Automated Diagnosis of Childhood Pneumonia in Chest Radiographs Using Modified Densely Residual Bottleneck-Layer Features
Abstract
:1. Introduction
1.1. Related Work
1.2. Limitations and Proposed Work
2. Proposed Method
2.1. Feature Extraction and Reduction
2.2. Prediction Model
2.3. Adaptive Score Fusion
| Algorithm 1: Adaboost ensemble weights learning. |
| initialize classifier weights as ; |
 |
3. Results and Discussion
3.1. SVM Optimization Using Bayesian Optimization
3.2. Normal vs. Bacterial vs. Viral Pneumonia Infected Lungs
3.3. Normal vs. Pneumonia Infected Lungs
3.4. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Viral vs. Bacterial Pneumonia: Understanding the Difference. 2020. Available online: https://www.pfizer.com/news/hot-topics/viral_vs_bacterial_pneumonia_understanding_the_difference (accessed on 28 April 2021).
- Popovsky, E.Y.; Florin, T.A. Community-Acquired Pneumonia in Childhood. Ref. Modul. Biomed. Sci. 2020. [Google Scholar] [CrossRef]
- WHO Director Generals Opening Remarks at the Media Briefing on COVID-19 2020. 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19—11-march-2020 (accessed on 28 April 2021).
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al-Emadi, N.; et al. Can AI help in screening viral and COVID-19 pneumonia? arXiv 2020, arXiv:2003.13145. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Saikia, P.; Baruah, R.D.; Singh, S.K.; Chaudhuri, P.K. Artificial Neural Networks in the domain of reservoir characterization: A review from shallow to deep models. Comput. Geosci. 2020, 135, 104357. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Alkassar, S.; Jebur, B.A.; Abdullah, M.A.; Al-Khalidy, J.H.; Chambers, J. Going deeper: Magnification-invariant approach for breast cancer classification using histopathological images. IET Comput. Vis. 2021, 15, 151–164. [Google Scholar] [CrossRef]
- Abdullah, M.A.; Alkassar, S.; Jebur, B.; Chambers, J. LBTS-Net: A fast and accurate CNN model for brain tumour segmentation. Healthc. Technol. Lett. 2021, 8, 31. [Google Scholar] [CrossRef]
- Torrey, L.; Shavlik, J. Transfer learning. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Hershey, PA, USA, 2010; pp. 242–264. [Google Scholar]
- Fooladgar, F.; Kasaei, S. Lightweight residual densely connected convolutional neural network. Multimed. Tools Appl. 2020, 79, 25571–25588. [Google Scholar] [CrossRef]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and < 0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Asnaoui, K.E.; Chawki, Y.; Idri, A. Automated methods for detection and classification pneumonia based on X-ray images using deep learning. arXiv 2020, arXiv:2003.14363. [Google Scholar]
- Saraiva, A.; Ferreira, N.; Sousa, L.; Carvalho da Costa, N.; Sousa, J.; Santos, D.; Soares, S. Classification of Images of Childhood Pneumonia using Convolutional Neural Networks. In Proceedings of the 6th International Conference on Bioimaging, Prague, Czech Republic, 22–24 February 2019; pp. 112–119. [Google Scholar]
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Liang, G.; Zheng, L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Programs Biomed. 2019, 187, 104964. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models. IRBM 2019, 41, 212–222. [Google Scholar] [CrossRef]
- Rajpurkar, P.; Irvin, J.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.; Shpanskaya, K.; et al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv 2017, arXiv:1711.05225. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Han, Y.; Chen, C.; Tewfik, A.; Ding, Y.; Peng, Y. Pneumonia detection on chest X-ray using radiomic features and contrastive learning. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 247–251. [Google Scholar]
- Shih, G.; Wu, C.C.; Halabi, S.S.; Kohli, M.D.; Prevedello, L.M.; Cook, T.S.; Sharma, A.; Amorosa, J.K.; Arteaga, V.; Galperin-Aizenberg, M.; et al. Augmenting the National Institutes of Health chest radiograph dataset with expert annotations of possible pneumonia. Radiol. Artif. Intell. 2019, 1, e180041. [Google Scholar] [CrossRef]
- Chouhan, V.; Singh, S.K.; Khamparia, A.; Gupta, D.; Tiwari, P.; Moreira, C.; Damaševičius, R.; De Albuquerque, V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl. Sci. 2020, 10, 559. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Rahman, T.; Chowdhury, M.E.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. Transfer learning with deep convolutional neural network (CNN) for pneumonia detection using chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Pang, G.; Liao, Z.; Verjans, J.; Li, W.; Sun, Z.; He, J.; Li, Y.; Shen, C.; et al. Viral pneumonia screening on chest x-rays using Confidence-Aware anomaly detection. IEEE Trans. Med. Imaging 2020, 40, 879–890. [Google Scholar] [CrossRef]
- Ayan, E.; Karabulut, B.; Ünver, H.M. Diagnosis of Pediatric Pneumonia with Ensemble of Deep Convolutional Neural Networks in Chest X-ray Images. Arab. J. Sci. Eng. 2021, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nahiduzzaman, M.; Goni, M.O.F.; Anower, M.S.; Islam, M.R.; Ahsan, M.; Haider, J.; Gurusamy, S.; Hassan, R.; Islam, M.R. A Novel Method for Multivariant Pneumonia Classification based on Hybrid CNN-PCA Based Feature Extraction using Extreme Learning Machine with Chest X-ray Images. IEEE Access 2021, 9, 147512–147526. [Google Scholar] [CrossRef]
- Gour, M.; Jain, S. Uncertainty-aware convolutional neural network for COVID-19 X-ray images classification. Comput. Biol. Med. 2021, 140, 105047. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Chen, M.; Shi, X.; Zhang, Y.; Wu, D.; Guizani, M. Deep features learning for medical image analysis with convolutional autoencoder neural network. IEEE Trans. Big Data 2017, 7, 750–758. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Czarnecki, W.M.; Podlewska, S.; Bojarski, A.J. Robust optimization of SVM hyperparameters in the classification of bioactive compounds. J. Cheminform. 2015, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical bayesian optimization of machine learning algorithms. arXiv 2012, arXiv:1206.2944. [Google Scholar]
- Wenzel, F.; Deutsch, M.; Galy-Fajou, T.; Kloft, M. Scalable Approximate Inference for the Bayesian Nonlinear Support Vector Machine. In Proceedings of the 30th Conference on Neural Information Processing Systems (NIPS 2016), Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Wenzel, F.; Galy-Fajou, T.; Deutsch, M.; Kloft, M. Bayesian nonlinear support vector machines for big data. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases; Springer: Berlin/Heidelberg, Germany, 2017; pp. 307–322. [Google Scholar]
- Freund, Y.; Schapire, R.; Abe, N. A short introduction to boosting. J.-Jpn. Soc. Artif. Intell. 1999, 14, 1612. [Google Scholar]
- Kermany, D.; Zhang, K.; Goldbaum, M. Labeled optical coherence tomography (oct) and chest X-ray images for classification. Mendeley Data 2018, 2. [Google Scholar] [CrossRef]
- Swingler, G.H. Radiologic differentiation between bacterial and viral lower respiratory infection in children: A systematic literature review. Clin. Pediatr. 2000, 39, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6848–6856. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8697–8710. [Google Scholar]
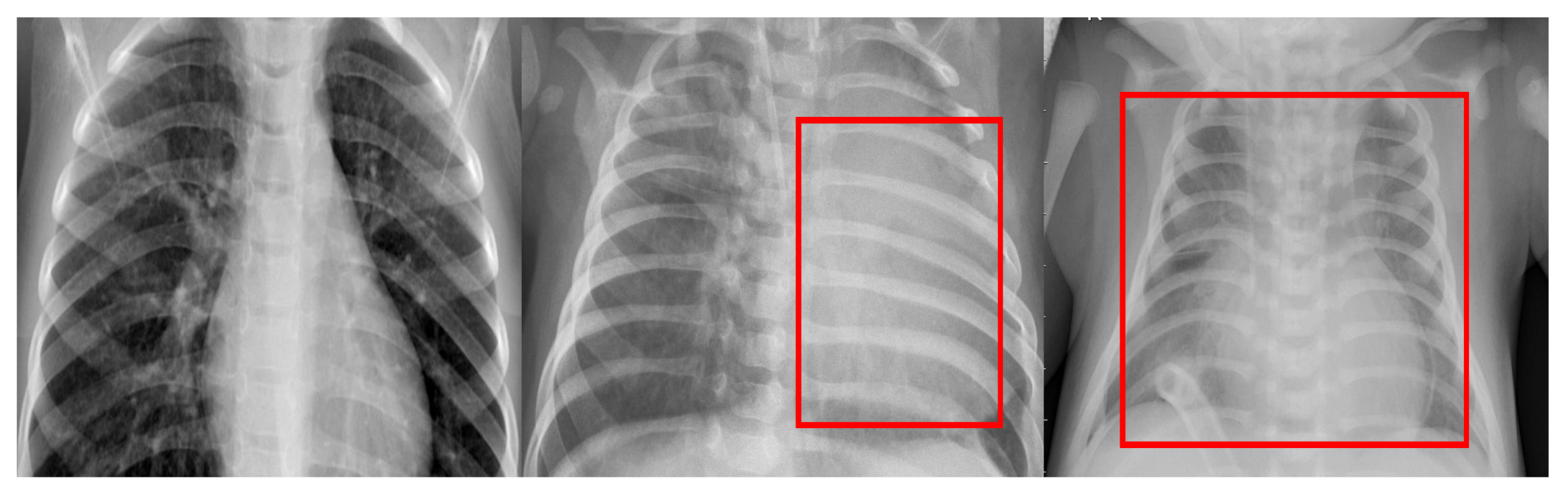







| Category | No. of Images | Training | Testing |
|---|---|---|---|
| Normal | 1349 | 1012 | 337 |
| Bacterial | 2538 | 1903 | 635 |
| Viral | 1345 | 1008 | 337 |
| Total | 5232 | 3922 | 1309 |
| Number of Bottleneck Blocks | Accuracy | Number of Features |
|---|---|---|
| 1 | 95.6 | 1 × 256 |
| 2 | 96.2 | 1 × 256 |
| 3 | 96.8 | 1 × 256 |
| 4 | 96.8 | 1 × 512 |
| 5 | 99.6 | 1 × 512 |
| 6 | 97.6 | 1 × 512 |
| 7 | 97.2 | 1 × 512 |
| Deep Network | Number of Layers | Accuracy (%) | |
|---|---|---|---|
| Deep Layer Features | Early Layer Features | ||
| AlexNet [5] | 8 | 96.0 | 97.3 |
| VGG [17] | 19 | 96.8 | 98.4 |
| SqueezeNet [13] | 14 | 96.7 | 96.0 |
| GoogleNet [29] | 27 | 96.2 | 97.7 |
| ShuffleNet [45] | 20 | 96.5 | 96.8 |
| NASNetMobile [46] | 913 | 95.8 | 96.9 |
| DenseNet [18] | 201 | 98.0 | 98.4 |
| Xception [19] | 36 | 96.4 | 98.4 |
| ResNet [6] | 50 | 97.6 | 97.1 |
| Proposed method | 35 | na | 99.6 |
| Pneumonia Diagnosis Method | Deep Learning Technique | Accuracy (%) |
|---|---|---|
| Chowdhury et al. [4] | Transfer Learning with SqueezeNet | 99.00 |
| Asnaoui et al. [14] | Transfer Learning with ResNet50 | 96.61 |
| Saraiva et al. [15] | CNN 10 Layers | 95.30 |
| Apostolopoulos et al. [16] | Transfer Learning with MobileNetv2 | 96.78 |
| Liang and Zheng [21] | CNN with 49 Residual Blocks | 95.30 |
| Kermany et al. [22] | Transfer Learning with AlexNet | 92.80 |
| Toğaçar et al. [23] | Deep Features Fused from AlexNet, VGG16, and VGG19 | 99.41 |
| Rajpurkar et al. [24] | Transfer Learning with ChexNet | 82.83 |
| Han et al. [26] | Contrastive Learning with ResNetAttention | 88.00 |
| Chouhan et al. [28] | Transfer Learning with 5 Deep Networks | 96.40 |
| Rahman et al. [30] | Transfer Learning with DenseNet201 | 98.00 |
| Zhang et al. [31] | One-Class Classification Based Anomaly Detection | 83.61 |
| Ayan et al. [32] | Transfer Learning with Ensemble Voting | 95.21 |
| Nahiduzzaman et al. [33] | CNN with EML and PCA | 99.83 |
| Gour and Jain [34] | fine-tuned EfficientNet-B3 | 99.83 |
| Proposed Method | Bottleneck Layer Features with 5 Densely-Connected Residual Building Blocks | 99.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkassar, S.; Abdullah, M.A.M.; Jebur, B.A.; Abdul-Majeed, G.H.; Wei, B.; Woo, W.L. Automated Diagnosis of Childhood Pneumonia in Chest Radiographs Using Modified Densely Residual Bottleneck-Layer Features. Appl. Sci. 2021, 11, 11461. https://doi.org/10.3390/app112311461
Alkassar S, Abdullah MAM, Jebur BA, Abdul-Majeed GH, Wei B, Woo WL. Automated Diagnosis of Childhood Pneumonia in Chest Radiographs Using Modified Densely Residual Bottleneck-Layer Features. Applied Sciences. 2021; 11(23):11461. https://doi.org/10.3390/app112311461
Chicago/Turabian StyleAlkassar, Sinan, Mohammed A. M. Abdullah, Bilal A. Jebur, Ghassan H. Abdul-Majeed, Bo Wei, and Wai Lok Woo. 2021. "Automated Diagnosis of Childhood Pneumonia in Chest Radiographs Using Modified Densely Residual Bottleneck-Layer Features" Applied Sciences 11, no. 23: 11461. https://doi.org/10.3390/app112311461
APA StyleAlkassar, S., Abdullah, M. A. M., Jebur, B. A., Abdul-Majeed, G. H., Wei, B., & Woo, W. L. (2021). Automated Diagnosis of Childhood Pneumonia in Chest Radiographs Using Modified Densely Residual Bottleneck-Layer Features. Applied Sciences, 11(23), 11461. https://doi.org/10.3390/app112311461








