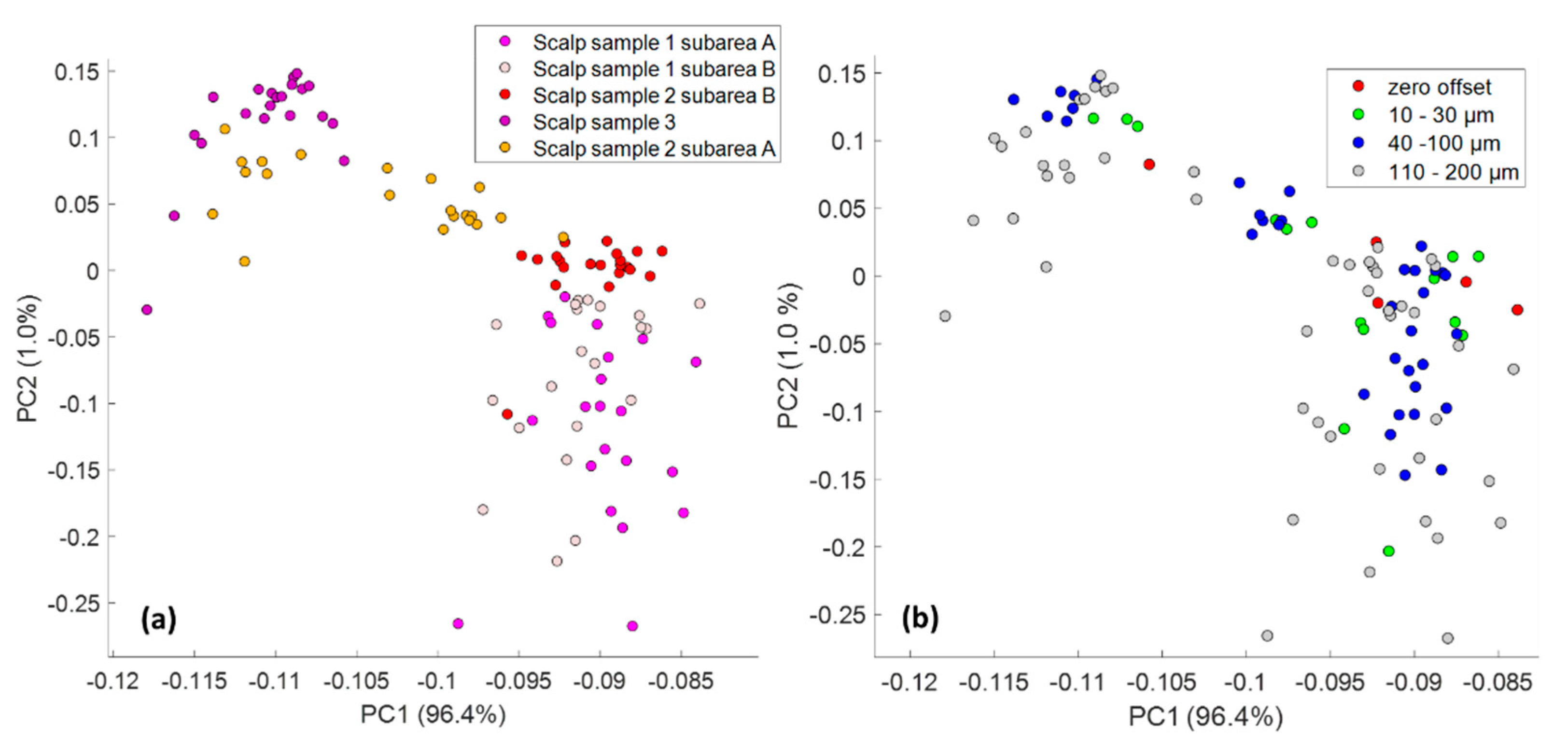
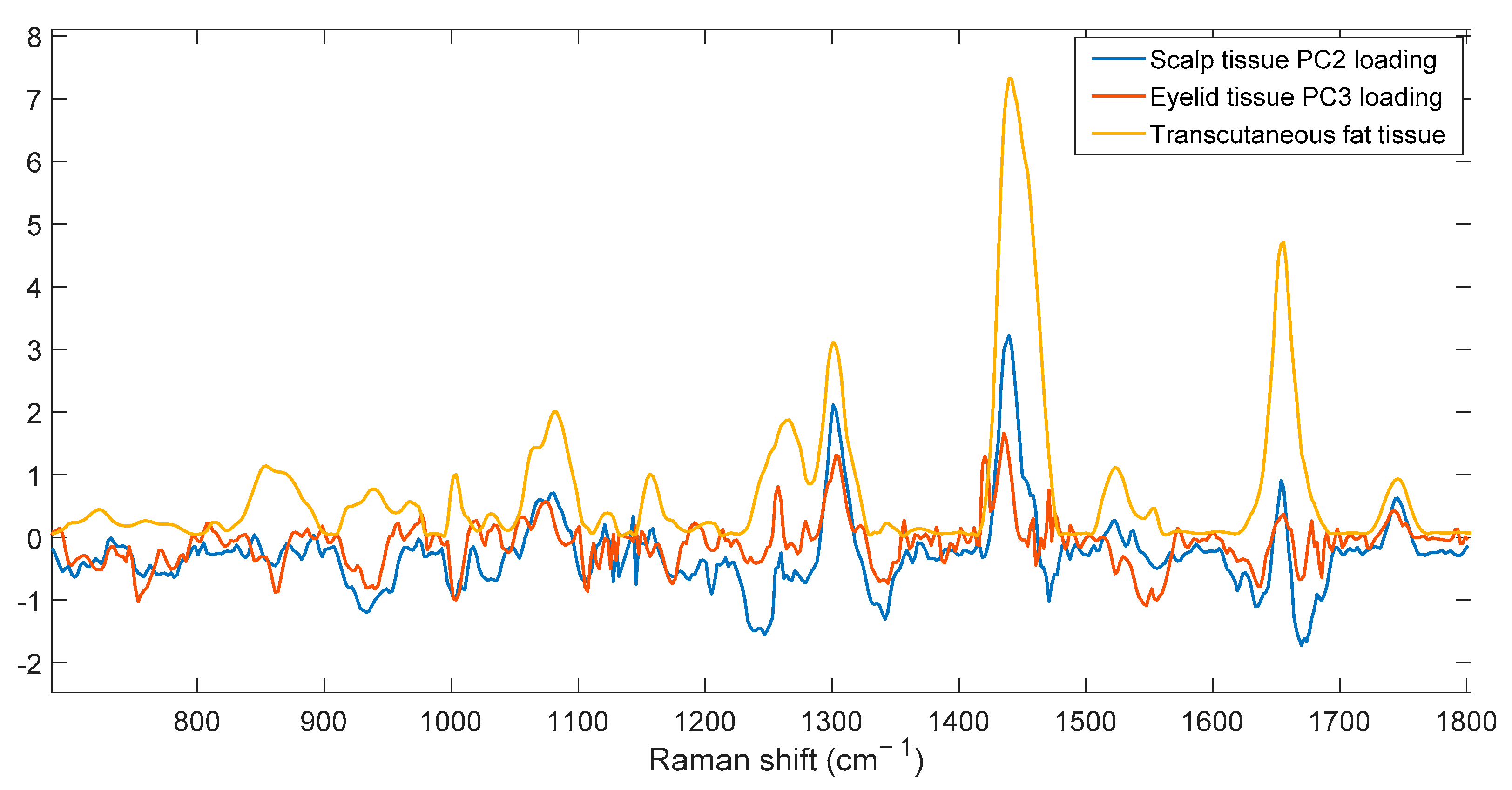
3.2. Human Skin Tissue
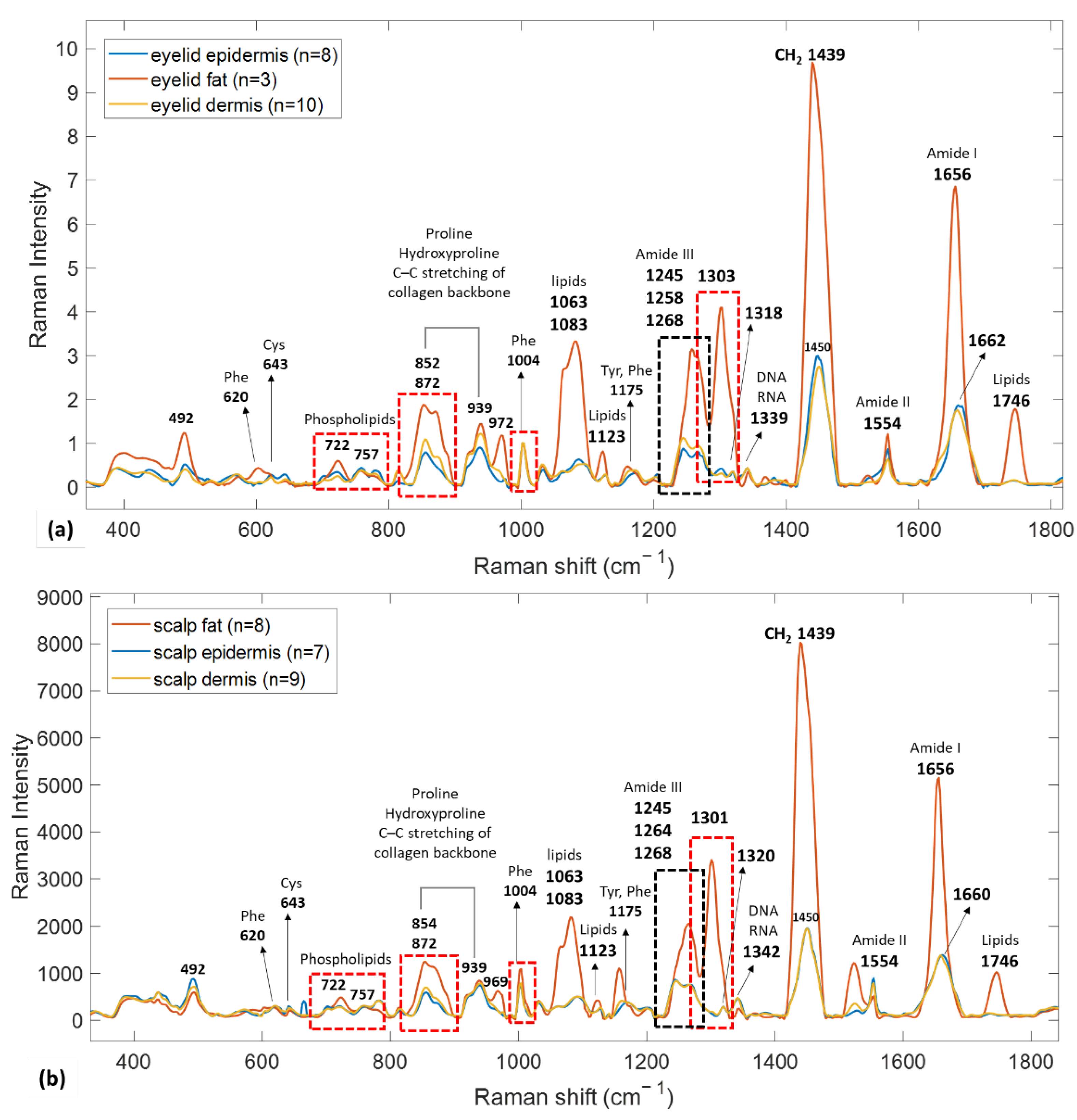
3.2.1. Reference Spectra
3.2.2. SORS Measurements
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leiter, U.; Keim, U.; Garbe, C. Epidemiology of Skin Cancer: Update 2019. Adv. Exp. Med. Biol. 2020, 1268, 123–139. [Google Scholar] [CrossRef]
- Seretis, K.; Thomaidis, V.; Karpouzis, A.; Tamiolakis, D.; Tsamis, I. Epidemiology of surgical treatment of nonmelanoma skin cancer of the head and neck in Greece. Derm. Surg. 2010, 36, 15–22. [Google Scholar] [CrossRef]
- Zaidi, M.R.; Fisher, D.E.; Rizos, H. Biology of melanocytes and primary melanoma. In Cutaneous Melanoma; Springer: Cham, Switzerland, 2020; pp. 3–40. [Google Scholar]
- English, D.R.; Del Mar, C.; Burton, R.C. Factors influencing the number needed to excise: Excision rates of pigmented lesions by general practitioners. Med. J. Aust. 2004, 180, 16–19. [Google Scholar] [CrossRef]
- Rangan, S.; Schulze, H.G.; Vardaki, M.Z.; Blades, M.W.; Piret, J.M.; Turner, R.F.B. Applications of Raman spectroscopy in the development of cell therapies: State of the art and future perspectives. Analyst 2020, 145, 2070–2105. [Google Scholar] [CrossRef]
- Petrokilidou, C.; Gaitanis, G.; Bassukas, I.D.; Velegraki, A.; Guevara, E.; Vardaki, M.Z.; Kourkoumelis, N. Emerging Optical Techniques for the Diagnosis of Onychomycosis. Appl. Sci. 2020, 10, 2340. [Google Scholar] [CrossRef] [Green Version]
- Philipsen, P.A.; Knudsen, L.; Gniadecka, M.; Ravnbak, M.H.; Wulf, H.C. Diagnosis of malignant melanoma and basal cell carcinoma by in vivo NIR-FT Raman spectroscopy is independent of skin pigmentation. Photochem. Photobiol. Sci. 2013, 12, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.M.; Zhang, Y.; Moy, A.J.; Feng, X.; Sebastian, K.R.; Reichenberg, J.S.; Fox, M.C.; Markey, M.K.; Tunnell, J.W. Characterization of Ex Vivo Nonmelanoma Skin Tissue Using Raman Spectroscopy. Photonics 2021, 8, 282. [Google Scholar] [CrossRef]
- Kourkoumelis, N.; Balatsoukas, I.; Moulia, V.; Elka, A.; Gaitanis, G.; Bassukas, I.D. Advances in the in Vivo Raman Spectroscopy of Malignant Skin Tumors Using Portable Instrumentation. Int. J. Mol. Sci. 2015, 16, 14554–14570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, I.P.; van Doorn, R.; Caspers, P.J.; Bakker Schut, T.C.; Barroso, E.M.; Nijsten, T.E.C.; Noordhoek Hegt, V.; Koljenovic, S.; Puppels, G.J. Improving clinical diagnosis of early-stage cutaneous melanoma based on Raman spectroscopy. Br. J. Cancer 2018, 119, 1339–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosca, S.; Conti, C.; Stone, N.; Matousek, P. Spatially offset Raman spectroscopy. Nat. Rev. Methods Primers 2021, 1, 22. [Google Scholar] [CrossRef]
- Vardaki, M.Z.; Gardner, B.; Stone, N.; Matousek, P. Studying the distribution of deep Raman spectroscopy signals using liquid tissue phantoms with varying optical properties. Analyst 2015, 140, 5112–5119. [Google Scholar] [CrossRef] [Green Version]
- Buckley, K.; Kerns, J.; Gikas, P.D.; Birch, H.; Vinton, J.; Keen, R.; Parker, A.W.; Matousek, P.; Goodship, A. Measurement of abnormal bone composition in vivo using noninvasive Raman spectroscopy. IBMS BoneKEy 2014, 11, 72178. [Google Scholar] [CrossRef]
- Lundsgaard-Nielsen, S.M.; Pors, A.; Banke, S.O.; Henriksen, J.E.; Hepp, D.K.; Weber, A. Critical-depth Raman spectroscopy enables home-use non-invasive glucose monitoring. PLoS ONE 2018, 13, e0197134. [Google Scholar] [CrossRef] [Green Version]
- Afseth, N.K.; Bloomfield, M.; Wold, J.P.; Matousek, P. A novel approach for subsurface through-skin analysis of salmon using spatially offset Raman spectroscopy (SORS). Appl. Spectrosc. 2014, 68, 255–262. [Google Scholar] [CrossRef]
- Pence, I.J.; O’Brien, C.M.; Masson, L.E.; Mahadevan-Jansen, A. Application driven assessment of probe designs for Raman spectroscopy. Biomed. Opt. Express 2021, 12, 852–871. [Google Scholar] [CrossRef]
- Schleusener, J.; Gluszczynska, P.; Reble, C.; Gersonde, I.; Helfmann, J.; Fluhr, J.W.; Lademann, J.; Rowert-Huber, J.; Patzelt, A.; Meinke, M.C. In vivo study for the discrimination of cancerous and normal skin using fibre probe-based Raman spectroscopy. Exp. Derm. 2015, 24, 767–772. [Google Scholar] [CrossRef]
- Lim, L.; Nichols, B.; Migden, M.R.; Rajaram, N.; Reichenberg, J.S.; Markey, M.K.; Ross, M.I.; Tunnell, J.W. Clinical study of noninvasive in vivo melanoma and nonmelanoma skin cancers using multimodal spectral diagnosis. J. Biomed. Opt. 2014, 19, 117003. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.; Baker, R.; Rogers, K.; Parker, A.W.; Matousek, P. Subsurface probing of calcifications with spatially offset Raman spectroscopy (SORS): Future possibilities for the diagnosis of breast cancer. Analyst 2007, 132, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Vardaki, M.Z.; Kourkoumelis, N. Tissue Phantoms for Biomedical Applications in Raman Spectroscopy: A Review. Biomed. Eng. Comput. Biol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Lai, P.X.; Xu, X.; Wang, L.H.V. Dependence of optical scattering from Intralipid in gelatin-gel based tissue-mimicking phantoms on mixing temperature and time. J. Biomed. Opt. 2014, 19, 035002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.; Garcia-Uribe, A.; Kothapalli, S.R.; Wang, L.H.V. Optical phantoms for ultrasound-modulated optical tomography. In Proceedings of the Design and Performance Validation of Phantoms Used in Conjunction with Optical Measurements of Tissue, San Jose, CA, USA, 19–24 January 2008; p. 6870. [Google Scholar] [CrossRef]
- Vardaki, M.Z.; Matousek, P.; Stone, N. Characterisation of signal enhancements achieved when utilizing a photon diode in deep Raman spectroscopy of tissue. Biomed. Opt. Express 2016, 7, 2130–2141. [Google Scholar] [CrossRef]
- Salomatina, E.; Jiang, B.; Novak, J.; Yaroslavsky, A.N. Optical properties of normal and cancerous human skin in the visible and near-infrared spectral range. J. Biomed. Opt. 2006, 11, 064026. [Google Scholar] [CrossRef]
- Lister, T.; Wright, P.A.; Chappell, P.H. Optical properties of human skin. J. Biomed. Opt. 2012, 17, 090901. [Google Scholar] [CrossRef]
- Jacques, S.L. Optical properties of biological tissues: A review. Phys. Med. Biol. 2013, 58, R37–R61. [Google Scholar] [CrossRef]
- Meglinski, I.V.; Matcher, S.J. Quantitative assessment of skin layers absorption and skin reflectance spectra simulation in the visible and near-infrared spectral regions. Physiol. Meas. 2002, 23, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, M.; Andrews, D.; Loeffen, P.; Tombling, C.; York, T.; Matousek, P. Non-invasive identification of incoming raw pharmaceutical materials using Spatially Offset Raman Spectroscopy. J. Pharm. Biomed. 2013, 76, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Nicolson, F.; Andreiuk, B.; Andreou, C.; Hsu, H.T.; Rudder, S.; Kircher, M.F. Non-invasive In Vivo Imaging of Cancer Using Surface-Enhanced Spatially Offset Raman Spectroscopy (SESORS). Theranostics 2019, 9, 5899–5913. [Google Scholar] [CrossRef]
- Matousek, P. Inverse spatially offset Raman spectroscopy for deep noninvasive probing of turbid media. Appl. Spectrosc. 2006, 60, 1341–1347. [Google Scholar] [CrossRef] [PubMed]
- Schulze, H.G.; Foist, R.B.; Okuda, K.; Ivanov, A.; Turner, R.F.B. A Small-Window Moving Average-Based Fully Automated Baseline Estimation Method for Raman Spectra. Appl. Spectrosc. 2012, 66, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Schulze, H.G.; Rangan, S.; Blades, M.W.; Piret, J.M.; Turner, R.F.B. Smoothing Raman Spectra with Contiguous Single-Channel Fitting of Voigt Distributions: An Automated, High-Quality Procedure. Appl. Spectrosc. 2019, 73, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Matousek, P.; Clark, I.P.; Draper, E.R.C.; Morris, M.D.; Goodship, A.E.; Everall, N.; Towrie, M.; Finney, W.F.; Parker, A.W. Subsurface probing in diffusely scattering media using spatially offset Raman spectroscopy. Appl. Spectrosc. 2005, 59, 393–400. [Google Scholar] [CrossRef]
- Matousek, P.; Morris, M.D.; Everall, N.; Clark, I.P.; Towrie, M.; Draper, E.; Goodship, A.; Parker, A.W. Numerical simulations of subsurface probing in diffusely scattering media using spatially offset Raman spectroscopy. Appl. Spectrosc. 2005, 59, 1485–1492. [Google Scholar] [CrossRef] [PubMed]
- Vardaki, M.Z.; Devine, D.V.; Serrano, K.; Simantiris, N.; Blades, M.W.; Piret, J.M.; Turner, R.F.B. Defocused Spatially Offset Raman Spectroscopy in Media of Different Optical Properties for Biomedical Applications Using a Commercial Spatially Offset Raman Spectroscopy Device. Appl. Spectrosc. 2020, 74, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Maher, J.R.; Berger, A.J. Determination of ideal offset for spatially offset Raman spectroscopy. Appl. Spectrosc. 2010, 64, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Konishi, E.; Yoshizawa, Y. Estimation of Depth of Basal-Cell Layer of Skin for Radiation Protection. Radiat. Prot. Dosim. 1985, 11, 29–33. [Google Scholar]
- Silveira, L., Jr.; Silveira, F.L.; Bodanese, B.; Zangaro, R.A.; Pacheco, M.T. Discriminating model for diagnosis of basal cell carcinoma and melanoma in vitro based on the Raman spectra of selected biochemicals. J. Biomed. Opt. 2012, 17, 077003. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Boffelli, M.; Miyamori, D.; Uemura, T.; Marunaka, Y.; Zhu, W.; Ikegaya, H. Raman spectroscopy of human skin: Looking for a quantitative algorithm to reliably estimate human age. J. Biomed. Opt. 2015, 20, 065008. [Google Scholar] [CrossRef]
- Ali, S.M.; Bonnier, F.; Tfayli, A.; Lambkin, H.; Flynn, K.; McDonagh, V.; Healy, C.; Lee, T.C.; Lyng, F.M.; Byrne, H.J. Raman spectroscopic analysis of human skin tissue sections ex-vivo: Evaluation of the effects of tissue processing and dewaxing. J. Biomed. Opt. 2013, 18, 061202. [Google Scholar] [CrossRef] [Green Version]
- Caspers, P.J.; Lucassen, G.W.; Wolthuis, R.; Bruining, H.A.; Puppels, G.J. In vitro and in vivo Raman spectroscopy of human skin. Biospectroscopy 1998, 4, S31–S39. [Google Scholar] [CrossRef]
- Kuzuhara, A. Analysis of structural change in keratin fibers resulting from chemical treatments using Raman spectroscopy. Biopolymers 2005, 77, 335–344. [Google Scholar] [CrossRef]
- Tfayli, A.; Piot, O.; Draux, F.; Pitre, F.; Manfait, M. Molecular characterization of reconstructed skin model by Raman microspectroscopy: Comparison with excised human skin. Biopolymers 2007, 87, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Baden, H.P.; Goldsmith, L.A.; Bonar, L. Conformational changes in the α-fibrous protein of epidermis. J. Investig. Dermatol. 1973, 60, 215–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tfaili, S.; Gobinet, C.; Josse, G.; Angiboust, J.F.; Manfait, M.; Piot, O. Confocal Raman microspectroscopy for skin characterization: A comparative study between human skin and pig skin. Analyst 2012, 137, 3673–3682. [Google Scholar] [CrossRef]
- Baden, H.P.; Bonar, L.; Katz, E. Fibrous proteins of epidermis. J. Investig. Derm. 1968, 51, 301–307. [Google Scholar] [CrossRef] [Green Version]
- Meksiarun, P.; Andriana, B.B.; Matsuyoshi, H.; Sato, H. Non-invasive Quantitative Analysis of Specific Fat Accumulation in Subcutaneous Adipose Tissues using Raman Spectroscopy. Sci. Rep. 2016, 6, 37068. [Google Scholar] [CrossRef] [Green Version]
- Mosca, S.; Dey, P.; Salimi, M.; Gardner, B.; Palombo, F.; Stone, N.; Matousek, P. Spatially Offset Raman Spectroscopy-How Deep? Anal. Chem. 2021, 93, 6755–6762. [Google Scholar] [CrossRef]
- Beattie, J.R.; Esmonde-White, F.W.L. Exploration of Principal Component Analysis: Deriving Principal Component Analysis Visually Using Spectra. Appl. Spectrosc. 2021, 75, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Thieu, K.; Ruiz, M.E.; Owens, D.M. Cells of origin and tumor-initiating cells for nonmelanoma skin cancers. Cancer Lett. 2013, 338, 82–88. [Google Scholar] [CrossRef] [Green Version]
- Vargis, E.; Byrd, T.; Logan, Q.; Khabele, D.; Mahadevan-Jansen, A. Sensitivity of Raman spectroscopy to normal patient variability. J. Biomed. Opt. 2011, 16, 117004. [Google Scholar] [CrossRef]
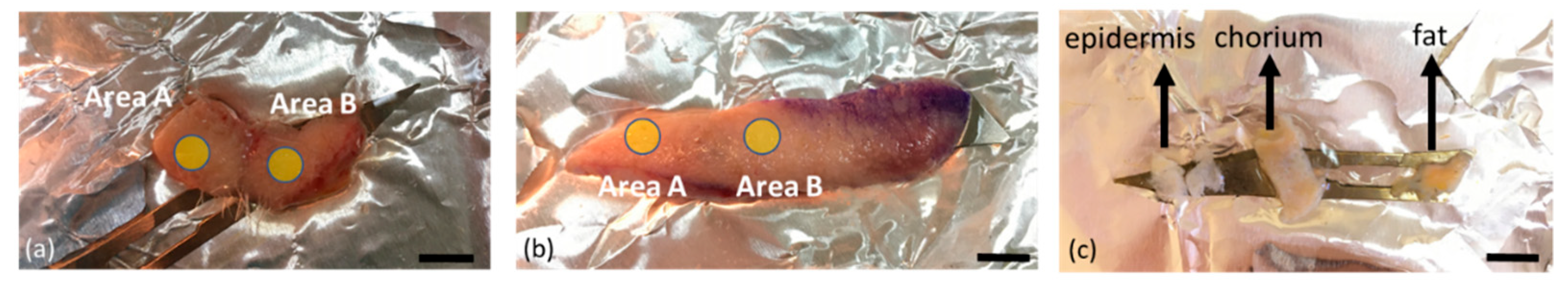

| Sample | Location | Subareas | Epidermis Thickness (mm) | Total Sample Thickness (mm) | Dissected |
|---|---|---|---|---|---|
| 1 | Scalp | A, B | 1 | 3.5 | Y |
| 2 | Scalp | A, B | 0.5 | 3 | N |
| 3 | Scalp | A | 1.3 | 6 | N |
| 4 | Eyelid | A, B | 0.4 | 2 | Y |
| 5 | Eyelid | A | 0.3 | 2 | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vardaki, M.Z.; Seretis, K.; Gaitanis, G.; Bassukas, I.D.; Kourkoumelis, N. Assessment of Skin Deep Layer Biochemical Profile Using Spatially Offset Raman Spectroscopy. Appl. Sci. 2021, 11, 9498. https://doi.org/10.3390/app11209498
Vardaki MZ, Seretis K, Gaitanis G, Bassukas ID, Kourkoumelis N. Assessment of Skin Deep Layer Biochemical Profile Using Spatially Offset Raman Spectroscopy. Applied Sciences. 2021; 11(20):9498. https://doi.org/10.3390/app11209498
Chicago/Turabian StyleVardaki, Martha Z., Konstantinos Seretis, Georgios Gaitanis, Ioannis D. Bassukas, and Nikolaos Kourkoumelis. 2021. "Assessment of Skin Deep Layer Biochemical Profile Using Spatially Offset Raman Spectroscopy" Applied Sciences 11, no. 20: 9498. https://doi.org/10.3390/app11209498
APA StyleVardaki, M. Z., Seretis, K., Gaitanis, G., Bassukas, I. D., & Kourkoumelis, N. (2021). Assessment of Skin Deep Layer Biochemical Profile Using Spatially Offset Raman Spectroscopy. Applied Sciences, 11(20), 9498. https://doi.org/10.3390/app11209498








