Establishment of the Optimal Common Data Model Environment for EMR Data Considering the Computing Resources of Medical Institutions
Abstract
:1. Introduction
2. Materials and Methods
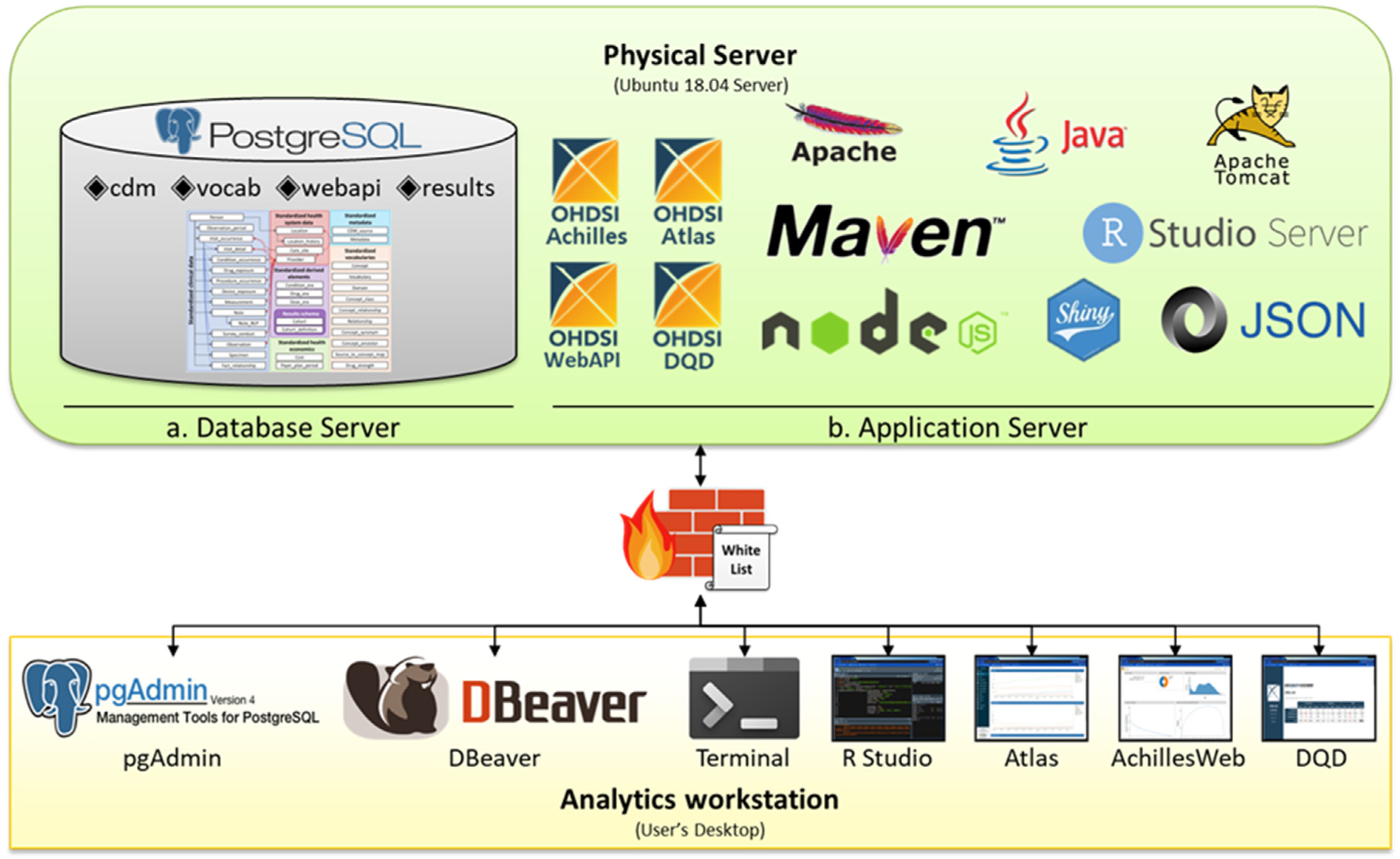
2.1. Server Design
2.1.1. Combined Server Design
2.1.2. Separated Server Design
2.2. Database Design
2.2.1. HW Resources Used in DB
2.2.2. Database Usage Type
2.2.3. DB Tuning
2.2.4. Performance Measurement
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Safran, C.; Bloomrosen, M.M.; Hammond, W.E.; Labkoff, S.; Markel-Fox, S.; Tang, P.C.; Detmer, D.E. Toward a national framework for the secondary use of health data: An American medical informatics association white paper. J. Am. Med. Inform. Assoc. 2007, 14, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Shortliffe, E.H. The evolution of electronic medical records. Acad. Med. 1999, 74, 414–419. [Google Scholar] [CrossRef]
- Bleich, H.L.; Slack, W.V. Reflections on electronic medical records: When doctors will use them and when they will not. Int. J. Med. Inform. 2010, 79, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Guttha, N.; Miao, Z.; Shamsuddin, R. Towards the development of a substance abuse index (SEI) through Informatics. Healthcare 2021, 9, 1596. [Google Scholar] [CrossRef]
- Observational Health Data Sciences and Informatics. The Book of OHDSI, 1st ed.; Observational Health Data Sciences and Informatics: Seoul, Korea, 2019; Available online: https://ohdsi.github.io/TheBookOfOhdsi/ (accessed on 30 March 2021).
- Lamer, A.; DePas, N.; Doutreligne, M.; Parrot, A.; Verloop, D.; Defebvre, M.-M.; Ficheur, G.; Chazard, E.; Beuscart, J.-B. Transforming french electronic health records into the observational medical outcome partnership’s common data model: A feasibility study. Appl. Clin. Inform. 2020, 11, 13–22. [Google Scholar] [CrossRef]
- Rho, M.J.; Kim, S.R.; Park, S.H.; Jang, K.S.; Park, B.J.; Choi, I.Y. Development common data model for adverse drug signal detection based on multi-center emr systems. In Proceedings of the 2013 International Conference on Information Science and Applications (ICISA), Pattaya, Thailand, 24–26 June 2013. [Google Scholar]
- Kent, S.; Burn, E.; Dawoud, D.; Jonsson, P.; Østby, J.T.; Hughes, N.; Rijnbeek, P.; Bouvy, J.C. Common problems, common data model solutions: Evidence generation for health technology assessment. PharmacoEconomics 2021, 39, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Klann, J.G.; Joss, M.A.H.; Embree, K.; Murphy, S.N. Data model harmonization for the all of US research program: Transforming i2b2 data into the OMOP common data model. PLoS ONE 2019, 14, e0212463. [Google Scholar] [CrossRef] [Green Version]
- Schwalm, M.; Raoul, T.; Chu, D.; Shah, U.; Potdar, M.; Van Zandt, M.; Coffin, G.; Jouaville, S. Conversion of a french electronic medical record (Emr) database into the observational medical outcomes partnership common data model. Value Health 2017, 20, A741. [Google Scholar] [CrossRef]
- Kim, Y.H.; Ko, Y.-H.; Kim, S.; Kim, K. How closely is COVID-19 related to HCoV, SARS, and MERS? Clinical comparison of coronavirus infections and identification of risk factors influencing the COVID-19 severity using common data model (CDM). medRxiv 2021. [Google Scholar] [CrossRef]
- Williams, R.D.; Markus, A.F.; Yang, C.; Duarte Salles, T.; Falconer, T.; Jonnagaddala, J. Seek COVER: Development and validation of a personalized risk calculator for COVID-19 outcomes in an international network. MedRxiv 2020. [Google Scholar] [CrossRef]
- Warner, J.L.; Dymshyts, D.; Reich, C.G.; Gurley, M.J.; Hochheiser, H.; Moldwin, Z.H.; Belenkaya, R.; Williams, A.E.; Yang, P.C. HemOnc: A new standard vocabulary for chemotherapy regimen representation in the OMOP common data model. J. Biomed. Inform. 2019, 96, 103239. [Google Scholar] [CrossRef]
- Maier, C.; Lang, L.; Storf, H.; Vormstein, P.; Bieber, R.; Bernarding, J.; Herrmann, T.; Haverkamp, C.; Horki, P.; Laufer, J.; et al. Towards implementation of OMOP in a German university hospital consortium. Appl. Clin. Inform. 2018, 9, 54–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overhage, J.M.; Ryan, P.B.; Reich, C.G.; Hartzema, A.G.; Stang, P.E. Validation of a common data model for active safety surveillance research. J. Am. Med. Inform. Assoc. 2012, 19, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griebel, L.; Prokosch, H.-U.; Köpcke, F.; Toddenroth, D.; Christoph, J.; Leb, I.; Engel, I.; Sedlmayr, M. A scoping review of cloud computing in healthcare. BMC Med. Inform. Decis. Mak. 2015, 15, 17. [Google Scholar] [CrossRef] [Green Version]
- Aceto, G.; Persico, V.; Pescapé, A. Industry 4.0 and health: Internet of things, big data, and cloud computing for healthcare 4.0. J. Ind. Inf. Integr. 2020, 18, 100129. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Hsu, P.-Y. An empirical investigation of organizations’ switching intention to cloud enterprise resource planning: A cost-benefit perspective. Inf. Dev. 2017, 35, 290–302. [Google Scholar] [CrossRef]
- Sudhakar, G.; Karmouch, A.; Georganas, N. Design and performance evaluation considerations of a multimedia medical database. IEEE Trans. Knowl. Data Eng. 1993, 5, 888–894. [Google Scholar] [CrossRef]
- OHDSI. Publications—OHDSI. Available online: https://www.ohdsi.org/resources/publications/ (accessed on 30 March 2021).
- OHDSI. Observational Health Data Sciences and Informatics Repositories. GitHub.com. 2020. Available online: https://github.com/OHDSI/ (accessed on 3 April 2021).
- Liu, J.B.; Kelz, R.R. Types of hospitals in the United States. JAMA 2018, 320, 1074. [Google Scholar] [CrossRef]
- Bi, Y.; Zhu, D.; Cheng, J.; Zhu, Y.; Xu, N.; Cui, S.; Li, W.; Cheng, X.; Wang, F.; Hu, Y. The status of glycemic control: A cross-sectional study of outpatients with type 2 diabetes mellitus across primary, secondary, and tertiary hospitals in the jiangsu province of China. Clin. Ther. 2010, 32, 973–983. [Google Scholar] [CrossRef] [PubMed]
- OHDSI. Releases OHDSI/WebAPI GitHub. Available online: https://github.com/OHDSI/WebAPI/releases (accessed on 29 April 2021).
- Cioloca, C. Increasing database performance using indexes. Database Syst. J. 2011, 2, 13–22. Available online: https://pdfs.semanticscholar.org/1b96/91b7957ec3418c2b088177940de07483ee21.pdf (accessed on 29 April 2021).
- Myalapalli, V.K.; Savarapu, P.R. High performance SQL. In Proceedings of the 2014 Annual IEEE India Conference (INDICON), Pune, India, 11–13 December 2014. [Google Scholar]
- Myalapalli, V.K.; Totakura, T.P.; Geloth, S. Augmenting database performance via SQL tuning. In Proceedings of the 2015 International Conference on Energy Systems and Applications, Pune, India, 30 October–1 November 2015; pp. 13–18. [Google Scholar]
- Duan, S.; Thummala, V.; Babu, S. Tuning database configuration parameters with iTuned. Proc. VLDB Endow. 2009, 2, 1246–1257. [Google Scholar] [CrossRef] [Green Version]
- Kwan, E.; Lightstone, S.; Storm, A.; Wu, L.; Automatic Configuration for IBM DB2 Universal Database. Performance Technical Report. 2002. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.84.8661&rep=rep1&type=pdf (accessed on 29 March 2021).
- PostgreSQL. PostgreSQL: Documentation: 12: 19.4. Resource Consumption. Available online: https://www.postgresql.org/docs/12/runtime-config-resource.html (accessed on 29 March 2021).
- PostgreSQL. PostgreSQL: Documentation: 12: 19.7. Query Planning. Available online: https://www.postgresql.org/docs/12/runtime-config-query.html (accessed on 29 April 2021).
- PostgreSQL. PostgreSQL: Documentation: 12: 19.3. Connections and Authentication. Available online: https://www.postgresql.org/docs/12/runtime-config-connection.html (accessed on 29 April 2021).
- PostgreSQL. PostgreSQL: Documentation: 12: 19.5. Write ahead Log. Available online: https://www.postgresql.org/docs/12/runtime-config-wal.html (accessed on 29 April 2021).
- Shaik, B. Configure Your Database for Better Performance; PostgreSQL Configuration Apress: Berkeley, CA, USA, 2020; pp. 29–59. [Google Scholar] [CrossRef]
- Vasiliev, A. PGTune—Calculate Configuration for PostgreSQL Based on the Maximum Performance for a Given Hardware Configuration. Available online: https://pgtune.leopard.in.ua/#/ (accessed on 3 April 2021).
- Pgconfig.org. PGConfig—PostgreSQL Configuration Builder. Available online: https://www.pgconfig.org/ (accessed on 3 April 2021).
- PostgresSQL. PostgreSQL: Documentation: 13: Pgbench. Available online: https://www.postgresql.org/docs/current/pgbench.html (accessed on 30 March 2021).
- GitHub—Gregs1104/Pgbench-Tools: PostgreSQL Benchmarking Toolset. Available online: https://github.com/gregs1104/pgbench-tools (accessed on 27 May 2021).
- Managing PostgreSQL Connections. Available online: https://cloud.ibm.com/docs/databases-for-postgresql?topic=databases-for-postgresql-managing-connections&locale=en (accessed on 26 May 2021).

| Parameters | Description |
|---|---|
| Memory shared_buffers effective_cache_size work_mem maintenance_work_mem | Amount of memory the DB server uses for shared memory buffers Effective size of the disk cache available to a single query Maximum amount of memory to be used by a query operation Maximum amount of memory to be used by maintenance operations |
| Storage random_page_cost effective_io_concurrency | Planner’s estimate of the cost of a non-sequentially-fetched disk page Number of concurrent disk I/O operations executed simultaneously |
| Query and Connection default_statistics_target max_connections | Default statistics target for table columns without a column-specific target set Maximum number of concurrent connections to the database server |
| Write-Ahead Logging (WAL) min_wal_size max_wal_size checkpoint_completion_target wal_buffers | Minimum size to let the WAL grow during automatic checkpoints Maximum size to let the WAL grow during automatic checkpoints Target of checkpoint completion Shared memory amount used for WAL data yet to be written to disk |
| Worker Processes max_worker_processes max_parallel_workers_per_gather max_parallel_workers max_parallel_maintenance_workers | Maximum number of background processes that the system can support Maximum number of workers that can be started by a single Gather Maximum number of workers that can support for parallel operations Maximum number of parallel workers started by a single utility command |
| DA | OLTP | DW | |
|---|---|---|---|
| Hospital Type | Primary | Secondary Tertiary | Tertiary |
| Data amount | Small | Medium | Large |
| Number of Servers | One | One | Two or more |
| Server Design | Combined | Combined | Separated |
| Memory shared_buffers effective_cache_size work_mem maintenance_work_mem (max = 2 GB) | RAM (GB)/16 RAM (GB)/4 RAM (GB)/6/MC RAM (GB)/16 | RAM (GB)/4 RAM (GB)/4 × 3 RAM (GB)/MC RAM (GB)/16 | RAM (GB)/4 RAM (GB)/4 × 3 RAM (GB)/2/MC RAM (GB)/8 |
| WStorage random_page_cost effective_io_concurrency | HDD = 4.0, SSD = SAN = 1.1 HDD = 2, SSD = 200, SAN = 300 | ||
| Query and Connection default_statistics_target max_connections | 100 15 | 100 30 | 500 20 |
| Write-Ahead Logging min_wal_size max_wal_size checkpoint_completion_target wal_buffers (max = 16 MB) | 100 MB 2 GB 0.5 SB × 0.03 | 2 GB 8 GB 0.9 SB × 0.03 | 4 GB 16 GB 0.9 SB × 0.03 |
| Worker Processes max_worker_processes max_parallel_workers_per_gather max_parallel_workers max_parallel_maintenance_workers (max = 4) | CPU CPU/2 (max = 4) CPU CPU/2 | CPU CPU/2 (max = 4) CPU CPU/2 | Number of CPU CPU/2 CPU CPU/2 |
| DA | OLTP | DW | |
|---|---|---|---|
| Data source | CDM 1000 sample data per table from Seoul St. hospital | Eight disease-related data from Seoul St. hospital | All data from Seoul St. hospital, Yeouido St. hospital, and St. Vincent hospital |
| Number of Patient Data | 1000 | 131,411 | 7,956,745 |
| DB Size | 12 GB | 76 GB | 634 GB |
| Number of Servers | One | One | Two |
| Server Format | Physical Desktop | Cloud Server | Physical Server |
| Server Design | Combined | Combined | Separated |
| Memory shared_buffers effective_cache_size work_mem maintenance_work_mem | 32 GB 2 GB 8 GB RAM (GB)/6/MC 2 GB | 8 GB 2 GB 6 GB RAM (GB)/MC 500 MB | 128 GB 32 GB 96 GB RAM (GB)/2/MC 2 GB |
| Storage random_page_cost effective_io_concurrency | SSD 1.1 200 | SSD 1.1 200 | HDD 4.0 2 |
| Query and Connection default_statistics_target max_connections | 100 15 | 100 30 | 500 20 |
| Write-Ahead Logging min_wal_size max_wal_size checkpoint_completion_target wal_buffers | 100 MB 2 GB 0.5 6 MB | 2 GB 8 GB 0.9 6 MB | 4 GB 16 GB 0.9 16 MB |
| Worker Processes max_worker_processes max_parallel_workers_per_gather max_parallel_workersmax_parallel_maintenance_workers | 20 4 20 4 | 36 4 36 4 | 16 8 16 4 |
| DB Type | Scale | TPS | Avg. Latency | 90%< | Max Latency |
|---|---|---|---|---|---|
| DA | 1 | 34,758 | 0.081 | 0.088 | 0.323 |
| 10 | 34,131 | 0.082 | 0.090 | 0.668 | |
| 100 | 33,985 | 0.083 | 0.089 | 0.405 | |
| 1000 | 31,161 | 0.091 | 0.100 | 2.086 | |
| OLTP | 1 | 67,752 | 0.126 | 0.160 | 6.104 |
| 10 | 66,829 | 0.128 | 0.162 | 7.385 | |
| 100 | 65,800 | 0.130 | 0.165 | 8.161 | |
| 1000 | 62,535 | 0.138 | 0.176 | 7.512 | |
| DW | 1 | 47,046 | 0.102 | 0.113 | 2.780 |
| 10 | 48,422 | 0.096 | 0.106 | 0.985 | |
| 100 | 48,172 | 0.096 | 0.106 | 1.059 | |
| 1000 | 45,492 | 0.103 | 0.113 | 34.36 |
| DB Type | Client | TPS | Avg_Latency | 90%< | Max_Latency |
|---|---|---|---|---|---|
| DA | 1 | 13,778 | 0.071 | 0.075 | 0.220 |
| 2 | 24,531 | 0.080 | 0.087 | 1.049 | |
| 3 | 33,916 | 0.086 | 0.09 | 0.874 | |
| 4 | 43,942 | 0.089 | 0.096 | 0.597 | |
| 5 | 51,376 | 0.095 | 0.110 | 1.612 | |
| OLTP | 1 | 13,839 | 0.071 | 0.076 | 0.262 |
| 5 | 51,880 | 0.094 | 0.108 | 1.638 | |
| 10 | 75,988 | 0.128 | 0.144 | 4.134 | |
| 15 | 91,536 | 0.159 | 0.217 | 10.829 | |
| 20 | 95,403 | 0.200 | 0.284 | 19.591 | |
| DW | 1 | 13,488 | 0.074 | 0.076 | 5.986 |
| 3 | 32,761 | 0.091 | 0.095 | 15.557 | |
| 5 | 50,663 | 0.097 | 0.111 | 12.840 | |
| 7 | 63,109 | 0.108 | 0.121 | 6.617 | |
| 10 | 76,395 | 0.127 | 0.143 | 7.979 |
| Virtual Memory Status | |
|---|---|
| DA |  |
| OLTP |  |
| DW |  |
| . | Disk I/O Status |
|---|---|
| DA |  |
| OLTP |  |
| DW |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.M.; Ko, T.; Yang, Y.-s.; Park, S.J.; Choi, I.-Y.; Chang, D.-J. Establishment of the Optimal Common Data Model Environment for EMR Data Considering the Computing Resources of Medical Institutions. Appl. Sci. 2021, 11, 12056. https://doi.org/10.3390/app112412056
Kim TM, Ko T, Yang Y-s, Park SJ, Choi I-Y, Chang D-J. Establishment of the Optimal Common Data Model Environment for EMR Data Considering the Computing Resources of Medical Institutions. Applied Sciences. 2021; 11(24):12056. https://doi.org/10.3390/app112412056
Chicago/Turabian StyleKim, Tong Min, Taehoon Ko, Yoon-sik Yang, Sang Jun Park, In-Young Choi, and Dong-Jin Chang. 2021. "Establishment of the Optimal Common Data Model Environment for EMR Data Considering the Computing Resources of Medical Institutions" Applied Sciences 11, no. 24: 12056. https://doi.org/10.3390/app112412056






