Multiple Instance Classification for Gastric Cancer Pathological Images Based on Implicit Spatial Topological Structure Representation
Abstract
:1. Introduction
- (1)
- We present an efficient framework composed of a feature extraction module based on ImageNet-trained CNNs and a multiple instance classification module based on GCNs for the classification task of gastric pathological WSIs;
- (2)
- We construct the graph structure according to the similarity between the patch embeddings by implicitly fusing the information on their spatial topological structural relationships between instances. The proposed MIC module based on GCNs achieves information fusion in both physical space and feature space for all instances;
- (3)
- We conduct experiments on two real high-resolution gastric pathological image datasets with different imaging mechanisms to prove the effectiveness and robustness of our proposed framework. To our knowledge, our work is the first to conduct experiments both on an H&E-stained pathological image dataset and a stimulated Raman scattering (SRS) microscope image dataset.
2. Background Knowledge
2.1. Multiple Instance Classification
2.2. Graph Convolutional Networks and Graph Classification
2.3. Differentiable Pooling
3. Materials and Methods
3.1. Datasets
3.2. Data Preprocessing
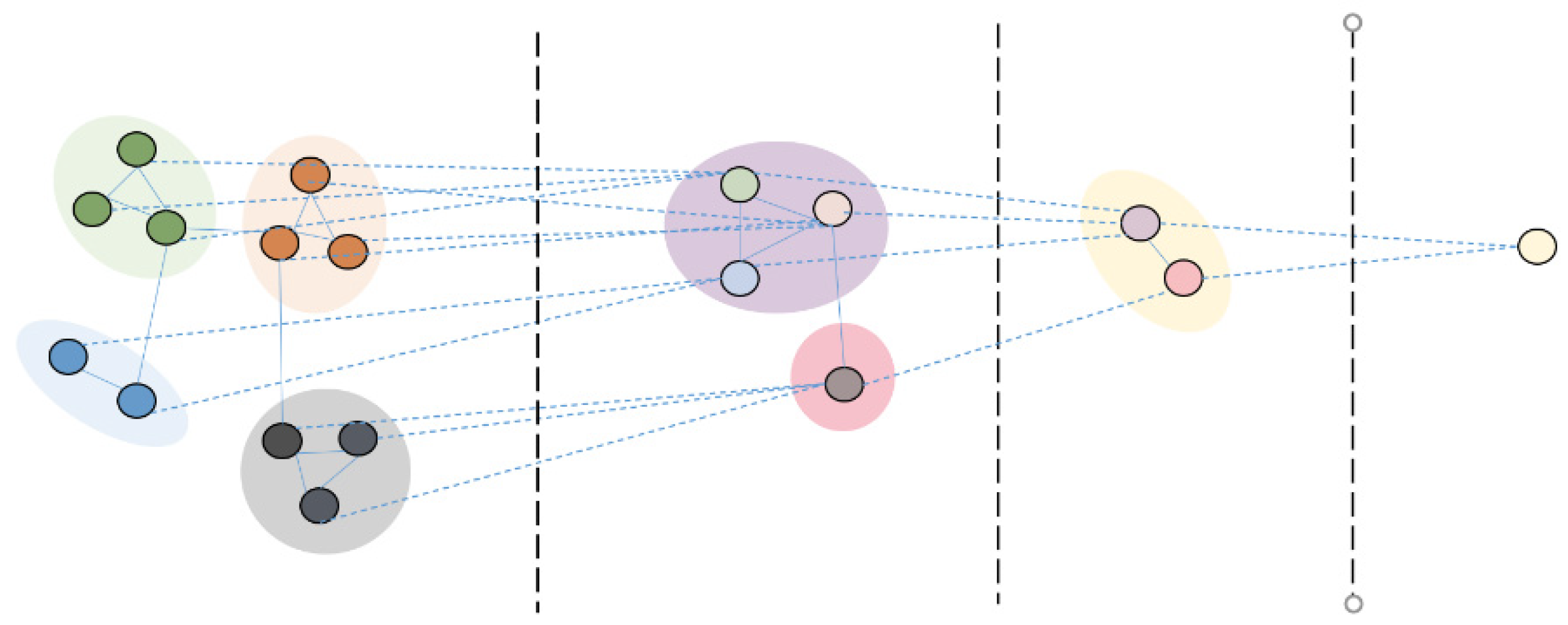
3.3. Proposed Model of Multiple Instance Classification Based on GCNs
4. Results and Discussion
4.1. Experimental Environment and Setup
4.2. Influence of Model Parameters
4.3. Performance Comparison of Different Feature Extractors
4.4. Comparisons with Other Multiple Instance Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, H.; Zerbe, N.; Klempert, I.; Hellwich, O.; Hufnagl, P. Deep Convolutional Neural Networks for Automatic Classification of Gastric Carcinoma Using Whole Slide Images in Digital Histopathology. Comput. Med Imaging Graph. 2017, 61, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, Y.; Yu, L.; Chen, H.; Lin, H.; Wan, X.; Fan, X.; Heng, P.-A. RMDL: Recalibrated Multi-Instance Deep Learning for Whole Slide Gastric Image Classification. Med. Image Anal. 2019, 58, 101549. [Google Scholar] [CrossRef] [PubMed]
- Geirhos, R.; Rubisch, P.; Michaelis, C.; Bethge, M.; Wichmann, F.A.; Brendel, W. ImageNet-trained CNNs are Biased Towards Texture; Increasing Shape Bias Improves Accuracy and Robustness. arXiv 2018, arXiv:1811.12231. [Google Scholar]
- Komura, D.; Ishikawa, S. Machine Learning Approaches for Pathologic Diagnosis. Virchows Arch. 2019, 475, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Shukla, S.; Gyanchandani, M. Pre-Trained Convolutional Neural Networks as Feature Extractors for Diagnosis of Breast Cancer Using Histopathology. Int. J. Imaging Syst. Technol. 2020, 30, 577–591. [Google Scholar] [CrossRef]
- Gupta, K.; Chawla, N. Analysis of Histopathological Images for Prediction of Breast Cancer Using Traditional Classifiers with Pre-Trained CNN. Procedia Comput. Sci. 2020, 167, 878–889. [Google Scholar] [CrossRef]
- Zhou, Z.-H. A Brief Introduction to Weakly Supervised Learning. Natl. Sci. Rev. 2018, 5, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Amores, J. Multiple Instance Classification: Review, Taxonomy and Comparative Study. Artif. Intell. 2013, 201, 81–105. [Google Scholar] [CrossRef]
- Conjeti, S.; Paschali, M.; Katouzian, A.; Navab, N. Deep Multiple Instance Hashing for Scalable Medical Image Retrieval. In Proceedings of the Medical Image Computing and Computer Assisted Intervention (MICCAI), Quebec City, QC, Canada, 10–14 September 2017; pp. 550–558. [Google Scholar]
- Das, K.; Conjeti, S.; Roy, A.G.; Chatterjee, J.; Sheet, D. Multiple Instance Learning of Deep Convolutional Neural Networks for Breast Histopathology whole Slide Classification. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI), Washington, DC, USA, 4–7 April 2018; pp. 578–581. [Google Scholar] [CrossRef]
- Ilse, M.; Tomczak, J.M.; Welling, M. Attention-based Deep Multiple Instance Learning. arXiv 2018, arXiv:1802.04712. [Google Scholar]
- Yao, J.; Zhu, X.; Jonnagaddala, J.; Hawkins, N.; Huang, J. Whole Slide Images Based Cancer Survival Prediction Using Attention Guided Deep Multiple Instance Learning Networks. Med. Image Anal. 2020, 65, 101789. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-Efficient and Weakly Supervised Computational Pathology on Whole-Slide Images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Carbonneau, M.-A.; Cheplygina, V.; Granger, E.; Gagnon, G. Multiple Instance Learning: A Survey of Problem Characteristics and Applications. Pattern Recognit. 2018, 77, 329–353. [Google Scholar] [CrossRef] [Green Version]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2017, arXiv:1609.02907. [Google Scholar]
- Ying, R.; You, J.; Morris, C.; Ren, X.; Hamilton, W.L.; Leskovec, J. Hierarchical Graph Representation Learning with Differentiable Pooling. In Proceedings of the Advances in Neural Information Processing Systems (NIPS), Montreal, MT, Canada, 3–8 December 2018; pp. 4805–4815. [Google Scholar]
- Cangea, C.; Velickovic, P.; Jovanovic, N.; Kipf, T.; Lio, P. Towards Sparse Hierarchical Graph Classifiers. arXiv 2018, arXiv:1811.01287. [Google Scholar]
- Diehl, F. Edge Contraction Pooling for Graph Neural Networks. arXiv 2019, arXiv:1905.10990. [Google Scholar]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive Representation Learning on Large Graphs. In Proceedings of the in Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017; pp. 1025–1035. [Google Scholar]
- Xu, B.; Wang, N.; Chen, T.; Li, M. Empirical Evaluation of Rectified Activations in Convolutional Network. arXiv 2015, arXiv:1505.00853. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on International Conference on Machine Learning, Lille, France, 6 July 2015; Volume 35, pp. 448–456. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning (ICML), Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]





| Dataset | WSI Number | Patch Resolution | Threshold | Patch Number |
|---|---|---|---|---|
| SRS | 185 | 224 × 224 pixels | 35,000 (17.3%) | 68,387 |
| Mars | 2032 | 450 × 450 pixels | 15,000 (29.9%) | 396,539 |
| Feature | Dimension | Recall (%) | Precision (%) | F1_Score (%) |
|---|---|---|---|---|
| TX_fea | 408 | 94.78 ± 0.013 | 94.99 ± 0.013 | 94.59 ± 0.012 |
| VGG-16_fea | 4096 | 97.24 ± 0.012 | 97.24 ± 0.013 | 97.26 ± 0.012 |
| ResNet-18_fea | 512 | 98.14 ± 0.005 | 98.14 ± 0.005 | 98.16 ± 0.005 |
| DenseNet-121_fea | 1024 | 98.34 ± 0.009 | 98.34 ± 0.009 | 98.35 ± 0.008 |
| EfficientNet-B0_fea | 1280 | 97.69 ± 0.010 | 97.69 ± 0.010 | 97.70 ± 0.010 |
| Method | Recall (%) | Precision (%) | F1_Score (%) |
|---|---|---|---|
| MIP (mean) | 82.10 ± 0.051 | 82.70 ± 0.051 | 81.83 ± 0.051 |
| MIP (max) | 83.65 ± 0.044 | 84.82 ± 0.041 | 83.81 ± 0.046 |
| MIP (attention) | 84.37 ± 0.042 | 85.34 ± 0.038 | 84.47 ± 0.041 |
| RMDL | 84.47 ± 0.044 | 85.29 ± 0.042 | 84.23 ± 0.045 |
| GCN (mean_pool) | 89.08 ± 0.038 | 89.88 ± 0.033 | 89.67 ± 0.033 |
| GCN (max_pool) | 89.13 ± 0.046 | 90.20 ± 0.042 | 88.96 ± 0.049 |
| GCN + DIFFPOOL | 90.40 ± 0.032 | 91.16 ± 0.032 | 90.75 ± 0.034 |
| Method | Recall (%) | Precision (%) | F1_Score (%) |
|---|---|---|---|
| MIP (mean) | 92.40 ± 0.013 | 92.45 ± 0.013 | 92.40 ± 0.014 |
| MIP (max) | 95.48 ± 0.011 | 95.52 ± 0.011 | 95.45 ± 0.011 |
| MIP (attention) | 93.11 ± 0.009 | 93.15 ± 0.009 | 93.13 ± 0.009 |
| RMDL | 93.76 ± 0.008 | 93.81 ± 0.008 | 93.74 ± 0.009 |
| GCN(mean_pool) | 95.81 ± 0.008 | 95.83 ± 0.008 | 95.81 ± 0.008 |
| GCN (max_pool) | 97.70 ± 0.007 | 96.73 ± 0.007 | 97.70 ± 0.007 |
| GCN + DIFFPOOL | 98.24 ± 0.004 | 98.26 ± 0.004 | 98.24 ± 0.004 |
| Dataset | Shuffle | Recall (%) | Precision (%) | F1_Score (%) |
|---|---|---|---|---|
| SRS | Before | 90.40 ± 0.032 | 91.16 ± 0.032 | 90.75 ± 0.034 |
| After | 88.86 ± 0.036 | 88.30 ± 0.032 | 88.33 ± 0.035 | |
| Mars | Before | 98.24 ± 0.004 | 98.26 ± 0.004 | 98.24 ± 0.004 |
| After | 96.54 ± 0.005 | 96.59 ± 0.005 | 96.51 ± 0.005 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, X.; Wu, X. Multiple Instance Classification for Gastric Cancer Pathological Images Based on Implicit Spatial Topological Structure Representation. Appl. Sci. 2021, 11, 10368. https://doi.org/10.3390/app112110368
Xiang X, Wu X. Multiple Instance Classification for Gastric Cancer Pathological Images Based on Implicit Spatial Topological Structure Representation. Applied Sciences. 2021; 11(21):10368. https://doi.org/10.3390/app112110368
Chicago/Turabian StyleXiang, Xu, and Xiaofeng Wu. 2021. "Multiple Instance Classification for Gastric Cancer Pathological Images Based on Implicit Spatial Topological Structure Representation" Applied Sciences 11, no. 21: 10368. https://doi.org/10.3390/app112110368





