Zero-Emission of Palm Oil Mill Effluent Final Discharge Promoted Bacterial Biodiversity Rebound in the Receiving Water System
Abstract
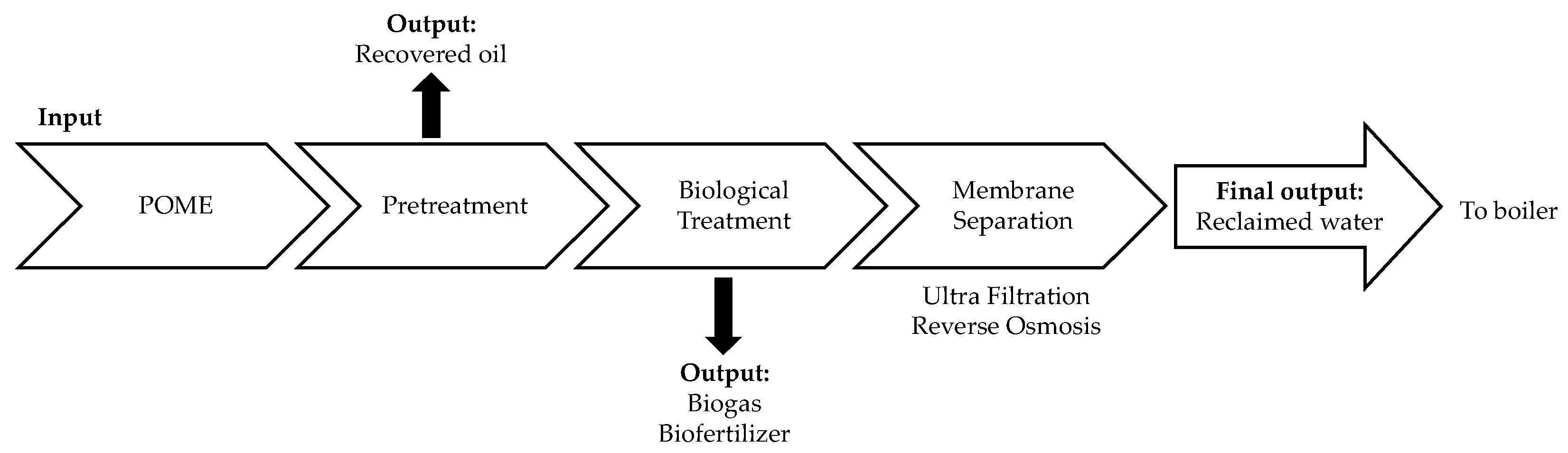
1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sample Collection
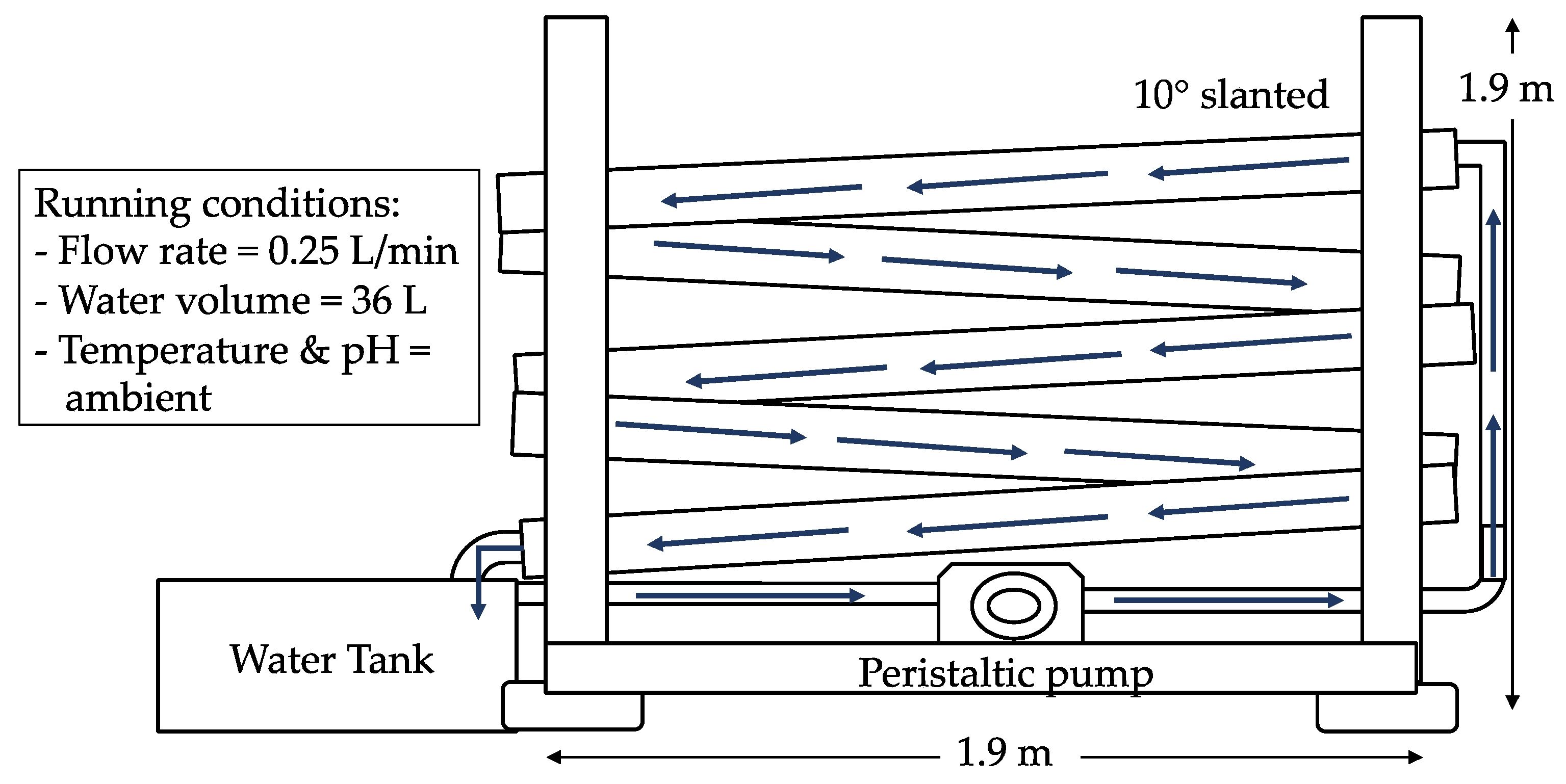
2.2. Construction of the Artificial River Water System
2.3. Water Feeding Exercise
2.4. Physicochemical Analyses
2.5. Nucleic Acid Double Staining Assay Based on Flow Cytometry
2.6. Nutrient Analyses
2.7. DNA Extraction
2.8. High Throughput 16S rRNA Sequencing
2.9. High-Throughput Data Processing and Analysis of Bacterial Community Composition
2.10. Statistical Analysis
3. Results and Discussion
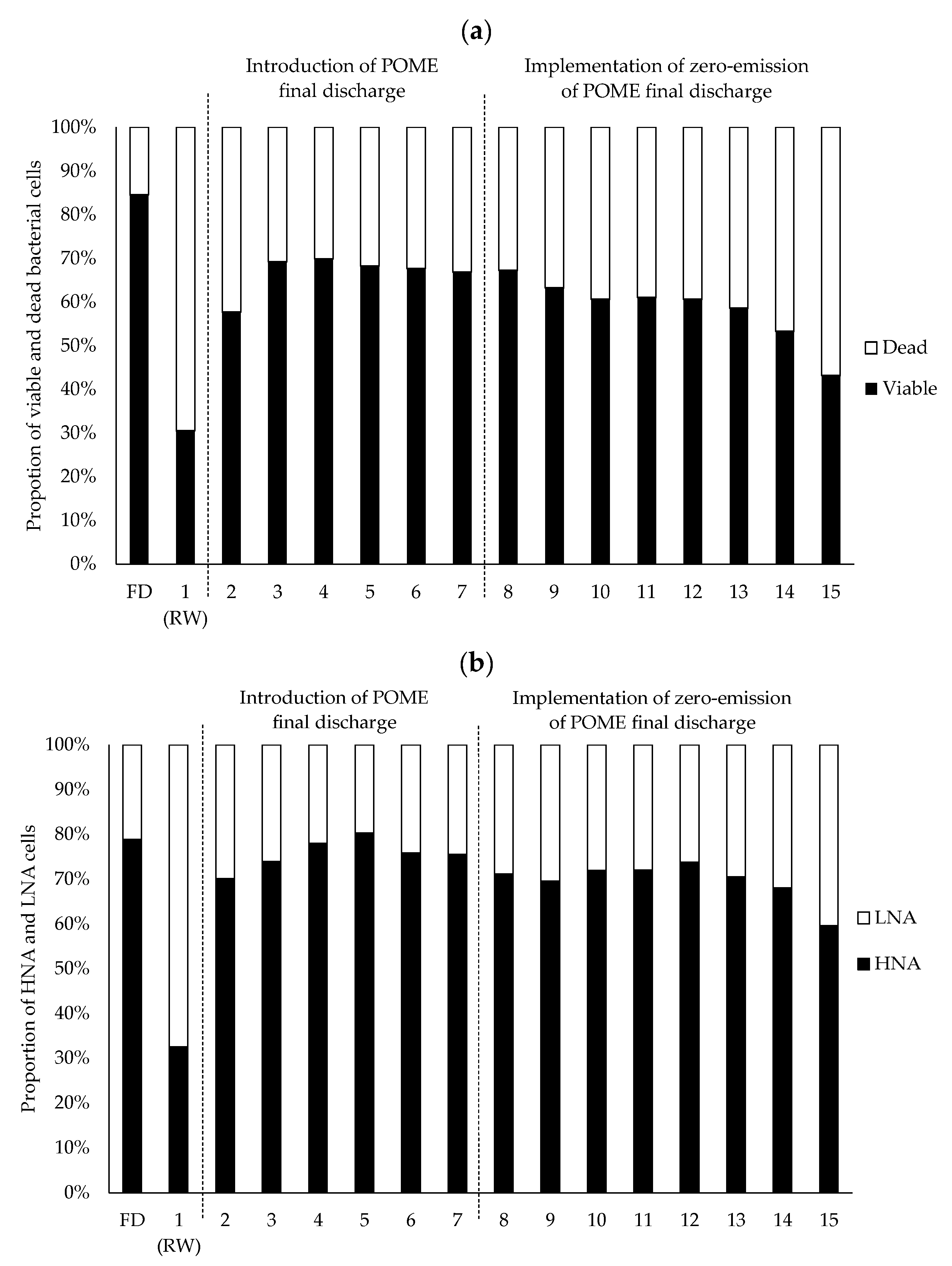
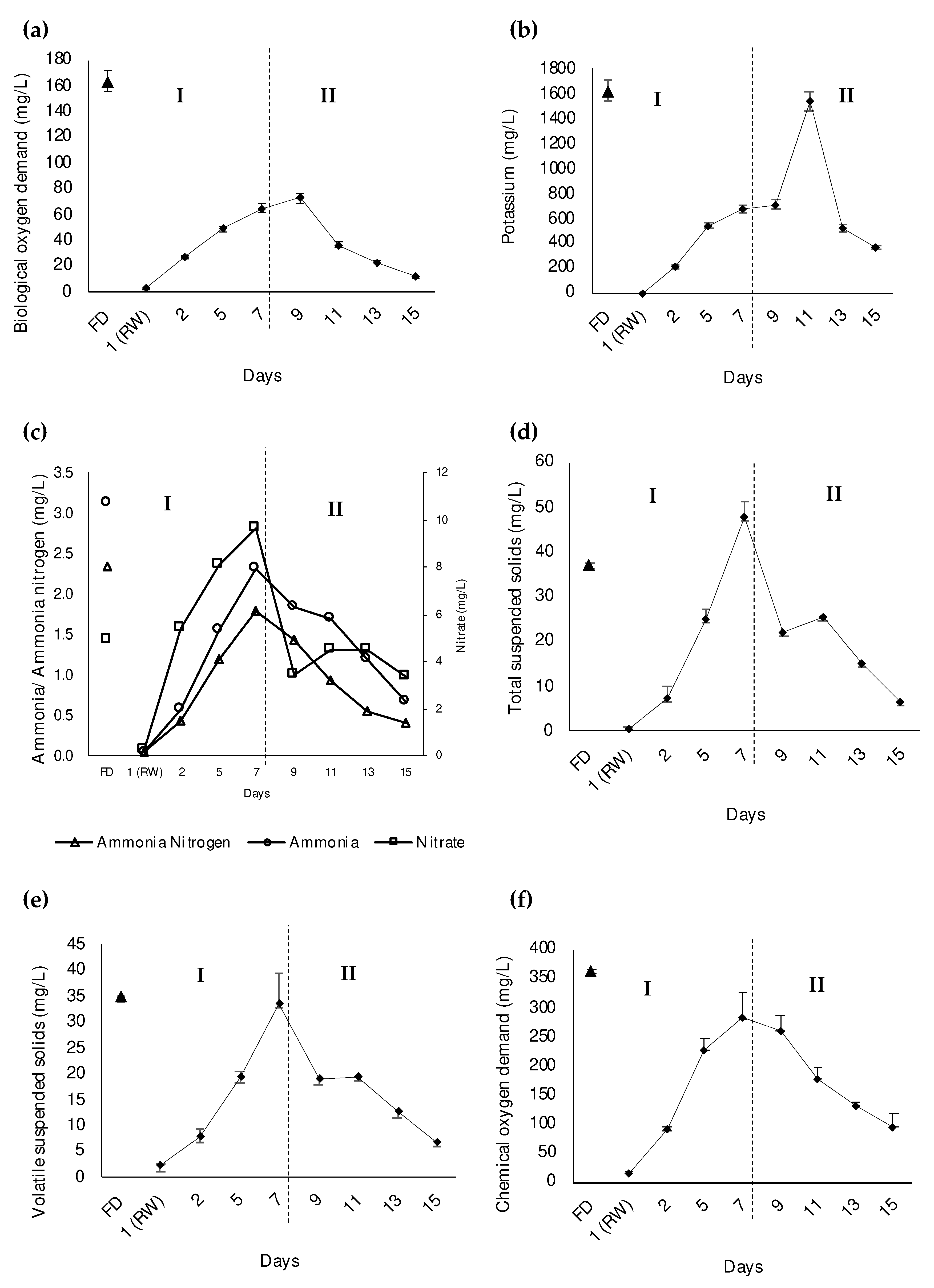
3.1. Improvement in the Functional Status of Bacterial Cells due to Environmental Factors in the Zero-Emission System of POME Final Discharge
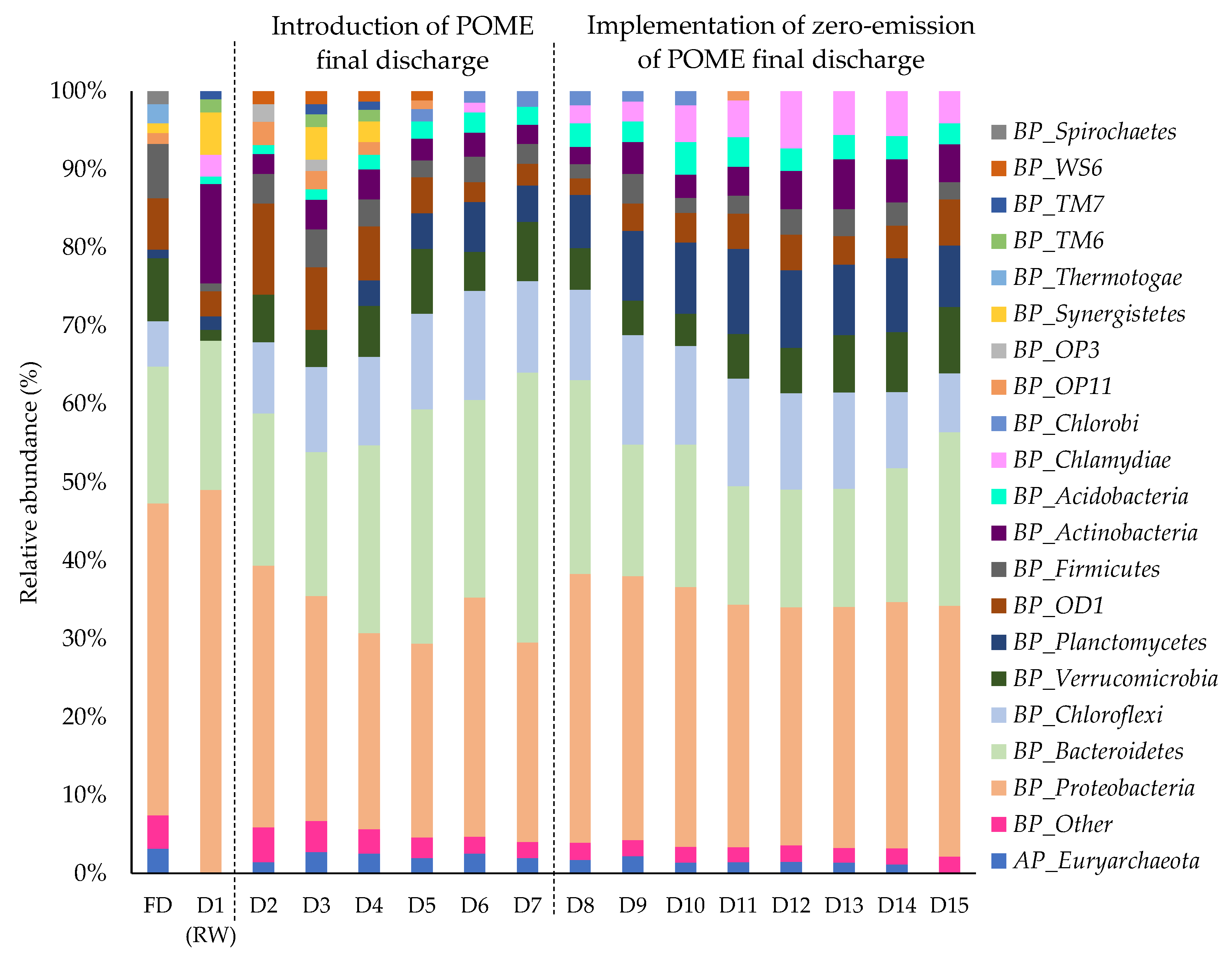
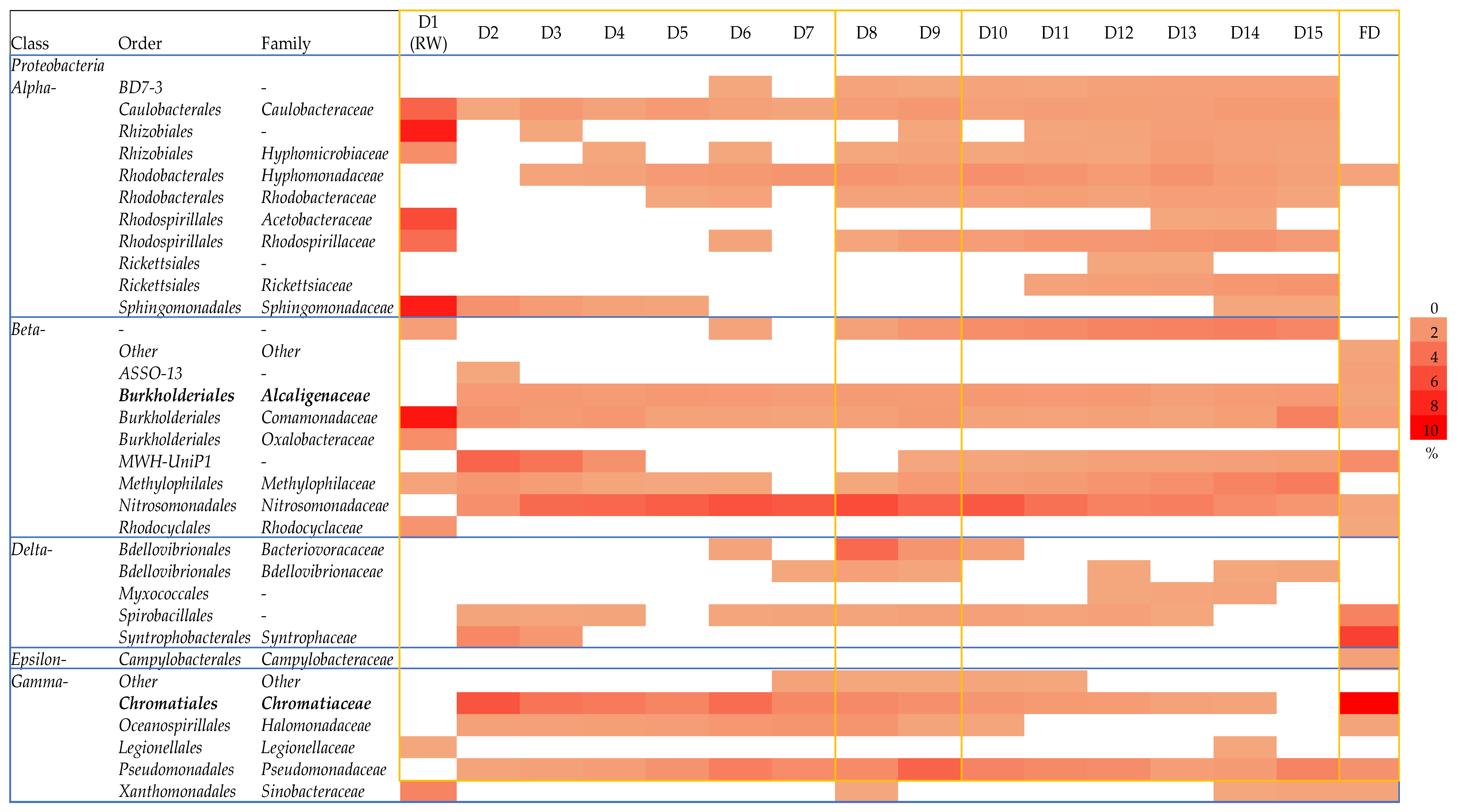
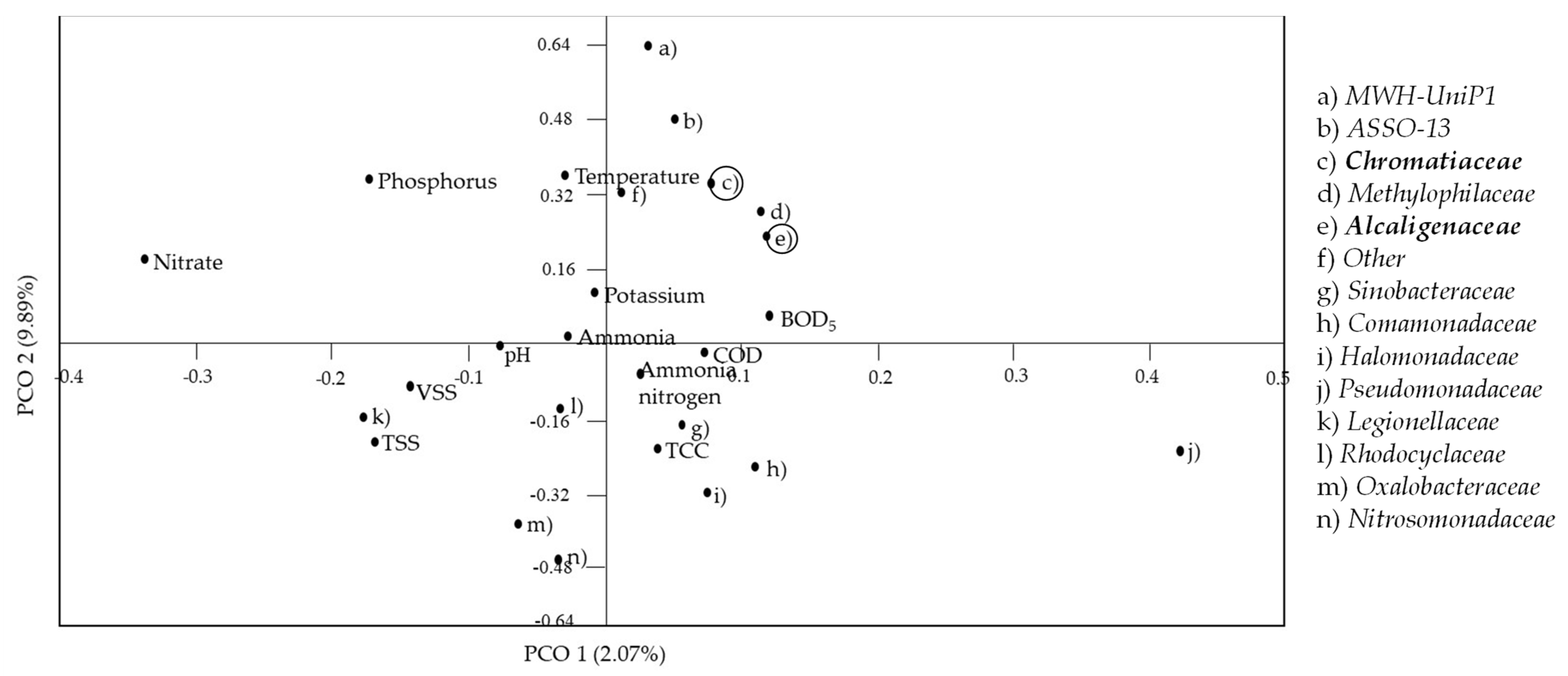
3.2. Effect of Zero-Emission of POME Final Discharge on the Composition of the Bacterial Community in the Receiving Water System
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Umana, U.S.; Ebong, M.S.; Godwin, E.O. Biomass production from oil palm and its value chain. J. Hum. Earth Future 2020, 1, 30–38. [Google Scholar]
- Ibrahim, I.; Hassan, M.A.; Abd-Aziz, S.; Shirai, Y.; Andou, Y.; Othman, M.R.; Mohd Ali, A.A.; Zakaria, M.R. Reduction of residual pollutants from biologically treated palm oil mill effluent final discharge by steam activated bioadsorbent from oil palm biomass. J. Clean. Prod. 2017, 141, 122–127. [Google Scholar] [CrossRef]
- Tabassum, S.; Zhang, Y.; Zhang, Z. An integrated method for palm oil mill effluent (POME) treatment for achieving zero liquid discharge—A pilot study. J. Clean. Prod. 2015, 95, 148–155. [Google Scholar] [CrossRef]
- Loh, S.K.; Lai, M.E.; Ngatiman, M.; Lim, W.S.; Choo, Y.M.; Zhang, Z.; Salimon, J. Zero discharge treatment technology of palm oil mill effluent. J. Oil Palm Res. 2013, 25, 273–281. [Google Scholar]
- Othman, M.R.; Hassan, M.A.; Shirai, Y.; Baharuddin, A.S.; Ali, A.A.M.; Idris, J. Treatment of effluents from palm oil mill process to achieve river water quality for reuse as recycled water in a zero-emission system. J. Clean. Prod. 2017, 67, 58–61. [Google Scholar] [CrossRef]
- Lam, M.K.; Lee, K.T. Renewable and sustainable bioenergies production from palm oil mill effluent (POME): Win–win strategies toward better environmental protection. Biotechnol. Adv. 2011, 29, 124–141. [Google Scholar] [CrossRef]
- Salihu, A.; Alam, M.Z. Palm oil mill effluent: A waste or a raw material? J. Appl. Sci. Res. 2012, 8, 466–473. [Google Scholar]
- Liew, W.L.; Kassim, M.A.; Muda, K.; Loh, S.K.; Affam, A.C. Conventional methods and emerging wastewater polishing technologies for palm oil mill effluent treatment: A review. J. Environ. Manag. 2015, 149, 222–235. [Google Scholar] [CrossRef]
- Tong, T.; Elimelech, M. The global rise of zero liquid discharge for wastewater management: Drivers, Technologies, and Future directions. Environ. Sci. Technol. 2016, 50, 6846–6855. [Google Scholar] [CrossRef]
- Wassen, M.J.; Barendregt, A.; Palczynski, A.; de Smidt, J.T.; de Mars, H. Hydro-ecological analysis of the Biebrza mire (Poland). Wetl. Ecol. Manag. 1992, 2, 119–134. [Google Scholar] [CrossRef]
- Quagraine, E.K.; Duncan, B.; Chi, M.; Lothian, A. 2 decades constructed wetland experience in treating municipal effluent for power plant cooling at the Shand Power Station, SaskPower Part III: Annual treatment performance on PO43- -P, TP, volatile and total suspended solids, inorganic constituents, and bacteria. J. Water Sustain. 2017, 7, 113. [Google Scholar] [CrossRef]
- Kandel, P.P.; Pasternak, Z.; Van Rijn, J.; Nahum, O.; Jurkevitch, E. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol. Ecol. 2014, 89, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Sharuddin, S.S.; Ramli, N.; Hassan, M.A.; Mustapha, N.A.; Amran, A.; Mohd-Nor, D.; Sakai, K.; Tashiro, Y.; Shirai, Y.; Maeda, T. Bacterial community shift revealed Chromatiaceae and Alcaligenaceae as potential bioindicators in the receiving river due to palm oil mill effluent final discharge. Ecol. Indic. 2017, 82, 526–529. [Google Scholar] [CrossRef]
- Mohd-Nor, D.; Ramli, N.; Sharuddin, S.S.; Hassan, M.A.; Mustapha, N.A.; Amran, A.; Sakai, K.; Shirai, Y.; Maeda, T. Alcaligenaceae and Chromatiaceae as reliable bioindicators present in palm oil mill effluent final discharge treated by different biotreatment processes. Ecol. Indic. 2018, 95, 468–473. [Google Scholar] [CrossRef]
- Zolkefli, N.; Ramli, N.; Mohamad-Zainal, N.S.L.; Mustapha, N.A.; Yusoff, M.Z.M.; Hassan, M.A.; Maeda, T. Alcaligenaceae and Chromatiaceae as pollution bacterial bioindicators in palm oil mill effluent (POME) final discharge polluted rivers. Ecol. Indic. 2020, 111, 106048. [Google Scholar] [CrossRef]
- Boeije, G.M.; Schowanek, D.R.; Vanrolleghem, P.A. Incorporation of biofilm activity in river biodegradation modeling: A case study for linear alkylbenzene sulphonate (LAS). Water Res. 2000, 34, 1479–1486. [Google Scholar] [CrossRef]
- Hodoki, Y. Direct and indirect effects of solar ultraviolet radiation on attached bacteria and algae in lotic systems. Hydrobiologia 2005, 549, 259–266. [Google Scholar] [CrossRef]
- Amneera, W.A.; Najib, N.W.A.Z.; Mohd Yusof, S.R.; Ragunathan, S. Water quality index of Perlis River, Malaysia. Int. J. Civ. Environ. 2013, 13, 1–6. [Google Scholar]
- American Public Health Association. APHA: Standard Methods Examination of Water Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Barbesti, S.; Citterio, S.; Labra, M.; Baroni, M.D.; Neri, M.G.; Sgorbati, S. Two and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry 2000, 40, 214–218. [Google Scholar] [CrossRef]
- Sharuddin, S.S.; Ramli, N.; Mohd-Nor, D.; Hassan, M.A.; Maeda, T.; Shirai, Y.; Sakai, K.; Tashiro, Y. Shift of low to high nucleic acid bacteria as a potential bioindicator for the screening of anthropogenic effects in a receiving river due to palm oil mill effluent final discharge. Ecol. Indic. 2018, 85, 79–84. [Google Scholar] [CrossRef]
- Foladori, P.; Laura, B.; Gianni, A.; Giuliano, Z. Effects of sonication on bacteria viability in wastewater treatment plants evaluated by flow cytometry-Fecal indicators, wastewater and activated sludge. Water Res. 2007, 41, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Ding, S.; Gai, S.; Xiao, R.; Wu, Y.; Geng, B.; Chu, W. Effect of oxoanions on oxidant decay, bromate and brominated disinfection by-product formation during chlorination in the presence of copper corrosion products. Water Res. 2019, 166, 115087. [Google Scholar] [CrossRef]
- Zainal, S.F.F.S.; Aziz, H.A. Potential of tin (IV) chloride for treatment in Alor Pongsu as stabilized landfill leachate. AIP Conf. Proc. 2017, 1892, 040003. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Mustapha, N.A.; Hu, A.; Yu, C.P.; Sharuddin, S.S.; Ramli, N.; Shirai, Y.; Maeda, T. Seeking key microorganisms for enhancing methane production in anaerobic digestion of waste sewage sludge. Appl. Microbiol. Biotechnol. 2018, 102, 5323–5334. [Google Scholar] [CrossRef]
- Hildebrand, F.; Tadeo, R.; Voigt, A.Y.; Bork, P.; Raes, J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome 2014, 2, 30. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1996, 53, 325–338. [Google Scholar] [CrossRef]
- Hu, W.; Murata, K.; Toyonaga, S.; Zhang, D. Bacterial abundance and viability in rainwater associated with cyclones, stationary fronts and typhoons in southwestern Japan. Atmos. Environ. 2017, 167, 104–115. [Google Scholar] [CrossRef]
- Tamaki, V.; Mercier, H. Effects of different ammoniacal nitrogen sources on N-metabolism of the atmospheric bromeliad Tillandsia pohliana Mez. Rev. Bras. De Bot. 2016, 24, 407–413. [Google Scholar] [CrossRef][Green Version]
- Lebaron, P.; Servais, P.; Agogué, H.; Courties, C.; Joux, F. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 2001, 67, 1775–1782. [Google Scholar] [CrossRef]
- Liu, G.; Bakker, G.L.; Li, S.; Vreeburg, J.H.G.; Verberk, J.Q.J.C.; Medema, G.J.; Liu, W.T.; Van Dijk, J.C. Pyrosequencing reveals bacterial communities in unchlorinated drinking water distribution system: An integral study of bulk water, suspended solids, loose deposits, and pipe wall biofilm. Environ. Sci. Technol. 2014, 48, 5467–5476. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Wen, D.; Shen, J.; and Wang, J. Zero discharge process for dyeing wastewater treatment. J. Water Process. Eng. 2016, 11, 98–103. [Google Scholar] [CrossRef]
- Das, S. Cleaning of the Ganga. J. Geol. Soc. India 2011, 78, 124–130. [Google Scholar] [CrossRef]
- Chidamba, L.; Korsten, L. Pyrosequencing analysis of roof-harvested rainwater and river water used for domestic purposes in Luthengele village in the Eastern Cape Province of South Africa. Environ. Monit. Assess. 2015, 187, 1–17. [Google Scholar] [CrossRef][Green Version]
- Salam, M.; Varma, A. Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electron. J. Biotechnol. 2019, 41, 72–80. [Google Scholar] [CrossRef]
- Cho, B.C.; Jang, G.I. Active and diverse rainwater bacteria collected at an inland site in spring and summer 2011. Atmos. Environ. 2014, 94, 409–416. [Google Scholar] [CrossRef]
- Psenner, R.; Alfreider, A.; Schwarz, A. Aquatic microbial ecology: Water desert, microcosm, ecosystem. What’s next? Int. Rev. Hydrobiol. 2008, 93, 606–623. [Google Scholar] [CrossRef]
- Hu, W.; Niu, H.; Murata, K.; Wu, Z.; Hu, M.; Kojima, T.; Zhang, D. Bacteria in atmospheric waters: Detection, characteristics and implications. Atmos. Environ. 2018, 179, 201–221. [Google Scholar] [CrossRef]
- Hiraoka, S.; Miyahara, M.; Fujii, K.; Machiyama, A.; Iwasaki, W. Seasonal analysis of microbial communities in precipitation in the greater Tokyo area, Japan. Front. Microbiol. 2017, 8, 1506. [Google Scholar] [CrossRef] [PubMed]
- Abu Bakar, S.N.H.; Abu Hasan, H.; Mohammad, A.W.; Abdullah, S.R.S.; Ngteni, R.; Yusof, K.M.M. Performance of a laboratory-scale moving bed biofilm reactor (MBBR) and its microbial diversity in palm oil mill effluent (POME) treatment. Process. Saf. Environ. Prot. 2020, 142, 325–335. [Google Scholar] [CrossRef]
- Mohd Udaiyappan, A.F.; Hasan, H.A.; Takriff, M.S.; Abdullah, S.R.S.; Maeda, T.; Mustapha, N.A.; Mohd Yasin, N.; Nazashida Mohd Hakimi, N.I. Microalgae-bacteria interaction in palm oil mill effluent treatment. J. Water Process. Eng. 2020, 35, 101203. [Google Scholar] [CrossRef]
- Šimek, K.; Horňák, K.; Jezbera, J.; Mašín, M.; Nedoma, J.; Gasol, J.M.; Schauer, M. Influence of top-down and bottom-up manipulations on the R-BT065 subcluster of β-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl. Environ. Microbiol. 2005, 71, 2381–2390. [Google Scholar] [CrossRef] [PubMed]
- Lemke, M.J.; Lienau, E.K.; Rothe, J.; Pagioro, T.A.; Rosenfeld, J.; Desalle, R. Description of freshwater bacterial assemblages from the upper Paraná river floodpulse system, Brazil. Microb. Ecol. 2009, 57, 94–103. [Google Scholar] [CrossRef]
- Elifantz, H.; Malmstrom, R.R.; Cottrell, M.T.; Kirchman, D.L. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware Estuary. Appl. Environ. Microbiol. 2005, 71, 7799–7805. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A Guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Res. 2011, 75, 14–49. [Google Scholar] [CrossRef]
- Amato, P.; Joly, M.; Besaury, L.; Oudart, A.; Taib, N.; Moné, A.I.; Deguillaume, L.; Delort, A.M.; Debroas, D. Active microorganisms thrive among extremely diverse communities in cloud water. PLoS ONE 2017, 12, e0182869. [Google Scholar] [CrossRef]
- Potočnjak, M.; Široka, M.; Rebić, D.; Gobin, I. The survival of Legionella in rainwater. Int. J. Sanit. Eng. Res. 2012, 6, 31–36. [Google Scholar]
- Caicedo, C.; Rosenwinkel, K.H.; Exner, M.; Verstraete, W.; Suchenwirth, R.; Hartemann, P.; Nogueira, R. Legionella occurrence in municipal and industrial wastewater treatment plants and risks of reclaimed wastewater reuse. Water Res. 2019, 149, 21–34. [Google Scholar] [CrossRef]
- Steinert, M.; Hentschel, U.; Hacker, J. Legionella pneumophila: An aquatic microbe goes astray. FEMS Microbiol. Rev. 2002, 26, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Karwautz, C.; Lueders, T. Impact of hydraulic well restoration on native bacterial communities in drinking water wells. Microbes Environ. 2014, 29, 363–369. [Google Scholar] [CrossRef]
- Webster, T.M.; Fierer, N. Microbial dynamics of biosand filters and contributions of the microbial food web to effective treatment of wastewater-impacted water sources. Appl. Environ. Microbiol. 2019, 85, e01142-19. [Google Scholar] [CrossRef] [PubMed]
- Liao, R.; Shen, K.; Li, A.M.; Shi, P.; Li, Y.; Shi, Q.; Wang, Z. High-nitrate wastewater treatment in an expanded granular sludge bed reactor and microbial diversity using 454 pyrosequencing analysis. Bioresour. Technol. 2013, 134, 190–197. [Google Scholar] [CrossRef]
- Ahmad, M.N.; Ali, S.R.A.; Hassan, M.A. Microbial succession in co-composting of chipped-ground oil palm frond and palm oil mill effluent. J. Oil Palm Res. 2016, 28, 191–197. [Google Scholar] [CrossRef][Green Version]
- Felföldi, T.; Székely, A.J.; Gorál, R.; Barkács, K.; Scheirich, G.; András, J.; Rácz, A.; Márialigeti, K. Polyphasic bacterial community analysis of an aerobic activated sludge removing phenols and thiocyanate from coke plant effluent. Bioresour. Technol. 2010, 101, 3406–3414. [Google Scholar] [CrossRef]
- Tsouko, E.; Alexandri, M.; Fernandes, K.V.; Freire, D.M.G.; Mallouchos, A.; Koutinas, A.A. Extraction of phenolic compounds from palm oil processing residues and their application as antioxidants. Food Technol. Biotechnol. 2019, 57, 29. [Google Scholar] [CrossRef] [PubMed]
- Anku, W.W.; Mamo, M.A.; Govender, P.P. Phenolic Compounds in Water: Sources, Reactivity, Toxicity and Treatment Methods; InTech Open: London, UK, 2017; pp. 419–443. [Google Scholar] [CrossRef]
- Benner, R.; Kaiser, K. Biological and photochemical transformations of amino acids and lignin phenols in riverine dissolved organic matter. Biogeochemistry 2011, 102, 209–222. [Google Scholar] [CrossRef]
- Price, J.R.; Ledford, S.H.; Ryan, M.O.; Toran, L.; Sales, C.M. Wastewater treatment plant effluent introduces recoverable shifts in microbial community composition in receiving streams. Sci. Total. Environ. 2018, 613–614, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamad-Zainal, N.S.L.; Ramli, N.; Zolkefli, N.; Jamari, N.A.; Mustapha, N.A.; Hassan, M.A.; Maeda, T. Zero-Emission of Palm Oil Mill Effluent Final Discharge Promoted Bacterial Biodiversity Rebound in the Receiving Water System. Appl. Sci. 2021, 11, 10814. https://doi.org/10.3390/app112210814
Mohamad-Zainal NSL, Ramli N, Zolkefli N, Jamari NA, Mustapha NA, Hassan MA, Maeda T. Zero-Emission of Palm Oil Mill Effluent Final Discharge Promoted Bacterial Biodiversity Rebound in the Receiving Water System. Applied Sciences. 2021; 11(22):10814. https://doi.org/10.3390/app112210814
Chicago/Turabian StyleMohamad-Zainal, Noor Shaidatul Lyana, Norhayati Ramli, Nurhasliza Zolkefli, Nur Azyani Jamari, Nurul Asyifah Mustapha, Mohd Ali Hassan, and Toshinari Maeda. 2021. "Zero-Emission of Palm Oil Mill Effluent Final Discharge Promoted Bacterial Biodiversity Rebound in the Receiving Water System" Applied Sciences 11, no. 22: 10814. https://doi.org/10.3390/app112210814
APA StyleMohamad-Zainal, N. S. L., Ramli, N., Zolkefli, N., Jamari, N. A., Mustapha, N. A., Hassan, M. A., & Maeda, T. (2021). Zero-Emission of Palm Oil Mill Effluent Final Discharge Promoted Bacterial Biodiversity Rebound in the Receiving Water System. Applied Sciences, 11(22), 10814. https://doi.org/10.3390/app112210814






