Dual Head and Dual Attention in Deep Learning for End-to-End EEG Motor Imagery Classification
Abstract
:1. Introduction
- -
- What’s the impact on the proposed attention strategy through end-to-end learning?
- -
- How to quantify or visualize the interpretability of deep DAC-Net?
- -
- How to accelerate the learning manifestation and effectiveness from the MI raw signals via DAC-Net for BMI recognition applications?
2. Data
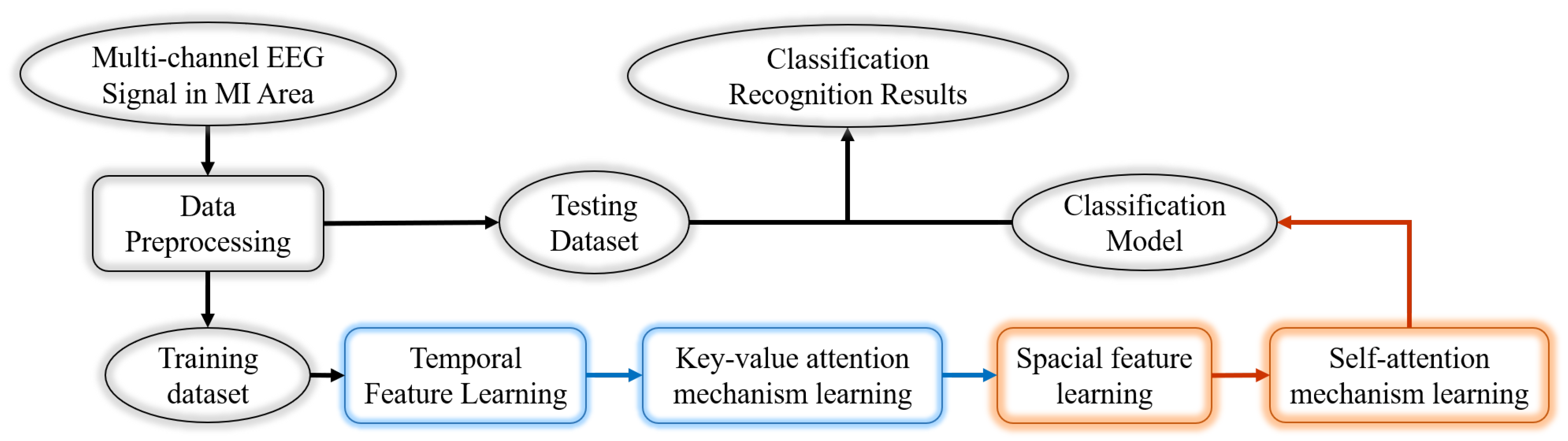
3. Method
3.1. Input Data
3.2. Attention Module
- The module is as simple and efficient as possible, relying on the combined operation of convolution, pooling, normalization, and anti-overfitting.
- The module has robust and nonlinear learning capabilities by enabling 1D CNNs in the temporal dimension and 2D CNNs in the spatial dimension.
- This module conducts attention learning in the temporal dimension firstly, which helps to improve the subsequent spatial dimension learning (see Section 5.1).
3.2.1. Key-Value Attention Mechanism
3.2.2. Spatial Attention
3.3. DHDANet
- TSA module: As for the characteristics of ERD/ERS phenomenon, the input heads perform three sets of time-domain wave amplitude feature learning. Each set of time-domain training includes one-dimensional convolution, maximum pooling, data normalization, and dropout operation. Then, the Key-value attention learning is performed, and the feature values of the dual input head correspond to the key and value in the attention mechanism. The three sets of time-domain feature extraction parameters are the same. It mainly performs neighborhood filtering. The parameter set and processing process of the network are shown in Figure 5. First, one-dimensional convolution is performed to extract different feature maps with a core of 32 and a time interval of 0.128s (32/250) because the down-sampling rate is 250 Hz. Then, MaxPool 1D continues. At this time, the learning processes volatility characteristic value at a time interval of 0.25s. Data normalization and dropout are conducted to prevent overflow and overfitting [41,42]. After three sets of time-domain features are extracted, each feature map covers 1s of EEG waveform feature information, and from the analysis in Section 3.1, the time period for an ERD/ERS peak or trough is generally between 500 ms to 1s [43,44]. It can be seen that, before entering the key-value attention calculation, a peak or trough of ERD/ERS exists in the two input feature maps.
- SSA module: After extracting the feature value of time domain, this module focuses on extracting the spectral features in spatial domains of the left and right hemisphere. For feature extraction in the spatial domain, the amplitude information of the ERD/ERS phenomenon cannot be extracted for convolution calculation that is too short or too long. In particular, if the triple feature extraction is performed on the input data of the network initially, the convolution and maximum pooling are used. The further reduction of computing will result in the loss of valuable information, which cannot be used for action recognition. To avoid this problem, the SSA module in this chapter first converts the 1D feature values output by the TSA module into a 2D tensor with a 4-column structure, Conv2D = (2,2), so that the symmetrical lead signals of the two brain regions can be convolved to calculate the weight of the feature map. Before and after the self-attention calculation, convolution and dropout calculations are added. This former is to obtain dynamic weights based on the feature information to prepare for self-attention calculations. In addition, the latter is to compress feature values to facilitate the calculation of the next module, as shown in Figure 6.
- Feature classification learning module: This module is to classify the temporal and spatial features learned in the training network and build a classifier. This module uses two fully connected layers, and the basic operation of fully connected is the matrix vector product. The first completely connected layer of the module aims to weight the probability of the existence of each neuron feature. After common machine learning operations with unique data and over-fitting, the second fully connected layer classifies the feature weights output by the previous connected layer absolutely.
4. Experiments and Results
4.1. Data PreProcessing
4.2. Result
- Validation loss rate must be lower than the previous iteration before this model is saved.
- If the test loss rate of the trained model does not decrease within 30 iterations, the training is automatically stopped.
5. Analysis and Discussion
5.1. Why Use the Attention Mechanism Algorithm?
5.2. Feature Works
6. Conclusions
- To learn the ERD/ERS features in the time domain, double-input EEG data are used. Meanwhile, the features are handled by the key value attention mechanism. Experimental results confirm that the key value attention mechanism is beneficial for both the recognition of motor imagery in the time domain and the follow-up learning of spatial EEG characteristics.
- Clever conversion methods are used to transform time domain features to spatial domain features. In addition, the EEG collection point information input into the network is combined into a two-dimensional matrix according to front-back and left-right symmetry in the brain area, to retain characteristics of the left and right brain activities when handling a three-dimensional matrix conversion.
- In the spatial feature learning module, a reasonable nonlinear computer system is constructed to extract features. In addition, a self-attention mechanism algorithm is introduced to further strengthen the features of motor imagery in the spatial dimension, see the comparison of the before and after feature maps of the key-value attention calculation in b and c in Figure 12.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagai, H.; Tanaka, T. Action Observation of Own Hand Movement Enhances Event-Related Desynchronization. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1407–1415. [Google Scholar] [CrossRef]
- Crk, I.; Kluthe, T.; Stefik, A. Understanding Programming Expertise: An Empirical Study of Phasic Brain Wave Changes. ACM Trans. Comput.-Hum. Interact. 2015, 23, 2–29. [Google Scholar] [CrossRef]
- Tariq, M.; Trivailo, P.M.; Simic, M. Detection of knee motor imagery by Mu ERD/ERS quantification for BCI based neurorehabilitation applications. In Proceedings of the 11th Asian Control Conference (ASCC), Gold Coast, QLD, Australia, 17–20 December 2017. [Google Scholar]
- Tang, Z.; Sun, S.; Zhang, S.; Chen, Y.; Li, C.; Chen, S. A brain–machine Interface Based on ERD/ERS for an Upper-Limb Exoskeleton Control. Sensors 2016, 16, 2050. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Yu, Z.L.; Gu, Z.; Li, Y. Classification of symmetric positive definite matrices based on bilinear isometric Riemannian embedding. Pattern Recognit. 2019, 87, 94–105. [Google Scholar] [CrossRef]
- Xie, X.; Yu, Z.L.; Gu, Z.; Zhang, J.; Cen, L.; Li, Y. Bilinear Regularized Locality Preserving Learning on Riemannian Graph for Motor Imagery BCI. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 1, 698–708. [Google Scholar] [CrossRef]
- Chu, Y.; Zhao, X.; Zou, Y.; Xu, W.; Song, G.; Han, J.; Zhao, Y. Decoding multiclass motor imagery EEG from the same upper limb by combining Riemannian geometry features and partial least squares regression. J. Neural Eng. 2020, 17, 046029. [Google Scholar] [CrossRef]
- Yger, F.; Berar, M.; Lotte, F. Riemannian approaches in Brain-Computer Interfaces: A review. IEEE Trans. Neural. Syst. Rehabil. Eng. 2017, 25, 1753–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, O.Y.; Lee, M.H.; Guan, C.; Lee, S.W. Subject-Independent Brain-Computer Interfaces Based on Deep Convolutional Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2019, 31, 3839–3852. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.; Zheng, Y.; Zhang, H.; Guan, C. Filter Bank Common Spatial Pattern (FBCSP) in Brain-Computer Interface. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 2390–2397. [Google Scholar]
- Ang, K.; Chin, Z.; Wang, C.; Guan, C.; Zhang, H. Filter bank common spatial pattern algorithm on BCI competition IV datasets 2a and 2b. Front. Neurosci. 2012, 6, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanini, P.; Congedo, M.; Jutten, C.; Said, S.; Berthoumieu, Y. Transfer Learning: A Riemannian geometry framework with applications to Brain-Computer Interfaces. IEEE Trans. Biomed. Eng. 2018, 65, 1107–1116. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Ma, Y.; Li, M.; Xiao, Y.; Feng, W.; Wu, X. Time-frequency decomposition-based weighted ensemble learning for motor imagery EEG classification. In Proceedings of the 2021 IEEE International Conference on Real-time Computing and Robotics (RCAR), Xining, China, 15–19 July 2021; pp. 620–625. [Google Scholar]
- TK, M.J.; Sanjay, M. Topography Based Classification for Motor Imagery BCI Using Transfer Learning. In Proceedings of the 2021 International Conference on Communication, Control and Information Sciences (ICCISc), Idukki, India, 16–18 June 2021; Volume 1, pp. 1–5. [Google Scholar]
- Xu, M.; Yao, J.; Zhang, Z.; Li, R.; Zhang, J. Learning EEG Topographical Representation for Classification via Convolutional Neural Network. Pattern Recognit. 2020, 105, 107390. [Google Scholar] [CrossRef]
- Sakhavi, S.; Guan, C.; Yan, S. Learning temporal information for brain-computer interface using convolutional neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 5619–5629. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, H.; Zhu, G.; You, F.; Kuang, S.; Sun, L. A Multi-branch 3D Convolutional Neural Network for EEG-Based Motor Imagery Classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 2164–2177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yao, L.; Chen, K.; Monaghan, J. A Convolutional Recurrent Attention Model for Subject-Independent EEG Signal Analysis. IEEE Signal Process. Lett. 2019, 26, 715–719. [Google Scholar] [CrossRef]
- Majoros, T.; Oniga, S. Comparison of Motor Imagery EEG Classification using Feedforward and Convolutional Neural Network. In Proceedings of the IEEE EUROCON 2021—19th International Conference on Smart Technologies, Lviv, Ukraine, 6–8 July 2021; pp. 25–29. [Google Scholar]
- Wang, P.; Jiang, A.; Liu, X.; Shang, J.; Zhang, L. LSTM-Based EEG Classification in Motor Imagery Tasks. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 2086–2095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Davoodnia, V.; Sepas-Moghaddam, A.; Zhang, Y.; Etemad, A. Classification of Hand Movements from EEG using a Deep Attention-based LSTM Network. IEEE Sens. J. 2019, 20, 3113–3122. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Yao, L.; Chen, K.; Wang, S. Ready for Use: Subject-Independent Movement Intention Recognition via a Convolutional Attention Model. In Proceedings of the 27th ACM International Conference on Information and Knowledge Management (CIKM), Torino, Italy, 22–26 October 2018; pp. 1763–1766. [Google Scholar]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A Compact Convolut- ional Neural Network for EEG-based Brain–Computer Interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Gu, X.; Lo, B. Semi-Supervised Contrastive Learning for Generalizable Motor Imagery EEG Classification. In Proceedings of the 2021 IEEE 17th International Conference on Wearable and Implantable Body Sensor Networks (BSN), Athens, Greece, 27–30 July 2021; pp. 1–4. [Google Scholar]
- Vařeka, L. Evaluation of convolutional neural networks using a large multi-subject P300 dataset. Biomed. Signal Process. Control 2020, 58, 101837. [Google Scholar] [CrossRef] [Green Version]
- Cecotti, H.; Graser, A. Convolutional Neural Networks for P300 Detection with Application to Brain-Computer Interfaces. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 433–445. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, X.; Zhang, W.; Chen, J. Learning Spatial–Spectral–Temporal EEG Features With Recurrent 3D Convolutional Neural Networks for Cross-Task Mental Workload Assessment. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Bashivan, P.; Rish, I.; Yeasin, M.; Codella, N. Learning Representations from EEG With Deep Recurrent-Convolutional Neural Networks. In Proceedings of the International Conference on Learning Representations(ICLR), San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Wanga, L.; Huanga, W.; Yanga, Z.; Zhang, C. Temporal-spatial-frequency depth extraction of brain-computerinterface based on mental tasks. Biomed. Signal Process. Control 2020, 58, 101845. [Google Scholar] [CrossRef]
- Khana, A.; Sung, J.E.; Kang, J.W. Multi-channel fusion convolutional neural network to classify syntactic anomaly from language-related ERP components. Inf. Fusion 2019, 52, 53–61. [Google Scholar] [CrossRef]
- Li, Y.; Guo, L.; Liu, Y.; Liu, J.; Meng, F. A Temporal-Spectral-Based Squeeze and Excitation Feature Fusion Network for Motor Imagery EEG Decoding. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1534–1545. [Google Scholar] [CrossRef]
- Lee, M.H.; Kwon, O.Y.; Kim, Y.J.; Kim, H.K.; Lee, Y.E.; Williamson, J.; Fazli, S.; Lee, S.W. EEG dataset and OpenBMI toolbox for three BCI paradigms: An investigation into BCI illiteracy. GigaScience 2019, 8, giz002. [Google Scholar] [CrossRef]
- Lal, T.N.; Schröder, M.; Hinterberger, T.; Weston, J.; Bogdan, M.; Birbaumer, N.; Schölkopf, B. Support Vector Channel Selection in BCI. IEEE Trans. Biomed. Eng. 2004, 51, 1003–1010. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Miao, Y.; Daly, I.; Zuo, C.; Hu, D.; Cichocki, A. Correlation-based channel selection and regularized feature optimization for MI-based BCI. Neural Netw. 2019, 118, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Bhimraj, K.; Haddad, R.J.; Ahad, M. Optimization of EEG-Based Imaginary Motion Classification Using Majority-Voting. In Proceedings of the SoutheastCon 2017, Concord, NC, USA, 30 March–2 April 2017; pp. 1–5. [Google Scholar]
- Ives-Deliperi, V.L.; Butler, J.T. Relationship between EEG electrode and functional cortex in the international 10 to 20 system. Clin. Neurophysiol. 2018, 35, 504–509. [Google Scholar] [CrossRef]
- Vaswan, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł. Attention Is All You Need. In Advances in Neural Information Processing Systems; Michael, I.J., Yann, L., Sara, A.S., Eds.; MIT Press: Cambridge, MA, USA, 2017; pp. 5998–6008. [Google Scholar]
- Zhang, D.; Yao, L.; Chen, K.; Wang, S.; Haghighi, P.D.; Bengio, Y. A Graph-based Hierarchical Attention Model for Movement Intention Detection from EEG Signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Zhang, W.; Zhang, M.; Song, J.; Sebe, N. Spatio-Temporal Attention Networks for Action Recognition and Detection. IEEE Trans. Multimed. 2020, 22, 2990–3001. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I.; Kang, J. Self-attention graph pooling. arXiv 2019, arXiv:1904.08082. [Google Scholar]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.A. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Pang, G.; Shen, C.; Cao, L.; Hengel, A.V.D. Deep learning for anomaly detection: A review. ACM Comput. Surv. (CSUR) 2021, 54, 38. [Google Scholar] [CrossRef]
- WolpawEmail, J.R.; Boulay, C.B. Brain signals for brain–computer interfaces. In Brain-Computer Interfaces; Springer: Berlin/Heidelberg, Germany, 2009; pp. 29–46. [Google Scholar]
- Pfurtscheller, G.; Neuper, C. Dynamics of sensorimotor oscillations in a motor task. In Brain-Computer Interfaces; Springer: Berlin/Heidelberg, Germany, 2009; pp. 47–64. [Google Scholar]
- Thomas, K.P.; Guan, C.; Tong, L.C.; Prasad, V.A. An Adaptive Filter Bank for Motor Imagery based Brain Computer Interface. In Proceedings of the 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC’08), Vancouver, BC, Canada, 20–25 August 2008; pp. 1104–1107. [Google Scholar]
- Zhang, Y.; Wang, Y.; Jin, J.; Wang, X. Sparse Bayesian Learning for Obtaining Sparsity of EEG Frequency Bands Based Feature Vectors in Motor Imagery Classification. Int. J. Neural Syst. 2017, 27, 537–552. [Google Scholar] [CrossRef]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep Learning with Convolutional Neural Networks for EEG Decoding and Visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mane, R.; Robinson, N.; Vinod, A.P.; Lee, S.W.; Guan, C. A Multi-view CNN with Novel Variance Layer for Motor Imagery Brain Computer Interface. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 2950–2953. [Google Scholar]
- Bai, X.; Yan, C.; Yang, H.; Bai, L.; Zhou, J.; Hancock, E.R. Adaptive Hash Retrieval with Kernel based Similarity. Pattern Recognit. 2018, 75, 136–148. [Google Scholar] [CrossRef] [Green Version]
- Kachenoura, A.; Albera, L.; Senhadji, L.; Comon, P. ICA: A Potential Tool for BCI Systems. IEEE Signal Process. Mag. 2007, 25, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Li, L.; Wang, Z.; Lv, H.; Song, J. The study of cortical lateralization and motor performance evoked by external visual stimulus during continuous training. IEEE Trans. Cogn. Dev. Syst. 2021. [Google Scholar] [CrossRef]
- Kim, H.S.; Ahn, M.H.; Min, B.K. Deep-Learning-Based Automatic Selection of Fewest Channels for brain–machine Interfaces. IEEE Trans. Cybern. 2021. [Google Scholar] [CrossRef]
- Papitto, G.; Friederici, A.D.; Zaccarella, E. The topographical organization of motor processing: An ALE meta-analysis on six action domains and the relevance of Broca’s region. NeuroImage 2020, 206, 116321. [Google Scholar] [CrossRef]
- Wei, L.; Zhou, C.; Chen, H.; Song, J.; Su, R. ACPred-FL: A sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 2018, 34, 4007–4016. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Hu, J.; Li, F.; Song, J.; Su, R.; Zou, Q. Comparative analysis and prediction of quorum-sensing peptides using feature representation learning and machine learning algorithms. Briefings Bioinform. 2020, 21, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Hu, J.; Zou, Q.; Manavalan, B.; Wei, L. Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools. Briefings Bioinform. 2020, 21, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Romero-Laiseca, M.A.; Delisle-Rodriguez, D.; Cardoso, V.; Gurve, D.; Loterio, F.; Nascimento, J.H.P.; Krishnan, S.; Frizera-Neto, A.; Bastos-Filho, T. Berlin Brain–Computer Interface—The HCI communication channel for discovery. Int. J. Hum.-Comput. Stud. 2007, 65, 460–477. [Google Scholar]













| Model | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| CSP_CV | 71.21 ± 14.79 | 73.69 ± 13.52 | 68.73 ± 16.47 |
| Deep ConvNet | 65.72 ± 13.96 | 65.89 ± 17.56 | 65.56 ± 17.88 |
| EEGNet | 66.75 ± 14.25 | 64.11 ± 16.95 | 69.39 ± 12.67 |
| FBCNet | 73.44 ± 14.37 | 76.37 ± 12.63 | 70.50 ± 18.47 |
| DHDANet | 75.52 ± 11.72 | 77.58 ± 10.85 | 73.46 ± 13.59 |
| SHNA | SHSA | SHDA | DHDA | DHTA | |
|---|---|---|---|---|---|
| Accuracy | 63.01% | 65.27% | 66.63% | 75.52% | 69.24% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Yao, J.; Ni, H. Dual Head and Dual Attention in Deep Learning for End-to-End EEG Motor Imagery Classification. Appl. Sci. 2021, 11, 10906. https://doi.org/10.3390/app112210906
Xu M, Yao J, Ni H. Dual Head and Dual Attention in Deep Learning for End-to-End EEG Motor Imagery Classification. Applied Sciences. 2021; 11(22):10906. https://doi.org/10.3390/app112210906
Chicago/Turabian StyleXu, Meiyan, Junfeng Yao, and Hualiang Ni. 2021. "Dual Head and Dual Attention in Deep Learning for End-to-End EEG Motor Imagery Classification" Applied Sciences 11, no. 22: 10906. https://doi.org/10.3390/app112210906
APA StyleXu, M., Yao, J., & Ni, H. (2021). Dual Head and Dual Attention in Deep Learning for End-to-End EEG Motor Imagery Classification. Applied Sciences, 11(22), 10906. https://doi.org/10.3390/app112210906






