Data Independent Acquisition Based Bi-Directional Deep Networks for Biometric ECG Authentication
Abstract
:1. Introduction
- Applied random segmentation and auto-correlation for various types of ECG data input independently, and to produce a reasonable quantity of training data from a raw signal [5].
- Proposed and compared the performance of generalization by designing 1D-CNN networks, bidirectional RNNs on both Long Short-Term Memory (LSTM) and Gated Recurrent Unit (GRU) cells.
2. Related Work
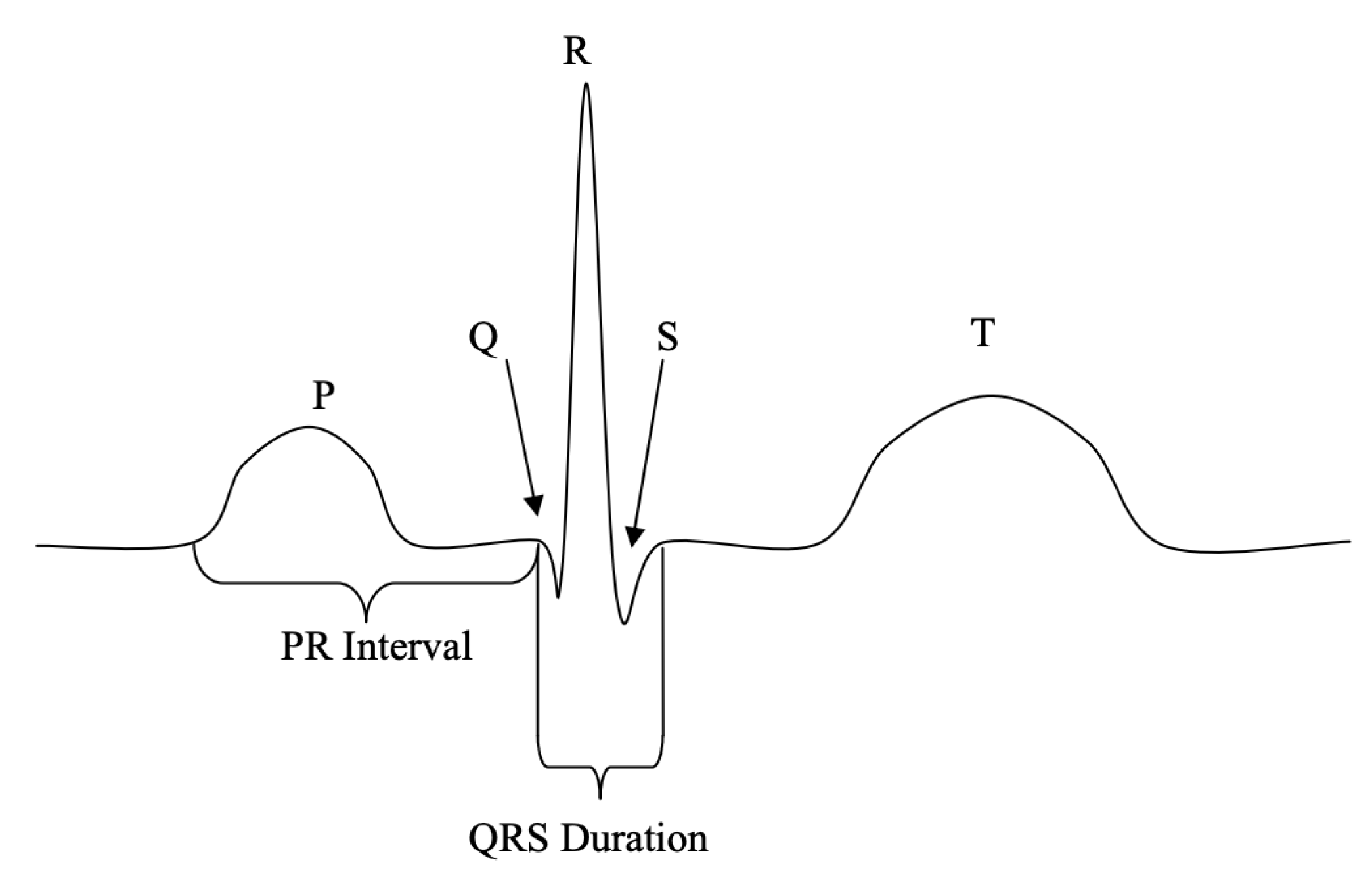
2.1. Fiducial Methods
2.2. Non-Fiducial Methods
2.2.1. Convolutional Neural Networks (CNN)
2.2.2. Recurrent Neural Networks (RNN)
2.2.3. Long Short-Term Memory (LSTM)
- Forget gate decides which part of long-term state should be omitted.
- Input gate controls which part should be added to long-term state.
- Output gate determines which part of should be read and outputs to and .
2.2.4. Gated Recurrent Unit(GRU)
3. Methodology
3.1. Data Argumentation Process
3.2. Extended Data Independent Acquisition
3.3. Models Overview
3.3.1. Proposed 1-D CNN Model
3.3.2. Proposed Bidirectional RNN Architectures
4. Experimental Results and Discussion
4.1. Network Training
4.2. System Evaluation
4.3. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stavroulakis, P.; Stamp, M. Handbook of Information and Communication Security; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Sufi, F.; Khalil, I.; Hu, J. ECG-based authentication. In Information and Communication Security; Springer: Berlin/Heidelberg, Germany, 2010; pp. 309–331. [Google Scholar]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M.; Gertych, A.; Tan, R.S. A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 2017, 89, 389–396. [Google Scholar] [CrossRef]
- Yildirim, Ö. A novel wavelet sequence based on a deep bidirectional LSTM network model for ECG signal classification. Comput. Biol. Med. 2018, 96, 189–202. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, D.; Zeng, X. HeartID: A multiresolution convolutional neural network for ECG-based biometric human identification in smart health applications. IEEE Access 2017, 5, 11805–11816. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. Real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef]
- Raman, P.; Ghosh, S.M. Classification of heart diseases based on ECG analysis using FCM and SVM methods. Int. J. Eng. Sci. 2016, 6, 673–6744. [Google Scholar]
- Sahoo, S.; Kanungo, B.; Behera, S.; Sabut, S. Multiresolution wavelet transform-based feature extraction and ECG classification to detect cardiac abnormalities. Measurement 2017, 108, 55–66. [Google Scholar] [CrossRef]
- Thomas, M.; Das, M.K.; Ari, S. Automatic ECG arrhythmia classification using dual tree complex wavelet-based features. AEU-Int. J. Electron. Commun. 2015, 69, 715–721. [Google Scholar] [CrossRef]
- Martis, R.J.; Acharya, U.R.; Min, L.C. ECG beat classification using PCA, LDA, ICA, and discrete wavelet transform. Biomed. Signal Process. Control. 2013, 8, 437–448. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning is applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, R.K.; Greff, K.; Schmidhuber, J. Highway networks. arXiv 2015, arXiv:1505.00387. [Google Scholar]
- Bengio, Y. Learning deep architectures for AI. Found. Trends Mach. Learn. 2009, 2, 1–127. [Google Scholar] [CrossRef]
- Yildirim, Ö.; Uçar, A.; Baloglu, U.B. Recognition of Real-world Texture Images under Challenging Conditions with Deep Learning. In Proceedings of the ASYU, Alanya, Turkey, 5–7 October 2017; Volume 52. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 60, 1097–1105. [Google Scholar] [CrossRef]
- Uçar, A.; Demir, Y.; Güzeliş, C. Object recognition and detection with deep learning for autonomous driving applications. Simulation 2017, 9, 759–769. [Google Scholar] [CrossRef]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Briefings Bioinf. 2017, 18, 851–869. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.H.; Hagiwara, Y.; Pang, W.; Lim, I.; Oh, S.L.; Adam, M.; San Tan, R.; Chen, M.; Acharya, U.R. Application of stacked convolutional and long short-term memory networks for accurate identification of CAD ECG signals. Comput. Biol. Med. 2018, 94, 19–26. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans. Biomed. Eng. 2016, 63, 664–675. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhang, L.; Wang, H.; Tang, J. Ballistocardiogram-based Person Identification and Authentication Using Recurrent Neural Networks. In Proceedings of the 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics, Beijing, China, 13–15 October 2018; pp. 1–5. [Google Scholar]
- Warrick, P.; Homsi, M.N. Cardiac Arrhythmia Detection from ECG Combining Convolutional and Long Short-term Memory Networks. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar]
- Zihlmann, M.; Perekrestenko, D.; Tschannen, M. Convolutional recurrent neural networks for electrocardiogram classification. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2018. [Google Scholar]
- Coşkun, M.; Uçar, A.; Yıldırım, Ö.; Demir, Y. Face recognition based on a CNN. In Proceedings of the 2017 International Conference on Modern Electrical and Energy Systems (MEES), Kremenchuk, Ukraine, 15–17 November 2017; pp. 376–379. [Google Scholar]
- Mostayed, A.; Luo, J.; Shu, X.; Wee, W. Classification of 12-Lead ECG signals with bi-directional LSTM network. arXiv 2018, arXiv:1811.02090. [Google Scholar]
- Tanaka, H.; Monahan, K.D.; Seals, D.R. Age-predicted maximal heart rate was revisited. J. Am. Coll. Cardiol. 2001, 37, 153–156. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Lugovaya, T.S. Biometric Human Identification Based on Electrocardiogram. Master’s Thesis, Faculty of Computing Technologies and Informatics, Electrotechnical University, Saint Petersburg, Russia, 2005. [Google Scholar]
- Moody, G.B.; Mark, R.G. Impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Biol. 2001, 2, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.P.; Pahlm, O.; Ringborn, M.; Warren, S.; Laguna, P.; Sornmo, L. The STAFF III Database: ECGs Recorded During Acutely Induced Myocardial Ischemia. Comput. Cardiol. 2017, 44, 1–4. [Google Scholar]
- Petrutiu, S.; Sahakian, A.V.; Swiryn, S. Abrupt changes in fibrillatory wave characteristics at the termination of paroxysmal atrial fibrillation in humans. Europace 2007, 9, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Moody, G.B. Spontaneous Termination of Atrial Fibrillation: A Challenge from PhysioNet and Computers. Comput. Cardiol. 2004, 31, 101–104. [Google Scholar]
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.C.; Mark, R.; Stanley, H.E. PhysioBank, PhysioToolkit, PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2010, 101, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Goldbergeret, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiological signals. Circulation 2000, 101, e215–e220. [Google Scholar]
- MathWorks. Remove Trends from Data. 2018. Available online: https://www.mathworks.com/help/signal/ug/remove-trends-from-data.html (accessed on 9 November 2020).
- Al Rahhal, M.M.; Bazi, Y.; AlHichri, H.; Alajlan, N.; Melgani, F.; Yager, R.R. Deep learning approach for the active classification of ECG signals. Inf. Sci. 2016, 345, 340–354. [Google Scholar] [CrossRef]
- Hamilton, P.S.; Tompkins, W.J. Quantitative investigation of QRS detection rules using the MIT/BIH Arrhythmia Database. IEEE Trans. Biomed. Eng. 1986, BME-33, 1157–1165. [Google Scholar] [CrossRef]
- Elman, J.L. Finding structure in time. Cognit. Sci. 1990, 14, 179–211. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.E.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Liu, F.; Zhou, X.; Wang, T.; Cao, J.; Wang, Z.; Wang, H.; Zhang, Y. Attention-based Hybrid LSTM-CNN Model for Arrhythmias Classification. In Proceedings of the 2019 International Joint Conference on Neural Networks, Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar]
- Nazi, Z.A.; Biswas, A.; Rayhan, M.A.; Azad Abir, T. Classification of ECG signals by dot residual LSTM network with data augmentation for anomaly detection. In Proceedings of the 22nd International Conference on Computer and Information Technology, Dhaka, Bangladesh, 18–20 December 2019. [Google Scholar]
- Ali Salah, A. Machine learning for biometrics. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Oxford, UK, 2010; Chapter 26; pp. 539–560. [Google Scholar]
- Yu, S.N.; Chou, K.T. Integration of independent component analysis and neural networks for ECG beat classification. Expert Syst. Appl. 2008, 34, 2841–2846. [Google Scholar] [CrossRef]
- Inan, O.T.; Giovangrandi, L.; Kovacs, G.T. Robust neural-network-based classification of premature ventricular contractions using wavelet transform and timing interval features. IEEE Trans. Biomed. Eng. 2006, 53, 2507–2515. [Google Scholar] [CrossRef] [PubMed]












| Layers | Kernel Size | Stride | Padding | Input Size | Output Size |
|---|---|---|---|---|---|
| 1 | 5 | 2 | 2 | 750 | 375 |
| 2 | 2 | 2 | 0 | 375 | 187 |
| 3 | 5 | 2 | 2 | 187 | 94 |
| 4 | 2 | 2 | 0 | 94 | 47 |
| Actual | Positives (1) | Negatives (0) | |
|---|---|---|---|
| Predicted | |||
| Positives (1) | TP | FP | |
| Negatives (0) | FN | TN | |
| Type of Model | Input Sequence Length (Number of Heartbeats) | Accuracy |
|---|---|---|
| Proposed 1D-CNN | 3 | 0.925 |
| 9 | 0.911 | |
| RNN + LSTM | 3 | 0.965 |
| 9 | 0.971 | |
| RNN + GRU | 3 | 0.952 |
| 9 | 0.978 | |
| Proposed BLSTM | 3 | 0.982 |
| 9 | 0.993 | |
| Proposed BGRU | 3 | 0.921 |
| 9 | 0.983 |
| Dataset | Method | Acc | Sen | Spe | Ppr | F1 | FM |
|---|---|---|---|---|---|---|---|
| ECG-ID | Zhang et al. [5] | 98.3 | 75.2 | 98.3 | 99.8 | 85.8 | 85.8 |
| Mostayed et al. [24] | 98.4 | 93 | 97.5 | 98.2 | 95.5 | 95.5 | |
| Zabir Al et al. [41] | 90.3 | 94.2 | 95.6 | 93.1 | 93.6 | 93.6 | |
| Fan Liu et al. [40] | 98.3 | 95.7 | 98.2 | 99.2 | 97.4 | 97.4 | |
| Proposed | 99.3 | 98.3 | 99.2 | 99.4 | 98.84 | 98.84 | |
| MIT-BIH ECG | Zhang et al. [5] | 98.6 | 95.2 | 97.3 | 89.5 | 92.2 | 92.2 |
| Mostayed et al. [24] | 99.4 | 95.8 | 99.7 | 97.8 | 96.8 | 96.8 | |
| Zabir Al et al. [41] | 80.1 | 82.8 | 89.1 | 84.4 | 83.59 | 83.59 | |
| Fan Liu et al. [40] | 80.1 | 82.8 | 89.1 | 84.4 | 83.59 | 83.59 | |
| Proposed | 99.5 | 99.2 | 98.8 | 99.2 | 99.2 | 99.2 | |
| STAFF-III | Zhang et al. [5] | 98.1 | 86.6 | 99.3 | 96.2 | 91.2 | 91.2 |
| Mostayed et al. [24] | 98.7 | 91.3 | 97.4 | 97.8 | 94.4 | 94.4 | |
| Zabir Al et al. [41] | 89.4 | 88.7 | 92.3 | 89.6 | 89.14 | 89.14 | |
| Fan Liu et al. [40] | 98.6 | 94.6 | 99.2 | 98.5 | 96.5 | 96 | |
| Proposed | 99.3 | 95.5 | 97.9 | 99.2 | 97.31 | 97.07 | |
| LT-AF | Zhang et al. [5] | 97.6 | 95.8 | 95.3 | 96.4 | 96 | 96 |
| Mostayed et al. [24] | 99.1 | 99.4 | 98.7 | 98.5 | 98.6 | 98.6 | |
| Zabir Al et al. [41] | 89.4 | 88.4 | 90.2 | 93.2 | 90.7 | 90.76 | |
| Fan Liu et al. [40] | 99.4 | 99.2 | 98.4 | 97.6 | 98.39 | 98.39 | |
| Proposed | 99.2 | 99.6 | 98.2 | 99.5 | 99.5 | 99.5 | |
| AFDB | Zhang et al. [5] | 89.6 | 91.3 | 90.5 | 88.7 | 90 | 90 |
| Mostayed et al. [24] | 83.1 | 88.4 | 89.3 | 88.3 | 88.34 | 88.34 | |
| Zabir Al et al. [41] | 79.1 | 81.2 | 88.4 | 86.5 | 83.8 | 83.8 | |
| Fan Liu et al. [40] | 90.2 | 89.8 | 92.5 | 90.3 | 90.4 | 90.4 | |
| Proposed | 98.5 | 97.3 | 99.1 | 98.6 | 97.94 | 97.94 | |
| AHA | Zhang et al. [5] | 84.3 | 81.5 | 83.6 | 88.4 | 84.87 | 84.87 |
| Mostayed et al. [24] | 85.4 | 86.4 | 88.3 | 83.4 | 84.8 | 84.8 | |
| Zabir Al et al. [41] | 76.5 | 81.2 | 83.2 | 88.2 | 84.62 | 84.62 | |
| Fan Liu et al. [40] | 87.5 | 88.3 | 89.2 | 83.6 | 85.91 | 85.91 | |
| Proposed | 97.3 | 96.4 | 97.2 | 98.4 | 97.39 | 97.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lynn, H.M.; Kim, P.; Pan, S.B. Data Independent Acquisition Based Bi-Directional Deep Networks for Biometric ECG Authentication. Appl. Sci. 2021, 11, 1125. https://doi.org/10.3390/app11031125
Lynn HM, Kim P, Pan SB. Data Independent Acquisition Based Bi-Directional Deep Networks for Biometric ECG Authentication. Applied Sciences. 2021; 11(3):1125. https://doi.org/10.3390/app11031125
Chicago/Turabian StyleLynn, Htet Myet, Pankoo Kim, and Sung Bum Pan. 2021. "Data Independent Acquisition Based Bi-Directional Deep Networks for Biometric ECG Authentication" Applied Sciences 11, no. 3: 1125. https://doi.org/10.3390/app11031125
APA StyleLynn, H. M., Kim, P., & Pan, S. B. (2021). Data Independent Acquisition Based Bi-Directional Deep Networks for Biometric ECG Authentication. Applied Sciences, 11(3), 1125. https://doi.org/10.3390/app11031125







