Ensemble Deep Learning for the Detection of COVID-19 in Unbalanced Chest X-ray Dataset
Abstract
:1. Introduction
1.1. Related Works
1.2. Our Contributions
- Performing lung segmentation before the classification of diseases;
- Applying five different approaches which are simple, easy to implement and reproduce, yet effective, to tackle unbalanced class distribution;
- Evaluating and validating the presented algorithms using the dataset with the largest number of COVID-19 CXRs;
- Visualizing the indicative regions that highly influence CNNs’ prediction, which are deemed useful for interpretability and explainability.
2. Dataset Description
3. Methodology
3.1. Image Preprocessing
3.2. Lung Segmentation
3.3. Classification of COVID-19, Normal and Pneumonia Using Deep Convolutional Neural Networks
3.3.1. Approach 1: Deep Learning with Weighted Loss on an Unbalanced Dataset
3.3.2. Approach 2: Deep Learning with Image Augmentation on an Unbalanced Dataset
3.3.3. Approach 3: Deep Learning with Undersampling on an Unbalanced Dataset
3.3.4. Approach 4: Deep Learning with Oversampling on an Unbalanced Dataset
3.3.5. Approach 5: Deep Learning with Hybrid Sampling on an Unbalanced Dataset
3.4. Ensemble Learning
3.5. Visualization
4. Experimental Setting, Results and Discussions
4.1. Experimental Setting and Training Strategy
4.2. Performance Metrics
- TruePositive denotes the number of correctly classified CXRs in which the true label is positive and prediction is positive for the particular class.
- TrueNegative refers to the number of CXRs in which the true label is negative and prediction is negative for the particular class.
- FalsePositive represents the number of CXRs which are negative but predicted as the positive for the particular class.
- FalseNegative (FN) denotes the number of CXRs which are positive but predicted as the negative for the particular class.
4.3. Experimental Results and Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
- The convolution layer is the core building block of a CNN, which is used to automatically extract the features from the input image and generate the feature map using the filters. Filters are applied to each training image at different resolutions, and the output of each convolved image is used as the input to the next layer. In CNN model, there are many convolutional layers that retrieve low to high-level features. Earlier layers extract the low-level features whereas the latter layers in the networks extract the high–level features. The output of the feature map relies on the input size, the filter size and its striding, and padding.
- Activation functions are non-linear functions that allow the network to learn non-linear mappings. It performs as the selection criteria to decide whether the selected neuron will activate. Only the activated features or neurons are carried forward into the next layer. Activation functions are usually embedded after the convolutional layer. The most common activation functions are sigmoid, Tanh, ReLU, and softmax.
- Pooling layer aims to simplify the output by applying nonlinear downsampling. Pooling can progressively reduce the spatial dimensions of the image, thereby reducing the number of parameters and computation in a network. It reduces the output feature map of the convolution layers by extracting important pixels and removing noise. The output of the pooling layer depends on the filter size and stride. The most common pooling layers used in CNN are max pooling and average pooling.
- The next-to-last layer of CNN is the fully connected layer, also known as the dense layer. It receives all extracted features from the previous layers as the input and used them to classify the image with the help of softmax or sigmoid function. The fully connected layer generates the vector that contains the probabilities for each class of any image being classified.
| Deep CNNs | Number of Layers | Number of Parameters | Input Size |
|---|---|---|---|
| XceptionNet | 71 | 22,910,480 | 299 × 299 |
| InceptionV3 | 48 | 23,851,784 | 299 × 299 |
| VGG16 | 16 | 138,357,544 | 224 × 224 |
| VGG19 | 19 | 143,667,240 | 224 × 224 |
| ResNet50 | 50 | 25,636,712 | 224 × 224 |
| ResNet152 | 152 | 60,419,944 | 224 × 224 |
| MobileNetV2 | 53 | 3,538,984 | 224 × 224 |
| DenseNet201 | 201 | 8,062,504 | 224 × 224 |
| InceptionResNetV2 | 164 | 55,873,736 | 299 × 299 |
| EfficientNetB7 | * | 66,658,687 | 600 × 600 |
| NasNetMobile | * | 5,326,716 | 224 × 224 |
Appendix A.1. InceptionV3

Appendix A.2. VGG-16 and VGG-19
Appendix A.3. Xception

Appendix A.4. ResNet50 and ResNet152

Appendix A.5. MobileNetV2

Appendix A.6. DenseNet-201

Appendix A.7. InceptionResNetV2
Appendix A.8. EfficientNetB7
Appendix A.9. NasNetMobile
References
- World Health Organization. Coronavirus Disease (COVID-19). Available online: https://www.who.int/health-topics/coronavirus (accessed on 1 September 2021).
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 1 September 2021).
- Axell-House, D.B.; Lavingia, R.; Rafferty, M.; Clark, E.; Amirian, E.S.; Chiao, E.Y. The estimation of diagnostic accuracy of tests for COVID-19: A scoping review. J. Infect. 2020, 81, 681–697. [Google Scholar] [CrossRef]
- Zu, Z.Y.; Jiang, M.D.; Xu, P.P.; Chen, W.; Ni, Q.Q.; Lu, G.M.; Zhang, L.J. Coronavirus disease 2019 (COVID-19): A perspective from China. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef] [Green Version]
- Cozzi, D.; Albanesi, M.; Cavigli, E.; Moroni, C.; Bindi, A.; Luvarà, S.; Lucarini, S.; Busoni, S.; Mazzoni, L.N.; Miele, V. Chest X-ray in new Coronavirus Disease 2019 (COVID-19) infection: Findings and correlation with clinical outcome. La Radiol. Med. 2020, 125, 730–737. [Google Scholar] [CrossRef]
- Mossa-Basha, M.; Meltzer, C.C.; Kim, D.C.; Tuite, M.J.; Kolli, K.P.; Tan, B.S. Radiology department preparedness for COVID-19:Radiology scientific expert panel. Radiology 2020, 296, E106–E112. [Google Scholar] [CrossRef] [Green Version]
- Bai, H.X.; Hsieh, B.; Xiong, Z.; Halsey, K.; Choi, J.W.; Tran, T.M.L.; Pan, I.; Shi, L.; Wang, D.; Mei, J.; et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology 2020, 296, 200823. [Google Scholar] [CrossRef]
- World Health Organization. Medical Doctors (per 10,000 Population). 2021. Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/medical-doctors-(per-10-000-population) (accessed on 1 September 2021).
- Rehman, A.; Iqbal, M.A.; Xing, H.; Ahmed, I. COVID-19 Detection Empowered with Machine Learning and Deep Learning Techniques: A Systematic Review. Appl. Sci. 2021, 11, 3414. [Google Scholar] [CrossRef]
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Sahlol, A.T.; Yousri, D.; Ewees, A.A.; Al-Qaness, M.A.; Damasevicius, R.; Abd Elaziz, M. COVID-19 image classification using deep features and fractional-order marine predators algorithm. Sci. Rep. 2020, 10, 15364. [Google Scholar] [CrossRef]
- Alazab, M.; Awajan, A.; Mesleh, A.; Abraham, A.; Jatana, V.; Alhyari, S. COVID-19 prediction and detection using deep learning. Int. J. Comput. Inf. Syst. Ind. Manag. Appl. 2020, 12, 168–181. [Google Scholar]
- Duran-Lopez, L.; Dominguez-Morales, J.P.; Corral-Jaime, J.; Vicente-Diaz, S.; Linares-Barranco, A. COVID-XNet: A custom deep learning system to diagnose and locate COVID-19 in chest X-ray images. Appl. Sci. 2020, 10, 5683. [Google Scholar] [CrossRef]
- Khasawneh, N.; Fraiwan, M.; Fraiwan, L.; Khassawneh, B.; Ibnian, A. Detection of COVID-19 from Chest X-ray Images Using Deep Convolutional Neural Networks. Sensors 2021, 21, 5940. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Ahmed, S.; Yap, M.H.; Tan, M.; Hasan, M.K. Reconet: Multi-level preprocessing of chest x-rays for covid-19 detection using convolutional neural networks. medRxiv 2020. [Google Scholar] [CrossRef]
- Yoo, S.H.; Geng, H.; Chiu, T.L.; Yu, S.K.; Cho, D.C.; Heo, J.; Choi, M.S.; Choi, I.H.; Cung Van, C.; Nhung, N.V.; et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020, 7, 427. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Ben Jabra, M.; Koubaa, A.; Benjdira, B.; Ammar, A.; Hamam, H. COVID-19 Diagnosis in Chest X-rays Using Deep Learning and Majority Voting. Appl. Sci. 2021, 11, 2884. [Google Scholar] [CrossRef]
- Shelke, A.; Inamdar, M.; Shah, V.; Tiwari, A.; Hussain, A.; Chafekar, T.; Mehendale, N. Chest X-ray classification using Deep learning for automated COVID-19 screening. SN Comput. Sci. 2021, 2, 1–9. [Google Scholar] [CrossRef]
- Oh, Y.; Park, S.; Ye, J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med. Imaging 2020, 39, 2688–2700. [Google Scholar] [CrossRef]
- Rajaraman, S.; Siegelman, J.; Alderson, P.O.; Folio, L.S.; Folio, L.R.; Antani, S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access 2020, 8, 115041–115050. [Google Scholar] [CrossRef]
- Bridge, J.; Meng, Y.; Zhao, Y.; Du, Y.; Zhao, M.; Sun, R.; Zheng, Y. Introducing the GEV activation function for highly unbalanced data to develop COVID-19 diagnostic models. IEEE J. Biomed. Health Inform. 2020, 24, 2776–2786. [Google Scholar] [CrossRef]
- Nishio, M.; Noguchi, S.; Matsuo, H.; Murakami, T. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: Combination of data augmentation methods. Sci. Rep. 2020, 10, 17532. [Google Scholar] [CrossRef]
- Rahman, T.; Khandakar, A.; Qiblawey, Y.; Tahir, A.; Kiranyaz, S.; Kashem, S.B.A.; Islam, M.T.; Al Maadeed, S.; Zughaier, S.M.; Khan, M.S.; et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021, 132, 104319. [Google Scholar] [CrossRef]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al Emadi, N.; et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Qin, C.; Yao, D.; Shi, Y.; Song, Z. Computer-aided detection in chest radiography based on artificial intelligence: A survey. Biomed. Eng. Online 2018, 17, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.; Yang, G.; Tang, G. A fast two-dimensional median filtering algorithm. IEEE Trans. Acoust. Speech Signal Process. 1979, 27, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Zuiderveld, K. Contrast limited adaptive histogram equalization. In Graphics Gems IV; Academic Press Professional, Inc.: Cambridge, MA, USA, 1994; pp. 474–485. [Google Scholar]
- Win, K.Y.; Maneerat, N.; Hamamoto, K.; Sreng, S. Hybrid Learning of Hand-Crafted and Deep-Activated Features Using Particle Swarm Optimization and Optimized Support Vector Machine for Tuberculosis Screening. Appl. Sci. 2020, 10, 5749. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the Springer on European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Soille, P. Morphological Image Analysis: Principles and Applications; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 4, p. 12. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 24 May 2019; pp. 6105–6114. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- Rajpurkar, P. AI for Medical Diagnosis, Coursera. Available online: https://www.coursera.org/learn/ai-for-medical-diagnosis (accessed on 1 September 2021).
- Brownlee, J. Imbalanced Classification with Python: Better Metrics, Balance Skewed Classes, Cost-Sensitive Learning; Machine Learning Mastery: Vermont, Australia, 2020. [Google Scholar]
- He, H.; Ma, Y. Imbalanced Learning: Foundations, Algorithms, and Applications; Wiley-IEEE Press: Hoboken, NJ, USA, 2013. [Google Scholar]
- Witten, I.H.; Frank, E.; Mark, A. Hall Data Mining: Practical Machine Learning; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 9780123748560. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the CVPR, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.X.J.; Lu, P.X.; Thoma, G. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef]




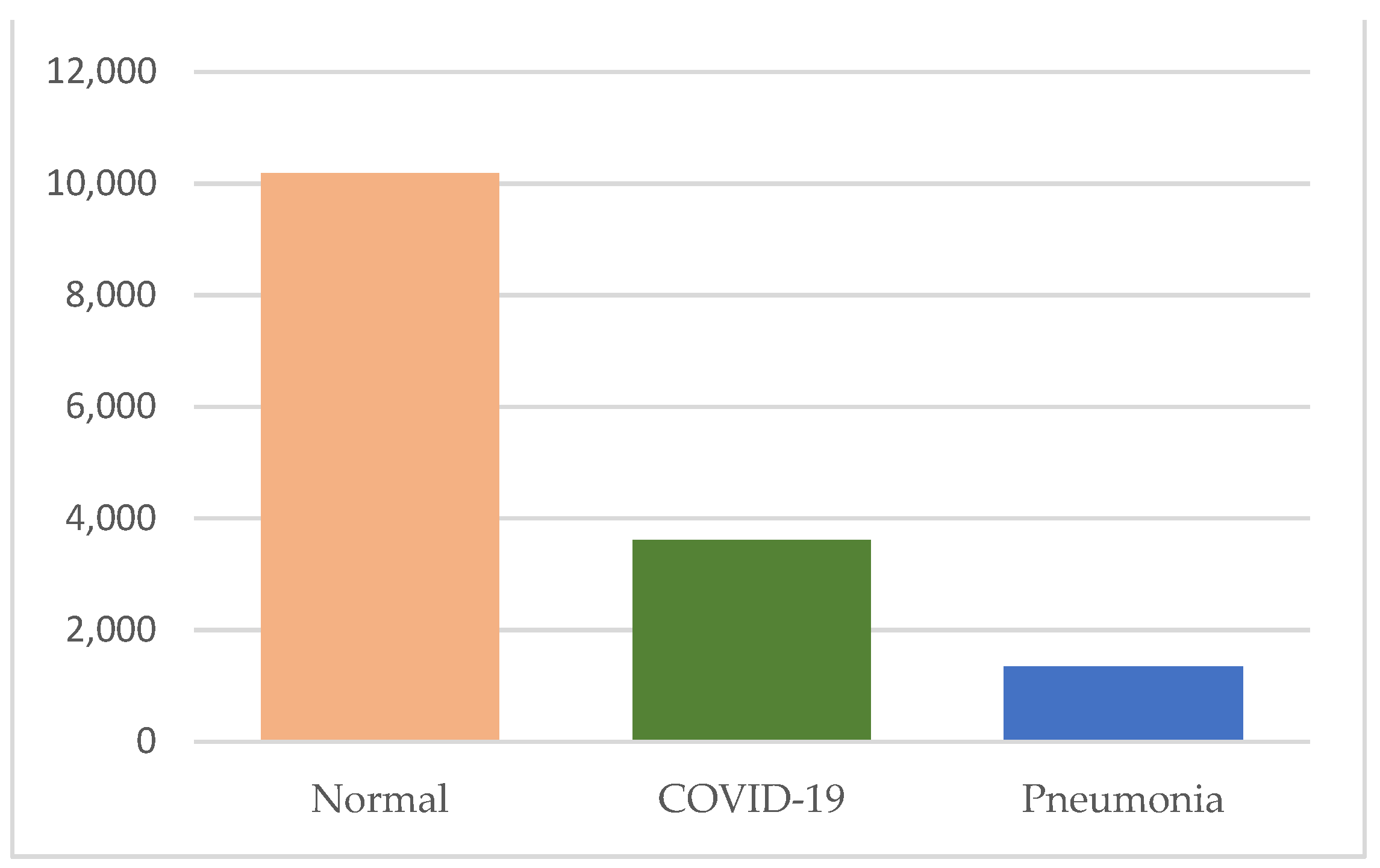











| Class | Number of Images | File Type | Image Resolution |
|---|---|---|---|
| Normal | 10,192 | PNG | 299 × 299 |
| COVID-19 | 3616 | PNG | 299 × 299 |
| Pneumonia | 1345 | PNG | 299 × 299 |
| Unbalanced Data Handling Approaches | Methods | Data | Hyperparameters of CNNs | ||||
|---|---|---|---|---|---|---|---|
| COVID-19 | Normal | Pneumonia | α | β_1 | β_2 | ||
| Approach 0 | Normal loss | 2893 | 8154 | 1076 | 0.001 | 0.9 | 0.999 |
| Approach 1 | Weighted loss | 2893 | 8154 | 1076 | 0.001 | 0.9 | 0.999 |
| Approach 2 | Image augmentation | 5393 | 4363 | 8154 | 0.001 | 0.9 | 0.999 |
| Approach 3 | Undersampling | 1013 | 1223 | 1076 | 0.001 | 0.9 | 0.999 |
| Approach 4 | Oversampling | 8100 | 8154 | 8070 | 0.001 | 0.9 | 0.999 |
| Approach 5 | Hybrid sampling | 2893 | 2854 | 2690 | 0.001 | 0.9 | 0.999 |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 97.07% | 97.13% | 92.77% | 99.80% | 96.20% |
| InceptionV3 | 95.10% | 98.57% | 93.63% | 99.57% | 91.60% |
| VGG-16 | 83.97% | 99.87% | 67.40% | 99.30% | 74.77% |
| VGG-19 | 87.60% | 97.27% | 79.87% | 97.80% | 82.80% |
| ResNet50 | 97.70% | 99.47% | 96.37% | 99.90% | 95.23% |
| ResNet152 | 95.83% | 97.27% | 91.53% | 99.53% | 93.40% |
| MobileNetV2 | 97.17% | 98.07% | 94.57% | 98.93% | 94.13% |
| DenseNet201 | 95.97% | 95.43% | 96.67% | 98.37% | 93.23% |
| InceptionResNetV2 | 96.53% | 98.67% | 94.53% | 99.70% | 93.97% |
| EfficientNetB7 | 88.43% | 89.37% | 91.50% | 98.23% | 83.80% |
| NasNetMobile | 97.97% | 98.97% | 96.03% | 99.90% | 94.90% |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 97.40% | 99.37% | 94.97% | 99.90% | 95.17% |
| InceptionV3 | 86.67% | 87.20% | 80.23% | 99.03% | 77.83% |
| VGG-16 | 83.90% | 99.33% | 74.07% | 98.60% | 72.77% |
| VGG-19 | 86.03% | 94.83% | 69.60% | 96.73% | 78.30% |
| ResNet50 | 86.63% | 98.83% | 76.53% | 98.87% | 83.57% |
| ResNet152 | 94.27% | 98.27% | 89.50% | 99.33% | 88.20% |
| MobileNetV2 | 97.23% | 98.33% | 95.43% | 99.67% | 94.63% |
| DenseNet201 | 95.47% | 99.40% | 89.63% | 99.67% | 93.63% |
| InceptionResNetV2 | 97.17% | 99.07% | 94.00% | 99.80% | 95.30% |
| EfficientNetB7 | 83.37% | 94.87% | 64.83% | 97.47% | 80.43% |
| NasNetMobile | 93.13% | 94.07% | 94.20% | 99.17% | 87.60% |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 98.17% | 98.33% | 98.53% | 99.93% | 97.07% |
| InceptionV3 | 93.50% | 95.13% | 95.23% | 99.67% | 88.47% |
| VGG-16 | 96.73% | 95.33% | 97.10% | 99.33% | 94.03% |
| VGG-19 | 91.60% | 85.50% | 88.70% | 98.07% | 83.90% |
| ResNet50 | 97.63% | 96.93% | 97.90% | 99.70% | 95.10% |
| ResNet152 | 97.40% | 96.03% | 97.53% | 99.47% | 95.33% |
| MobileNetV2 | 97.57% | 97.50% | 97.53% | 99.63% | 95.20% |
| DenseNet201 | 98.00% | 97.90% | 98.50% | 99.80% | 95.67% |
| InceptionResNetV2 | 96.63% | 95.53% | 96.10% | 99.30% | 94.17% |
| EfficientNetB7 | 97.13% | 98.10% | 97.70% | 99.87% | 92.10% |
| NasNetMobile | 95.93% | 97.13% | 92.47% | 99.77% | 93.30% |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 97.60% | 98.23% | 98.07% | 99.90% | 95.43% |
| InceptionV3 | 95.80% | 97.00% | 96.83% | 99.83% | 92.33% |
| VGG-16 | 95.17% | 95.90% | 96.53% | 99.50% | 92.23% |
| VGG-19 | 95.67% | 94.47% | 94.03% | 99.07% | 92.03% |
| ResNet50 | 97.13% | 97.40% | 97.57% | 99.70% | 94.97% |
| ResNet152 | 96.43% | 97.03% | 95.77% | 99.27% | 93.50% |
| MobileNetV2 | 98.43% | 98.20% | 98.23% | 99.83% | 96.20% |
| DenseNet201 | 98.57% | 98.17% | 97.40% | 99.87% | 97.70% |
| InceptionResNetV2 | 98.50% | 98.50% | 98.50% | 99.87% | 97.43% |
| EfficientNetB7 | 93.87% | 95.40% | 95.50% | 98.67% | 85.50% |
| NasNetMobile | 98.20% | 98.33% | 98.47% | 99.87% | 96.73% |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 97.80% | 97.93% | 97.87% | 99.77% | 96.075 |
| InceptionV3 | 96.63% | 93.13% | 94.13% | 99.37% | 93.93% |
| VGG-16 | 95.57% | 95.50% | 96.77% | 99.07% | 92.27% |
| VGG-19 | 94.67% | 93.77% | 95.37% | 98.70% | 90.60% |
| ResNet50 | 97.77% | 97.80% | 98.13% | 99.83% | 96.33% |
| ResNet152 | 96.67% | 96.67% | 97.60% | 99.60% | 93.43% |
| MobileNetV2 | 96.33% | 94.97% | 96.07% | 99.30% | 91.63% |
| DenseNet201 | 96.33% | 97.17% | 96.73% | 99.57% | 93.87% |
| InceptionResNetV2 | 96.77% | 96.67% | 97.17% | 98.50% | 94.73% |
| EfficientNetB7 | 97.57% | 95.67% | 96.20% | 99.57% | 95.83% |
| NasNetMobile | 97.40% | 95.47% | 97.17% | 99.50% | 95.50% |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 98.63% | 97.57% | 98.23% | 99.90% | 97.47% |
| InceptionV3 | 96.27% | 97.57% | 96.43% | 99.53% | 93.43% |
| VGG-16 | 96.27% | 96.20% | 96.83% | 99.37% | 93.73% |
| VGG-19 | 92.93% | 94.20% | 95.00% | 98.70% | 88.77% |
| ResNet50 | 97.77% | 97.27% | 97.70% | 99.63% | 96.17% |
| ResNet152 | 97.37% | 97.77% | 97.73% | 99.77% | 95.43% |
| MobileNetV2 | 97.90% | 96.10% | 97.40% | 99.77% | 95.83% |
| DenseNet201 | 98.20% | 98.07% | 97.23% | 99.77% | 97.27% |
| InceptionResNetV2 | 98.67% | 98.17% | 98.23% | 99.90% | 97.63% |
| EfficientNetB7 | 89.03% | 90.00% | 92.07% | 98.97% | 84.90% |
| NasNetMobile | 97.80% | 98.03% | 99.83% | 99.83% | 96.17% |
| Deep CNNs | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|
| XceptionNet | 97.36% | 97.86% | 97.70% | 99.63% | 95.80% |
| InceptionV3 | 93.23% | 95.86% | 94.70% | 99.60% | 86.40% |
| VGG-16 | 95.93% | 95.23% | 96.37% | 99.03% | 92.43% |
| VGG-19 | 95.97% | 96.13% | 96.63% | 99.30% | 93.30% |
| ResNet50 | 93.63% | 94.30% | 94.67% | 99.00% | 90.90% |
| ResNet152 | 93.60% | 90.37% | 91.70% | 94.90% | 89.50% |
| MobileNetV2 | 97.23% | 98.33% | 95.43% | 99.67% | 94.63% |
| DenseNet201 | 95.80% | 96.43% | 97.07% | 99.70% | 93.73% |
| InceptionResNetV2 | 92.23% | 92.40% | 93.83% | 96.47% | 83.73% |
| EfficientNetB7 | 93.03% | 92.93% | 94.83% | 99.67% | 88.50% |
| NasNetMobile | 96.67% | 97.40% | 97.43% | 99.77% | 94.63% |
| Deep CNNs | Voting Strategy | Accuracy | Sensitivity | Specificity | AUC | F-Measure |
|---|---|---|---|---|---|---|
| Ensemble of Approach 0 | Soft Voting | 97.93% | 98.43% | 98.40% | 99.90% | 96.70% |
| Hard Voting | 96.63% | 97.97% | 98.44% | N/A | 96.62% | |
| Ensemble of Approach 1 | Soft Voting | 99.03% | 98.97% | 99.20% | 99.97% | 98.23% |
| Hard Voting | 98.35% | 98.66% | 99.11% | N/A | 97.88% | |
| Ensemble of Approach 2 | Soft Voting | 99.23% | 99.27% | 99.27% | 99.97% | 98.30% |
| Hard Voting | 98.84% | 99.05% | 99.38% | N/A | 98.41% | |
| Ensemble of Approach 3 | Soft Voting | 98.00% | 98.17% | 98.37% | 99.87% | 96.53% |
| Hard Voting | 96.53% | 97.63% | 98.33% | N/A | 96.13% | |
| Ensemble of Approach 4 | Soft Voting | 99.17% | 98.87% | 98.80% | 99.93% | 98.57% |
| Hard Voting | 98.61% | 98.28% | 98.90% | N/A | 98.43% | |
| Ensemble of Approach 5 | Soft Voting | 98.00% | 99.33% | 95.93% | 99.90% | 96.63% |
| Hard Voting | 98.75% | 98.41% | 99.07% | N/A | 98.59% |
| Methods | False-Negative of COVID-19 |
|---|---|
| Ensemble of CNNs with normal loss | 14 |
| Ensemble of CNNs with weighted loss | 8 |
| Ensemble of CNNs with image augmentation | 8 |
| Ensemble of CNNs with undersampling | 7 |
| Ensemble of CNNs with oversampling | 9 |
| Ensemble of CNNs with hybrid sampling | 3 |
| Authors | Classification Methods | Classification Strategy | Data | Performance |
|---|---|---|---|---|
| Hemdan et al. [10] | COVIDX-Net using six CNNs | Binary classification of normal and COVID-19 | COVID-19: 25 Normal: 25 | F-Measure 81% (normal) 91% (COVID-19) |
| Sahlol et al. [11] | Hybrid of InceptionNet as a feature extractor and Marine Predator as a feature selection method | Binary classification between COVID-19 and Normal | COVID-19: 200 Normal: 1675 | Accuracy 98.7% |
| COVID-19: 219 Normal: 1341 | Accuracy 99.6% | |||
| Alazab et al. [12] | VGG-16 based classification with and without data augmentation | Binary classification of healthy and COVID-19 | COVID-19: 70 Healthy: 28 (original dataset) | F-measure 95% |
| COVID-19: 500 Healthy: 500 (augmented) | F-measure 99% | |||
| Duran-Lopez et al. [13] | A customized deep learning model | Binary classification of non-findings and COVID-19 | COVID-19: 2589 Normal: 4337 | Accuracy 94.43% |
| Khasawneh et al. [14] | 2D CNN, VGG16, Mobile Nets | Binary classification of normal and COVID-19 | COVID-19: 1210 Normal: 1583 | Accuracy 98.7% |
| Wang et al. [15] | A custom deep learning algorithm | Multi-classification of normal, COVID-19 and pneumonia | COVID-19: 266 Normal: 8066 Pneumonia: 5526 | Sensitivity 80% |
| Brunese et al. [16] | VGG16 based transfer learning | Two staged binary classification:
| (i) Healthy: 3520 lung diseases: 3003 | Accuracy 96% |
| (ii) COVID-19: 250 lung diseases: 2753 | Accuracy 98% | |||
| Ahmed et al. [17] | A custom deep learning model with multiple feature extraction layers | Multi-classification of normal, COVID-19 and pneumonia | COVID-19: 238 Normal: 8851 Pneumonia: 6045 | Accuracy 97.48% |
| Yoo et al. [18] | Dee learning based decision tree classifier | Step by step binary classification:
| Normal: 558 Abnormal: 558 | Accuracy 98% |
| TB: 492 non TB: 492 | Accuracy 80% | |||
| COVID-19: 142 TB: 142 | Accuracy 95% | |||
| Ozturk et al. [19] | Modified Darknet-19 model | Binary classification of non-findings and COVID-19 | COVID-19: 125 Normal: 500 | Accuracy 98.08% |
| Multi classification of normal, COVID-19 and pneumonia | COVID-19: 125 Normal: 500 Pneumonia: 500 | Accuracy 87.02% | ||
| Ben Jabra et al. [20] | 16 state-of-the-art CNNs model and ensemble of those models using voting | Multi-classification of normal, COVID-19, and pneumonia | COVID-19: 237 Normal: 1338 Pneumonia: 1336 | Accuracy 99.31 % |
| Shelke et al. [21] | VGG-19, DenseNet-161 and ResNet-18 | Three-staged classification
| Normal: 526 Pneumonia: 605 TB: 382 | Accuracy 95.9% |
| COVID-19: 735 Pneumonia: 650 | Accuracy 98.9% | |||
| Mild: 80 Medium: 80 Severe: 80 | Accuracy 76% | |||
| Oh et al. [22] | A patch-based ResNet18 model | Multi-classification of normal, COVID-19, pneumonia and TB | COVID-19: 180 Normal: 191 Pneumonia: 74 TB: 57 | Sensitivity 92.5% |
| Rajaraman et al. [23] | Ensemble of iteratively pruned deep learning models | Multi-classification of normal, COVID-19 and pneumonia | COVID-19: 313 Normal: 7595 Pneumonia: 8792 | Accuracy 99.01% |
| Bridge et al. [24] | InceptionNet with GEV activation function | Binary classification of non-findings and COVID-19 | COVID-19: 129 Normal: 62,267 Pneumonia: 5689 | AUC 82% |
| Multi classification of normal, pneumonia and COVID-19 | AUC 73.1% | |||
| Nishio et al. [25] | Transfer learning of VGG-16 with a combination of data augmentation methods | Multi-classification of healthy, COVID-19 pneumonia, and non-COVID-19 pneumonia | COVID-19: 215 Healthy: 500 Pneumonia: 533 | Accuracy 83.6% |
| Our proposed study |
| Multi-classification of normal, COVID-19, and pneumonia | COVID-19: 3616 Normal: 10,192 Pneumonia: 1345 | Accuracy 99.23% Sensitivity 99.27% AUC 99.97 F-measure 98.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Win, K.Y.; Maneerat, N.; Sreng, S.; Hamamoto, K. Ensemble Deep Learning for the Detection of COVID-19 in Unbalanced Chest X-ray Dataset. Appl. Sci. 2021, 11, 10528. https://doi.org/10.3390/app112210528
Win KY, Maneerat N, Sreng S, Hamamoto K. Ensemble Deep Learning for the Detection of COVID-19 in Unbalanced Chest X-ray Dataset. Applied Sciences. 2021; 11(22):10528. https://doi.org/10.3390/app112210528
Chicago/Turabian StyleWin, Khin Yadanar, Noppadol Maneerat, Syna Sreng, and Kazuhiko Hamamoto. 2021. "Ensemble Deep Learning for the Detection of COVID-19 in Unbalanced Chest X-ray Dataset" Applied Sciences 11, no. 22: 10528. https://doi.org/10.3390/app112210528
APA StyleWin, K. Y., Maneerat, N., Sreng, S., & Hamamoto, K. (2021). Ensemble Deep Learning for the Detection of COVID-19 in Unbalanced Chest X-ray Dataset. Applied Sciences, 11(22), 10528. https://doi.org/10.3390/app112210528








