Workflow for Segmentation of Caenorhabditis elegans from Fluorescence Images for the Quantitation of Lipids
Abstract
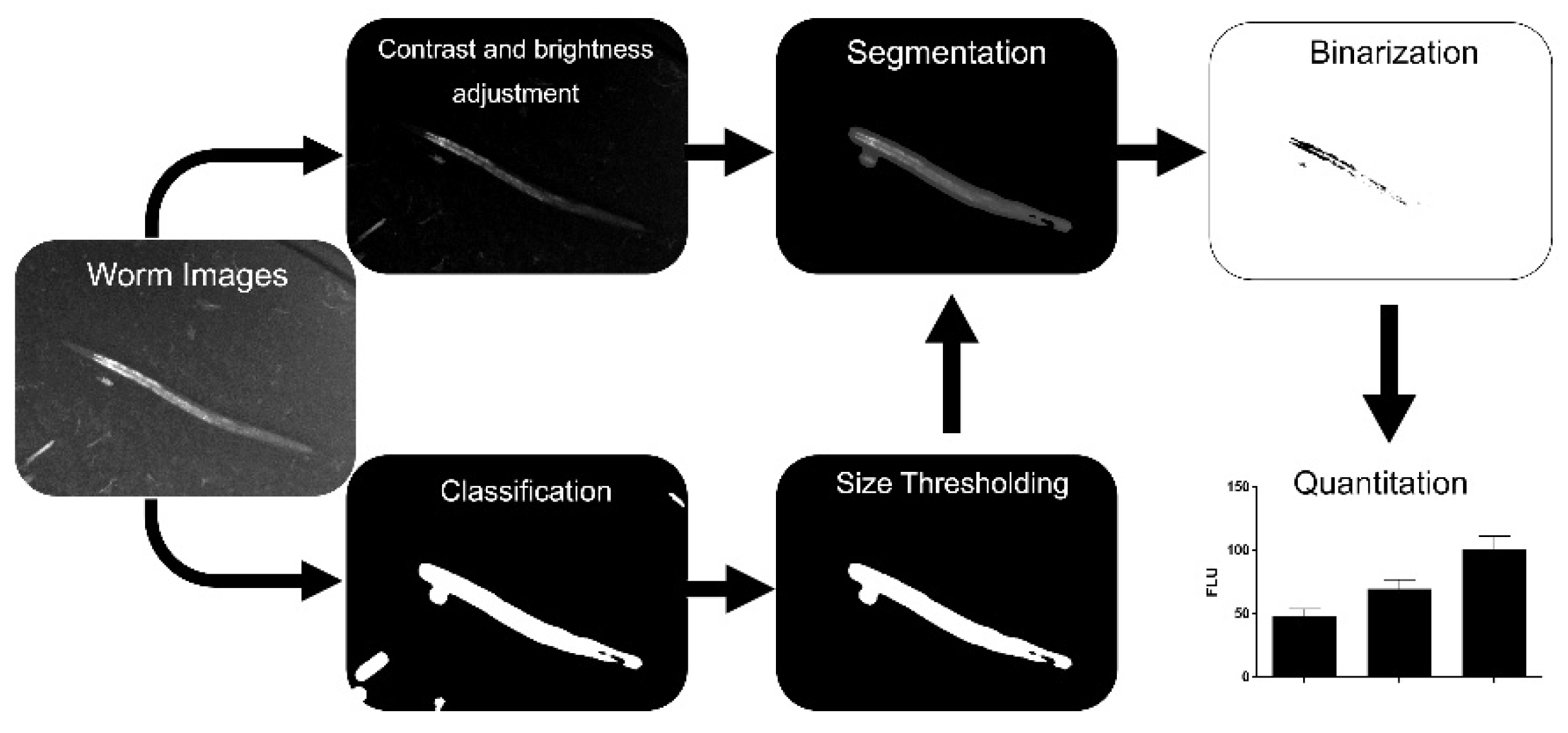
:1. Introduction
2. Materials and Methods
2.1. Nile Red Assay
2.2. Data Sets
2.3. Image Enhancement
2.4. Training of Classifier
2.5. Selection of Algorithm and Attributes
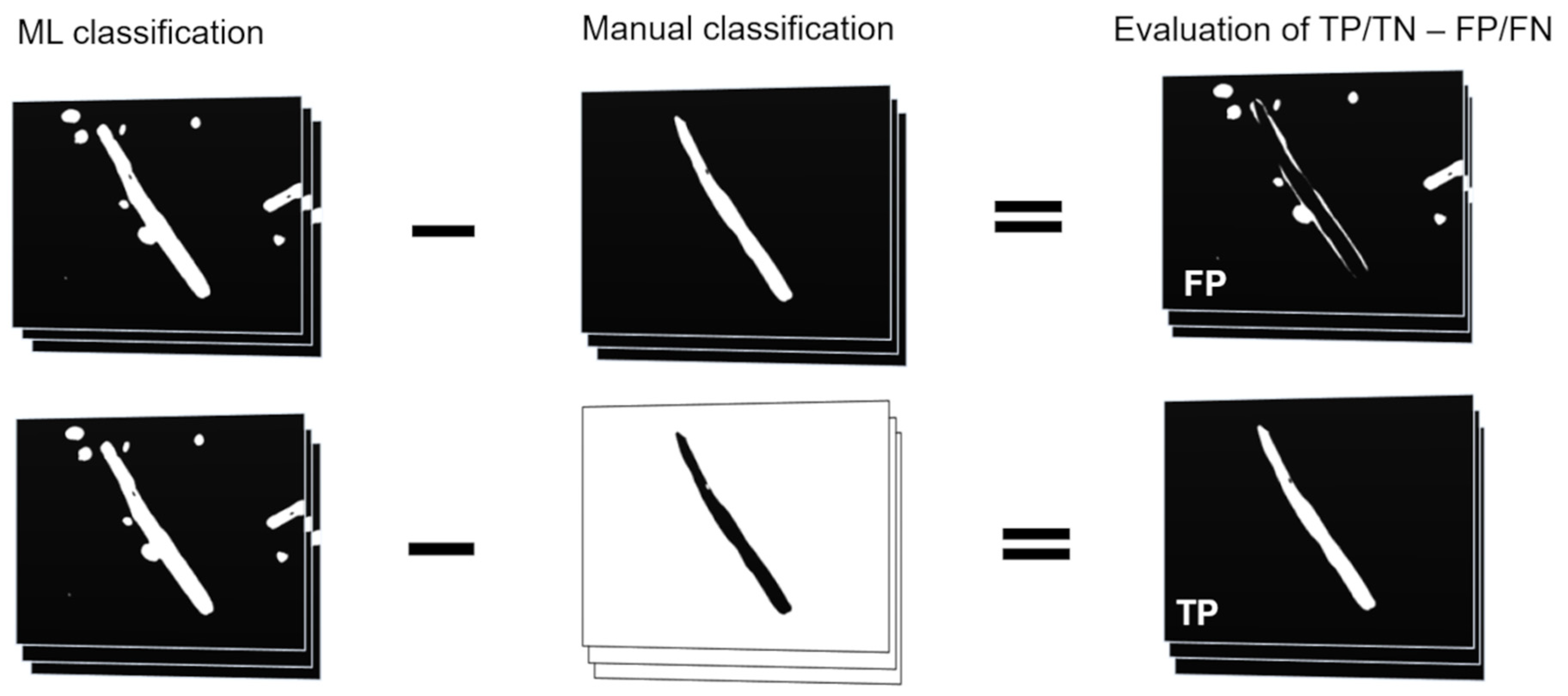
2.6. Evaluation of Attributes on Test Set
2.7. Evaluation of Size-Thresholding
2.8. Binarization
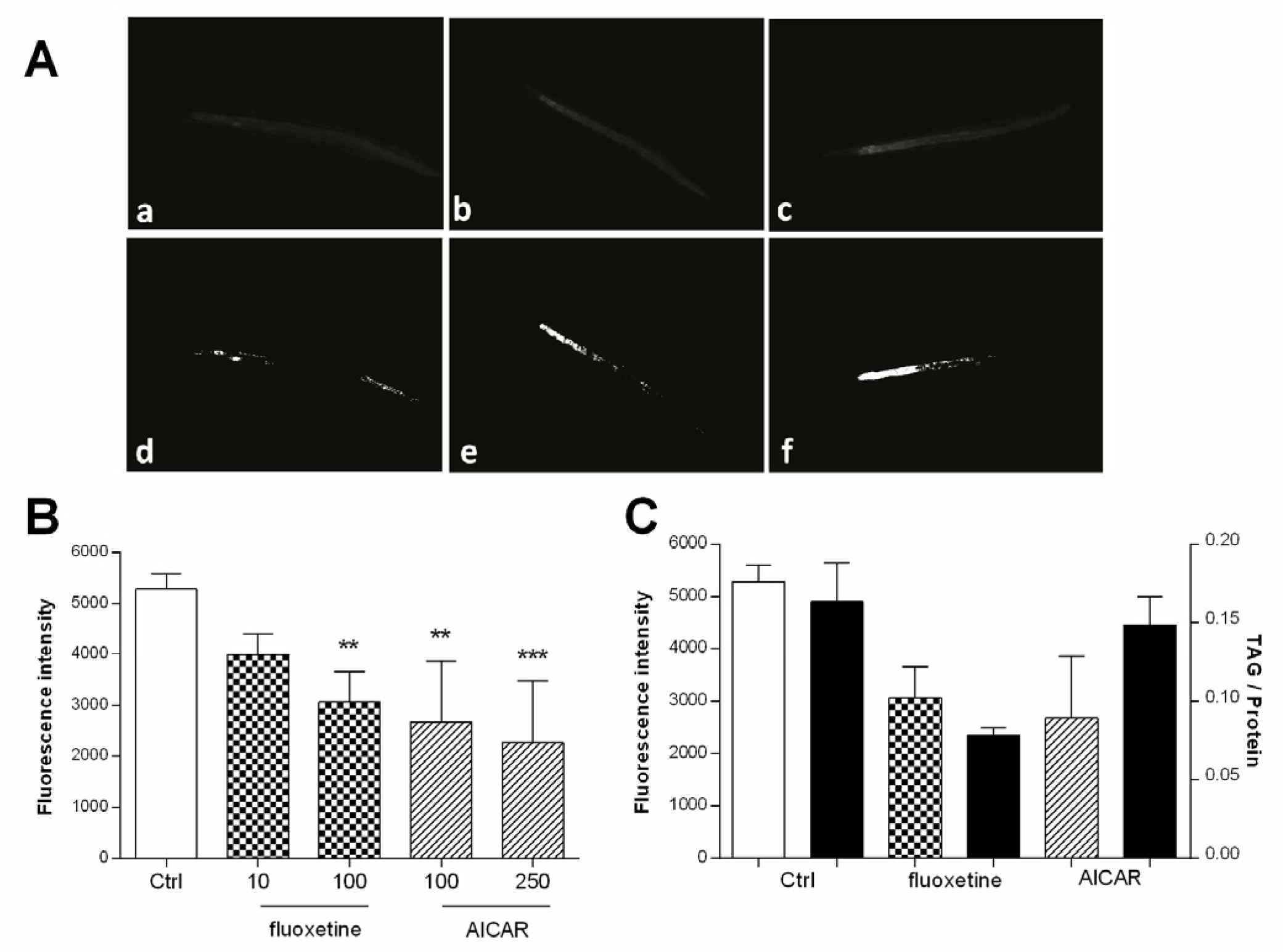
2.9. Experimental Validation, Nile Red Assay
2.10. Experimental Validation, Triacyl Glyceride Assay
3. Results
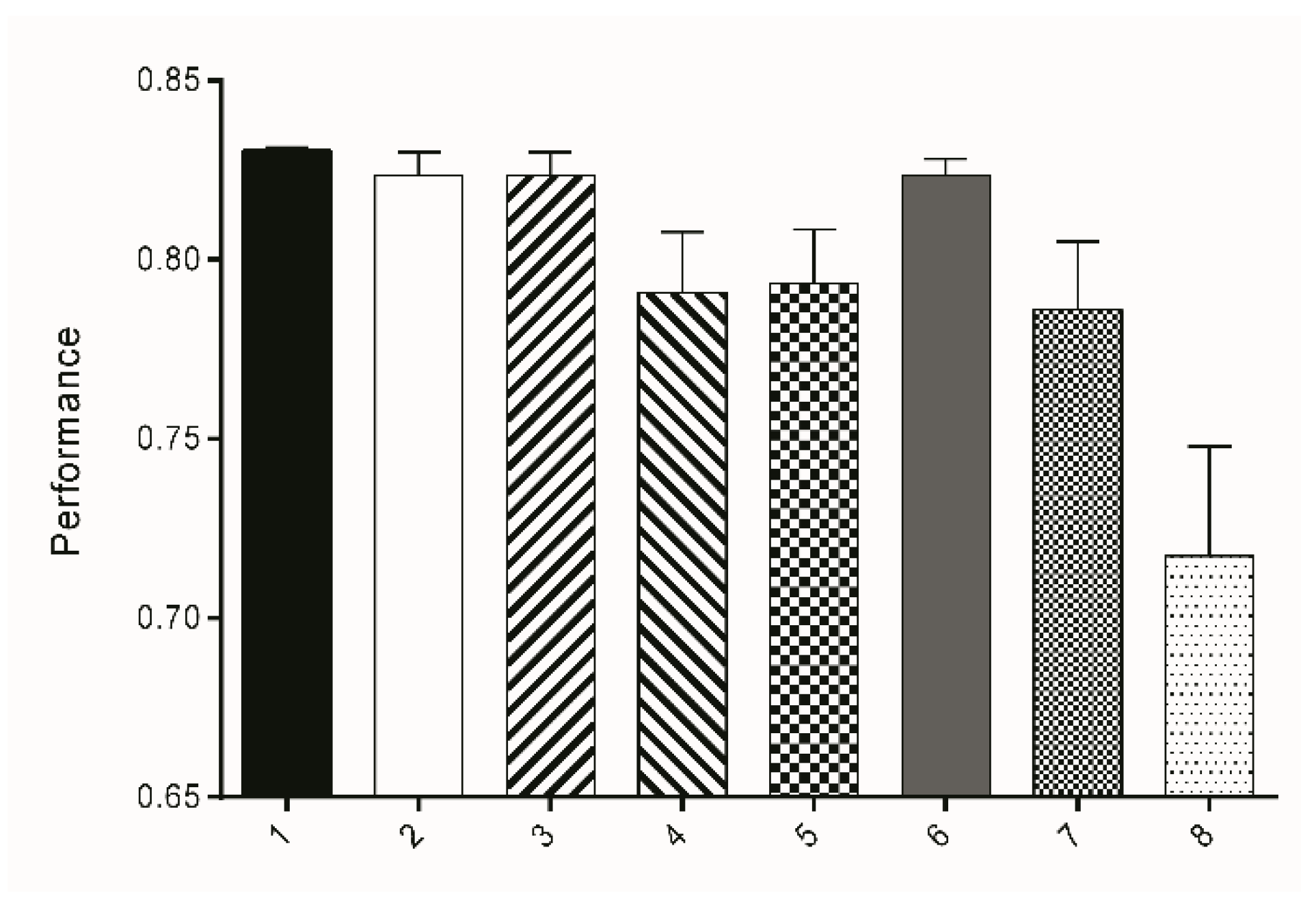
3.1. Segmentation/Selection of the Machine Learning Algorithm and Attribute Subset
3.2. Segmentation/Size-Thresholding Settings
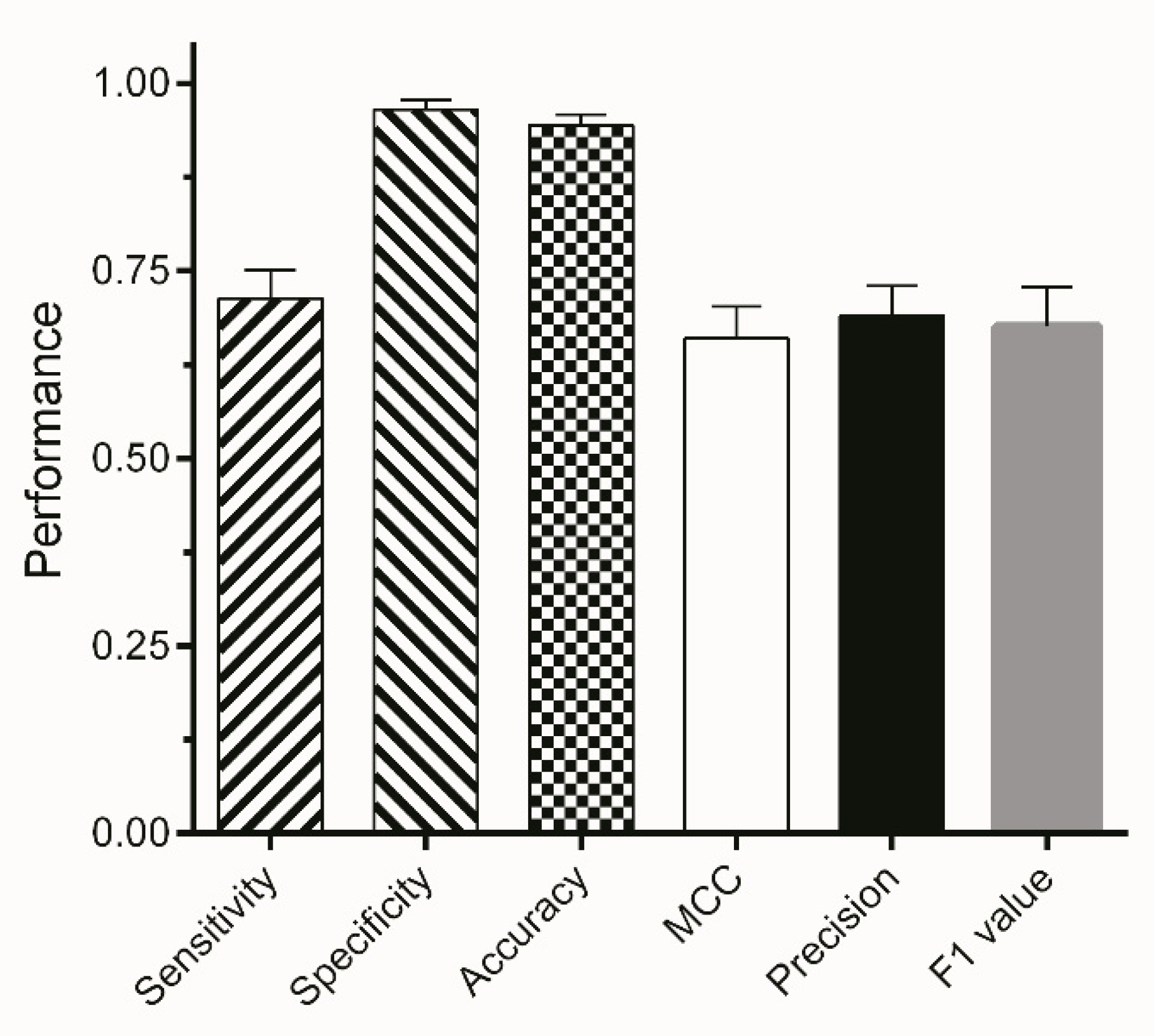
3.3. Validation
3.4. Binarization
3.5. Experimental Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hulme, S.E.; Whitesides, G.M. Chemistry and the worm: Caenorhabditis elegans as a platform for integrating chemical and biological research. Angew. Chem. Int. Ed. Engl. 2011, 50, 4774–4807. [Google Scholar] [CrossRef] [Green Version]
- Schulenburg, H.; Félix, M.-A. The Natural Biotic Environment of Caenorhabditis elegans. Genetics 2017, 206, 55–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Reilly, L.P.; Luke, C.J.; Perlmutter, D.H.; Silverman, G.A.; Pak, S.C. C. elegans in high-throughput drug discovery. Adv. Drug Deliv. Rev. 2014, 69–70, 247–253. [Google Scholar] [CrossRef] [Green Version]
- Lemieux, G.A.; Ashrafi, K. Insights and challenges in using C. elegans for investigation of fat metabolism. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.T.; Ashrafi, K. Caenorhabditis elegans as an emerging model for studying the basic biology of obesity. Dis. Model. Mech. 2009, 2, 224–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, P.; Yue, Y.; Park, Y. A living model for obesity and aging research: Caenorhabditis elegans. Crit. Rev. Food Sci. Nutr. 2018, 58, 741–754. [Google Scholar] [CrossRef]
- Soukas, A.A.; Kane, E.A.; Carr, C.E.; Melo, J.A.; Ruvkun, G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009, 23, 496–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezcurra, M.; Benedetto, A.; Sornda, T.; Gilliat, A.F.; Au, C.; Zhang, Q.; van Schelt, S.; Petrache, A.L.; Wang, H.; de la Guardia, Y.; et al. C. elegans eats its own intestine to make yolk leading to multiple senescent pathologies. Curr. Biol. 2018, 28, 2544–2556. [Google Scholar] [CrossRef]
- Salzer, L.; Witting, M. Quo vadis Caenorhabditis elegans metabolomics—A review of current methods and applications to explore metabolism in the nematode. Metabolites 2021, 11, 284. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.; Hirsh, D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 1999, 10, 4311–4326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Na, H.; Liu, Z.; Zhang, S.; Xue, P.; Chen, Y.; Pu, J.; Peng, G.; Huang, X.; Yang, F. Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol. Cell. Proteom. 2012, 11, 317–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.W.; Lemieux, G.A.; Camp, C.H., Jr.; Chang, T.C.; Ashrafi, K.; Cicerone, M.T. Spectroscopic coherent Raman imaging of Caenorhabditis elegans reveals lipid particle diversity. Nat. Chem. Biol. 2020, 16, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Yi, Y.H.; Chien, C.H.; Hsiung, K.C.; Ma, T.H.; Lin, Y.C.; Lo, S.J.; Chang, T.C. Specific polyunsaturated fatty acids modulate lipid delivery and oocyte development in C. elegans revealed by molecular-selective label-free imaging. Sci. Rep. 2016, 6, 32021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellerer, T.; Axäng, C.; Brackmann, C.; Hillertz, P.; Pilon, M.; Enejder, A. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 14658–14663. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, E.J.; Soukas, A.A.; Carr, C.E.; Ruvkun, G. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009, 10, 430–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, A.; Holdorf, A.D.; Walhout, A.J.M. Many transcription factors contribute to C. elegans growth and fat storage. Genes Cells 2017, 22, 770–784. [Google Scholar] [CrossRef] [Green Version]
- Lemieux, G.A.; Liu, J.; Mayer, N.; Bainton, R.J.; Ashrafi, K.; Werb, Z. A whole-organism screen identifies new regulators of fat storage. Nat. Chem. Biol. 2011, 7, 206–213. [Google Scholar] [CrossRef] [Green Version]
- Pino, E.C.; Webster, C.M.; Carr, C.E.; Soukas, A.A. Biochemical and high throughput microscopic assessment of fat mass in Caenorhabditis elegans. J. Vis. Exp. 2013, 30, e50180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, S.; Lynn, D.A.; Lo, J.Y.; Paek, J.; Curran, S.P. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 2014, 5, 5048. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.M.; Li, Z.Y.; Xu, Y.J.; Wang, W.; Zhou, M.G.; Zhang, P.; Liu, P.S.; Xu, T.; Wu, Z.X. PKG and NHR-49 signalling co-ordinately regulate short-term fasting-induced lysosomal lipid accumulation in C. elegans. Biochem. J. 2014, 461, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Zwirchmayr, J.; Kirchweger, B.; Lehner, T.; Tahir, A.; Pretsch, D.; Rollinger, J.M. A robust and miniaturized screening platform to study natural products affecting metabolism and survival in Caenorhabditis elegans. Sci. Rep. 2020, 10, 12323. [Google Scholar] [CrossRef]
- Rizwan, I.; Haque, I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Husson, S.J.; Costa, W.S.; Schmitt, C.; Gottschalk, A. Keeping Track of Worm Trackers; WormBook: Pasadena, CA, USA, 2013; pp. 1–17. [Google Scholar]
- Kabra, M.; Conery, A.; O’Rourke, E.; Xie, X.; Ljosa, V.; Jones, T.; Ausubel, F.; Ruvkun, G.; Carpenter, A.; Freund, Y. Towards automated high-throughput screening of C. elegans on agar. arXiv 2010, arXiv:1003.4287. [Google Scholar]
- Hernando-Rodríguez, B.; Erinjeri, A.P.; Rodríguez-Palero, M.J.; Millar, V.; González-Hernández, S.; Olmedo, M.; Schulze, B.; Baumeister, R.; Muñoz, M.J.; Askjaer, P.; et al. Combined flow cytometry and high-throughput image analysis for the study of essential genes in Caenorhabditis elegans. BMC Biol. 2018, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Hakim, A.; Mor, Y.; Toker, I.A.; Levine, A.; Neuhof, M.; Markovitz, Y.; Rechavi, O. WorMachine: Machine learning-based phenotypic analysis tool for worms. BMC Biol. 2018, 16, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, B.T.; Jordan, J.M.; Baugh, L.R. WormSizer: High-throughput Analysis of Nematode Size and Shape. PLoS ONE 2013, 8, e57142. [Google Scholar]
- Bornhorst, J.; Nustede, E.J.; Fudickar, S. Mass Surveilance of C. elegans-Smartphone-Based DIY Microscope and Machine-Learning-Based Approach for Worm Detection. Sensors 2019, 19, 1468. [Google Scholar] [CrossRef] [Green Version]
- Fudickar, S.; Nustede, E.J.; Dreyer, E.; Bornhorst, J. Mask R-CNN Based C. Elegans Detection with a DIY Microscope. Biosensors 2021, 11, 257. [Google Scholar] [CrossRef]
- Wählby, C.; Kamentsky, L.; Liu, Z.H.; Riklin-Raviv, T.; Conery, A.L.; O’Rourke, E.J.; Sokolnicki, K.L.; Visvikis, O.; Ljosa, V.; Irazoqui, J.E.; et al. An image analysis toolbox for high-throughput C. elegans assays. Nat. Methods 2012, 9, 714–716. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escorcia, W.; Ruter, D.L.; Nhan, J.; Curran, S.P. Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. J. Vis. Exp. 2018, 133, 57352. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kong, S.; Pincus, Z.; Fowlkes, C. Celeganser: Automated analysis of nematode morphology and age. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 968–969. [Google Scholar]
- Chen, L.; Strauch, M.; Daub, M.; Jiang, X.; Jansen, M.; Luigs, H.-G.; Schultz-Kuhlmann, S.; Krussel, S.; Merhof, D. A CNN Framework Based on Line Annotations for Detecting Nematodes in Microscopic Images. In Proceedings of the 2020 IEEE 17th International symposium on biomedical imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 508–512. [Google Scholar]
- Rastogi, S.; Borgo, B.; Pazdernik, N.; Fox, P.; Mardis, E.R.; Kohara, Y.; Havranek, J.; Schedl, T. Caenorhabditis elegans glp-4 Encodes a Valyl Aminoacyl tRNA Synthetase. G3 2015, 5, 2719–2728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiernagle, T. Maintenance of C. elegans; Worm Book: Pasadena, CA, USA, 2006; pp. 1–11. [Google Scholar]
- Porta-de-la-Riva, M.; Fontrodona, L.; Villanueva, A.; Cerón, J. Basic Caenorhabditis elegans methods: Synchronization and observation. J. Vis. Exp. 2012, 64, e4019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Wu, X.; Kumar, V.; Quinlan, J.R.; Ghosh, J.; Yang, Q.; Motoda, H.; McLachlan, G.J.; Ng, A.; Liu, B.; Yu, P.S.; et al. Top 10 algorithms in data mining. Knowl. Inf. Syst. 2008, 14, 1–37. [Google Scholar] [CrossRef] [Green Version]
- Bechara, B.P.; Leckie, S.K.; Bowman, B.W.; Davies, C.E.; Woods, B.I.; Kanal, E.; Sowa, G.A.; Kang, J.D. Application of a semiautomated contour segmentation tool to identify the intervertebral nucleus pulposus in MR images. Am. J. Neuroradiol. 2010, 31, 1640–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millioni, R.; Sbrignadello, S.; Tura, A.; Iori, E.; Murphy, E.; Tessari, P. The inter- and intra-operator variability in manual spot segmentation and its effect on spot quantitation in two-dimensional electrophoresis analysis. Electrophoresis 2010, 31, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Shahedi, M.; Cool, D.W.; Romagnoli, C.; Bauman, G.S.; Bastian-Jordan, M.; Gibson, E.; Rodrigues, G.; Ahmad, B.; Lock, M.; Fenster, A.; et al. Spatially varying accuracy and reproducibility of prostate segmentation in magnetic resonance images using manual and semiautomated methods. Med. Phys. 2014, 41, 113503. [Google Scholar] [CrossRef] [PubMed]
- Maes, T.; Jessop, R.; Wellner, N.; Haupt, K.; Mayes, A.G. A rapid-screening approach to detect and quantify microplastics based on fluorescent tagging with Nile Red. Sci. Rep. 2017, 7, 44501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lithgow, G.J.; Driscoll, M.; Phillips, P. A long journey to reproducible results. Nature 2007, 548, 387–388. [Google Scholar] [CrossRef] [Green Version]
- Jia, R.; Zhang, J.; Jia, K. Neuroendocrine regulation of fat metabolism by autophagy gene atg-18 in C. elegans dauer larvae. FEBS Open Bio 2019, 9, 1623–1631. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Herrera, D.; Aranaz, P.; Eder-Azanza, L.; Zabala, M.; Hurtado, C.; Romo-Hualde, A.; Martínez, J.A.; González-Navarro, C.J.; Vizmanos, J.L. Dihomo-gamma-linolenic acid induces fat loss in C. elegans in an omega-3-independent manner by promoting peroxisomal fatty acid β-oxidation. Food Funct. 2018, 9, 1621–1637. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.S.; Fujioka, K.; Devine, W.; Bray, G.A. A randomized double-blind clinical trial of fluoxetine in obese diabetics. Int. J. Obes. Relat. Metab. Disord. 1992, 16 (Suppl. 4), S67–S72. [Google Scholar] [PubMed]
- Goldstein, D.J.; Rampey, A.H., Jr.; Enas, G.G.; Potvin, J.H.; Fludzinski, L.A.; Levine, L.R. Fluoxetine: A randomized clinical trial in the treatment of obesity. Int. J. Obes. Relat. Metab. Disord. 1994, 18, 129–135. [Google Scholar] [PubMed]
- Levine, L.R.; Enas, G.G.; Thompson, W.L.; Byyny, R.L.; Dauer, A.D.; Kirby, R.W.; Kreindler, T.G.; Levy, B.; Lucas, C.P.; McIlwain, H.H. Use of fluoxetine, a selective serotonin-uptake inhibitor, in the treatment of obesity: A dose-response study (with a commentary by Michael Weintraub). Int. J. Obes. 1989, 13, 635–645. [Google Scholar]
- Giri, S.; Rattan, R.; Haq, E.; Khan, M.; Yasmin, R.; Won, J.-s.; Key, L.; Singh, A.K.; Singh, I. AICAR inhibits adipocyte differentiation in 3T3L1 and restores metabolic alterations in diet-induced obesity mice model. Nutr. Metab. 2006, 3, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, S.; Sadegh, L.; Elle, I.C.; Christensen, A.G.; Faergeman, N.J.; Ashrafi, K. Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab. 2008, 7, 533–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almotayri, A.; Thomas, J.; Munasinghe, M.; Weerasinghe, M.; Heydarian, D.; Jois, M. Metabolic and behavioral effects of olanzapine and fluoxetine on the model organism Caenorhabditis elegans. Saudi Pharm. J. 2021, 29, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, G.A.; Ashrafi, K. Investigating connections between metabolism, longevity, and behavior in Caenorhabditis elegans. Trends Endocrinol. Metab. 2016, 27, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Ranking | Attributes | Ranking | Attributes |
|---|---|---|---|
| 0.9717 | Entropy_16_256 | 0.692 | Membrane_projections_0_19_1 |
| 0.9717 | Entropy_16_64 | 0.692 | Membrane_projections_5_19_1 |
| 0.9717 | Entropy_16_128 | 0.692 | Membrane_projections_3_19_1 |
| 0.8915 | Entropy_32_256 | 0.6905 | Entropy_32_64 |
| 0.8915 | Entropy_32_128 | 0.6862 | Gabor_2_1.0_0.5_0_2.0 |
| 0.8845 | Variance_16.0 | 0.6832 | Gabor_1_4.0_1.0_2_2.0 |
| 0.8287 | Hessian_Eigenvalue_2_32.0 | 0.6828 | Hessian_Normalized_Eigenvalue_ Difference_16.0 |
| 0.8265 | Variance_32.0 | 0.6827 | Median_16.0 |
| 0.8186 | Laplacian_16.0 | 0.6656 | Gabor_1_1.0_1.0_0_2.0 |
| 0.7814 | Laplacian_32.0 | 0.6654 | Hessian_32.0 |
| 0.7714 | Gabor_1_1.0_0.25_0_2.0 | 0.6642 | Membrane_projections_1_19_1 |
| 0.759 | Entropy_16_32 | 0.6469 | Gabor_2_1.0_0.25_0_2.0 |
| 0.7534 | Gabor_1_2.0_1.0_0_2.0 | 0.6446 | Sobel_filter_16.0 |
| 0.7509 | Gabor_1_4.0_2.0_0_2.0 | 0.6127 | Hessian_Trace_16.0 |
| 0.7473 | Mean_16.0 | 0.6091 | Entropy_32_32 |
| 0.7434 | Hessian_Trace_32.0 | 0.6077 | Gabor_2_4.0_2.0_2_2.0 |
| 0.741 | Gabor_1_1.0_0.5_0_2.0 | 0.6074 | Gabor_2_4.0_1.0_2_2.0 |
| 0.7384 | Hessian_16.0 | 0.599 | Hessian_Eigenvalue_2_16.0 |
| 0.7277 | Gabor_1_4.0_1.0_0_2.0 | 0.5965 | Membrane_projections_4_19_1 |
| 0.7213 | Maximum_16.0 | 0.5858 | Hessian_Determinant_32.0 |
| 0.7106 | Membrane_projections_2_19_1 | 0.5853 | Gabor_2_1.0_1.0_0_2.0 |
| 0.7088 | Gabor_1_4.0_2.0_2_2.0 | 0.5836 | Structure_smallest_16.0_3.0 |
| 0.6956 | Hessian_Square_Eigenvalue_ Difference_16.0 | 0.5789 | Hessian_Normalized_Eigenvalue_ Difference_32.0 |
| 0.6948 | Gabor_1_2.0_2.0_0_2.0 | 0.5717 | Hessian_Determinant_16.0 |
| 0.6935 | Hessian_Eigenvalue_1_32.0 | 0.5614 | Mean_32.0 |
| Subset | Attribute Subsets | Performance | No. of Attributes | Instances | ||
|---|---|---|---|---|---|---|
| TS1 | TS2 | TS3 | ||||
| 1 | ENT 1-16 | 0.8296 | 0.8312 | 0.8342 | 22 | 237 |
| 2 | ENT 16-32 | 0.8187 | 0.8208 | 0.8308 | 10 | 86 |
| 3 | ENTVAR 1-16 | 0.8187 | 0.8208 | 0.8308 | 27 | 98 |
| 4 | ENTVAR 16-32 | 0.7734 | 0.7919 | 0.8070 | 12 | 55 |
| 5 | ENTVARHES 1-16 | 0.7822 | 0.7869 | 0.8107 | 75 | 132 |
| 6 | ENVARHES 16-32 | 0.8202 | 0.8285 | 0.8221 | 28 | 225 |
| 7 | ENTAVRHESLAP 1-16 | 0.7708 | 0.7798 | 0.8073 | 80 | 127 |
| 8 | ENTVARHESLAP 16-32 | 0.6921 | 0.7085 | 0.7513 | 30 | 98 |
| ETS | Sensitivity | Specificity | ACC | MCC | Precision |
|---|---|---|---|---|---|
| ETS1 | 1.0000 | 0.9980 | 0.9980 | 0.8592 | 0.7627 |
| ETS2 | 0.9999 | 0.9977 | 0.9977 | 0.7952 | 0.6817 |
| ETS3 | 1.0000 | 0.9993 | 0.9993 | 0.8453 | 0.7602 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lehner, T.; Pum, D.; Rollinger, J.M.; Kirchweger, B. Workflow for Segmentation of Caenorhabditis elegans from Fluorescence Images for the Quantitation of Lipids. Appl. Sci. 2021, 11, 11420. https://doi.org/10.3390/app112311420
Lehner T, Pum D, Rollinger JM, Kirchweger B. Workflow for Segmentation of Caenorhabditis elegans from Fluorescence Images for the Quantitation of Lipids. Applied Sciences. 2021; 11(23):11420. https://doi.org/10.3390/app112311420
Chicago/Turabian StyleLehner, Theresa, Dietmar Pum, Judith M. Rollinger, and Benjamin Kirchweger. 2021. "Workflow for Segmentation of Caenorhabditis elegans from Fluorescence Images for the Quantitation of Lipids" Applied Sciences 11, no. 23: 11420. https://doi.org/10.3390/app112311420
APA StyleLehner, T., Pum, D., Rollinger, J. M., & Kirchweger, B. (2021). Workflow for Segmentation of Caenorhabditis elegans from Fluorescence Images for the Quantitation of Lipids. Applied Sciences, 11(23), 11420. https://doi.org/10.3390/app112311420







