Early Effect of Pine Biochar on Peach-Tree Planting on Microbial Community Composition and Enzymatic Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Soil Sampling
2.2. Soil Chemical Analysis
2.3. Soil Water-Stable Aggregate Analysis and Calculation
2.4. Fatty Acid Methyl Ester (EL-FAME) Analysis
2.5. Soil Enzymatic Activities
2.6. Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties
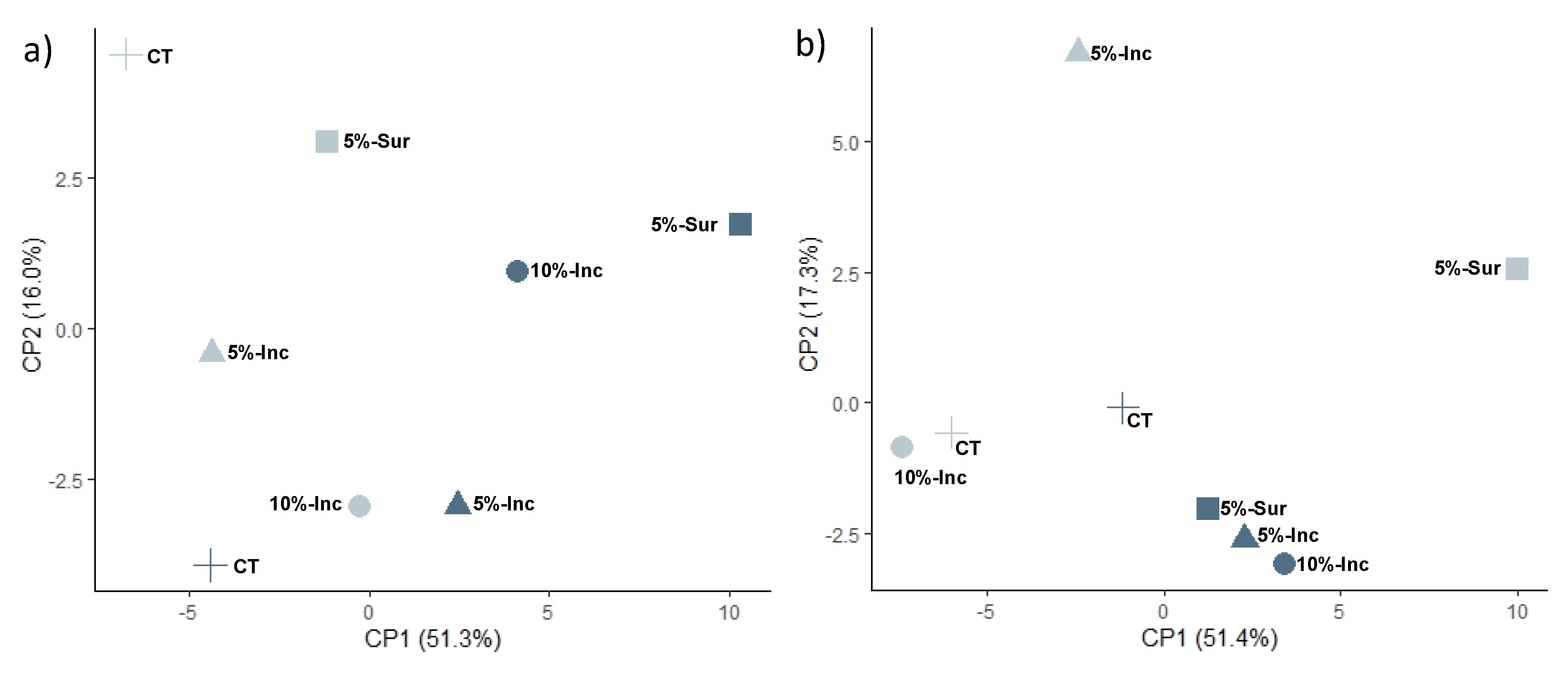
3.2. Soil Microbial Community Structure Based on FAME Analysis
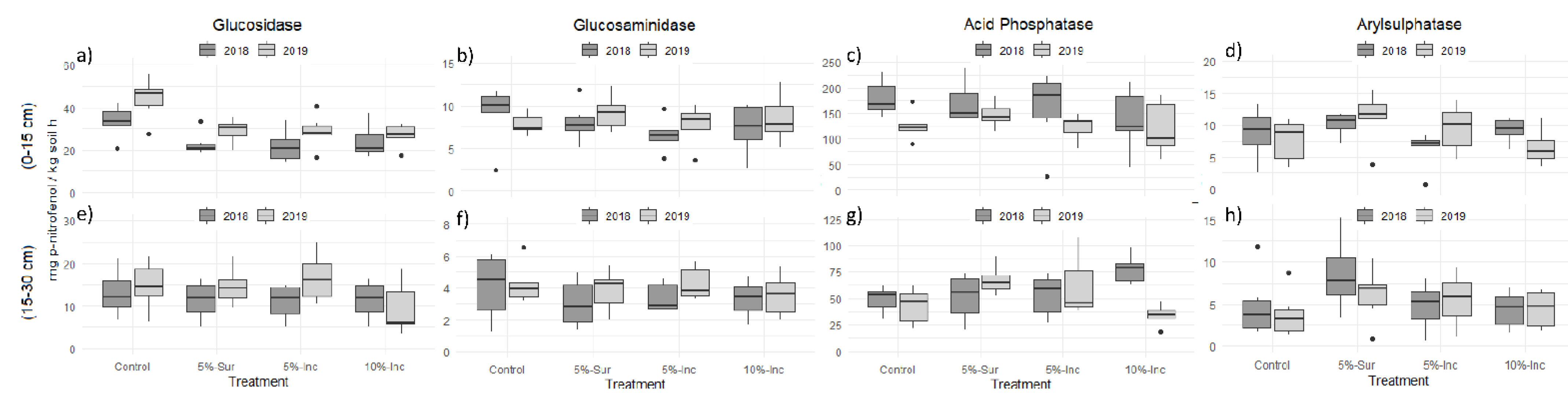
3.3. Soil Nutrient Cycling Based on Enzymes Activities
3.4. Correlations and RDA Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lehmann, J.; Joseph, S. Biochar for Environmental Management: Science and Technology; Earthscan Books Ltd.: London, UK, 2009; pp. 1–12. [Google Scholar]
- Palansooriya, K.N.; Wong, J.T.; Hashimoto, Y.; Huang, L.; Rinklebe, J.; Chang, S.X.; Bolan, N.; Wang, H.; Ok, Y.S. Response of microbial communities to biochar-amended soils: A critical review. Biochar 2019, 1, 3–22. [Google Scholar] [CrossRef] [Green Version]
- Guo, M. The 3R Principles for Applying Biochar to Improve Soil Health. Soil Syst. 2020, 4, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Chen, B.; Zhu, L.; Xing, B. Effects and mechanisms of biochar-microbe interactions in soil improvement and pollution remediation: A review. Environ. Pollut. 2017, 227, 98–115. [Google Scholar] [CrossRef] [PubMed]
- Verheijen, F.; Jeffery, A.C.; Bastos, A.C.; van der Velde, M.; Diafas, I. Biochar Application to Soil: A Critical Scientific Review of Effects on Soil Properties, Processes and Functions; EUR 24099 EN; Office for the Official Publications of the European Communities: Luxembourg, 2010. [Google Scholar]
- Yuan, P.; Wang, J.; Pan, Y.; Shen, B.; Wu, C. Review of biochar for the management of contaminated soil: Preparation, application, and prospect. Sci. Total Environ. 2019, 659, 473–490. [Google Scholar] [CrossRef]
- Sun, K.; Ro, K.; Guo, M.X.; Novak, J.; Mashayekhi, H.; Xing, B.S. Sorption of bisphenol A, 17α-ethinyl estradiol and phenanthrene on thermally and hydrothermally produced biochars. Bioresour. Technol. 2011, 102, 5757–5763. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.; Rillig, M.C.; Thies, J.; Masiello, C.A.; Hockaday, W.C.; Crowley, D. Biochar effects on soil biota—A review. Soil Biol. Biochem. 2011, 43, 1812–1836. [Google Scholar] [CrossRef]
- Liang, Q.; Chen, H.Q.; Gong, Y.S.; Yang, H.F.; Fan, M.S.; Kuzyakov, Y. Effects of 15 years of manure and mineral fertilizers on enzyme activities in particle-size fractions in a North China Plain soil. Eur. J. Soil Biol. 2014, 60, 112–119. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Subbotina, I.; Chen, H.; Bogomolova, I.; Xu, X. Black carbon decomposition and incorporation into soil microbial biomass estimated by 14C labeling. Soil Biol. Biochem. 2009, 41, 210–219. [Google Scholar] [CrossRef]
- Wang, D.; Fonte, S.; Parikh, S.; Six, J.; Scow, K. Biochar additions can enhance soil structure and the physical stabilization of C in aggregates. Geoderma 2017, 303, 110–117. [Google Scholar] [CrossRef] [Green Version]
- Aciego Pietri, J.C.; Brookes, P.C. Relationships between soil pH and microbial properties in a UK arable soil. Soil Biol. Biochem. 2008, 40, 1856–1861. [Google Scholar] [CrossRef]
- Laird, D.A. The charcoal vision: A win-win scenario for simultaneously producing bioenergy, permanently sequestering carbon, while improving soil and water quality. Agron. J. 2008, 100, 178–181. [Google Scholar]
- Farrell, M.; Kuhn, T.K.; Macdonald, L.M.; Maddern, T.M.; Murphy, D.V.; Hall, P.A.; Singh, B.P.; Baumann, K.; Krull, E.S.; Baldock, J.A. Microbial utilization of biochar-derived carbon. Sci. Total Environ. 2013, 465, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Quilliam, R.; Glanville, H.; Wade, S.; Jones, D. Life in the ‘charosphere’—Does biochar in agricultural soil provide a significant habitat for microorganisms? Soil Biol. Biochem. 2013, 65, 287–293. [Google Scholar] [CrossRef]
- Li, C.; Cano, A.; Acosta-Martinez, V.; Veum, K.; Moore-Kucera, J. A comparison between fatty acid methyl ester profiling methods (PLFA and EL-FAME) as soil health indicators. Soil Sci. Soc. Am. J. 2020, 84, 1153–1169. [Google Scholar] [CrossRef]
- Karlinski, L.; Ravnskov, S.; Rudawska, M. Soil Microbial Biomass and Community Composition Relates to Poplar Genotypes and Environmental Conditions. Forests 2020, 11, 262. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Martínez, V.; Dowd, S.E.; Bell, C.W.; Lascano, R.; Booker, J.D.; Zobeck, T.M.; Upchurch, D.R. Microbial Community Composition as Affected by Dryland Cropping Systems and Tillage in a Semiarid Sandy Soil. Diversity 2010, 2, 910–931. [Google Scholar] [CrossRef] [Green Version]
- Brinton, W.F. Phospholipid Fatty Acid (PLFA) Analysis: A Robust Indicator for Soil Health? Agri. Res. Technol. Open Access J. 2020, 24, 556281. [Google Scholar]
- Gardner, T.; Acosta-Martinez, V.; Senwo, Z.; Dowd, S.E. Soil Rhizosphere Microbial Communities and Enzyme Activities under Organic Farming in Alabama. Diversity 2011, 3, 308–328. [Google Scholar] [CrossRef]
- Luo, D.; Chen, R.-M.; Liu, S.; Shi, Z.-M.; Feng, Q.-H. Responses of Soil Microbial Community Composition and Enzyme Activities to Land-Use Change in the Eastern Tibetan Plateau, China. Forests 2020, 11, 483. [Google Scholar] [CrossRef]
- Cavigelli, M.A.; Robertson, G.P.; Klug, M.J. Fatty acid methyl ester (FAME) profiles as measures of soil microbial community structure. Plant Soil 1995, 170, 99–113. [Google Scholar] [CrossRef]
- Chen, J.; Li, S.; Liang, C.; Xu, Q.; Li, Y.; Qin, H.; Fuhrmann, J. Response of microbial community structure and function to short-term biochar amendment in an intensively managed bamboo (Phyllostachys praecox) plantation soil: Effect of particle size and addition rate. Sci. Total Environ. 2017, 574, 24–33. [Google Scholar] [CrossRef]
- Ameloot, N.; De Neve, S.; Jegajeevagan, K.; Yildiz, G.; Buchan, D.; Funkuin, Y.N.; Prins, W.; Bouckaert, L.; Sleutel, S. Short-term CO2 and N2O emissions and microbial properties of biochar amended sandy loam soils. Soil Biol. Biochem. 2013, 57, 401–410. [Google Scholar] [CrossRef]
- Mitchell, P.J.; Simpson, A.J.; Soong, R.; Simpson, M.J. Shifts in microbial community and water-extractable organic matter composition with biochar amendment in a temperate forest soil. Soil Biol. Biochem. 2015, 81, 244–254. [Google Scholar] [CrossRef]
- Zhang, L.; Jing, Y.; Xiang, Y.; Zhang, R.; Lu, H. Response of soil microbial community structure changes and activities to biochar addition: A meta-analysis. Sci. Total Environ. 2018, 643, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Martinez, V.; Moore-Kucera, J.; Cotton, J.; Gardner, T.; Wester, D. Soil enzyme activities during the 2011 Texas record drought/heatwave and implications to biogeochemical cycling and organic matter dynamics. Appl. Soil Ecol. 2014, 75, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Cotrufo, M.F.; Wallenstein, M.D.; Boot, C.M.; Denef, K.; Paul, E. The Microbial Efficiency-Matrix Stabilization (MEMS) framework integrates plant litter decomposition with soil organic matter stabilization: Do labile plant inputs form stable soil organic matter? Glob. Chang. Biol. 2013, 19, 988–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallenbach, C.M.; Frey, S.D.; Grandy, A.S. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 2016, 7, 13630. [Google Scholar] [CrossRef]
- Dick, R.P. (Ed.) Methods of Soil Enzymology; Soil Science Society of America: Madison, WI, USA, 2011; p. 395. [Google Scholar]
- Foster, E.; Hansen, N.; Wallenstein, M.; Cotrufo, M.F. Biochar and manure amendments impacts soil nutrients and microbial enzymatic activities in a semi-arid irrigated maize cropping system. Agric. Ecosyst. Environ. 2016, 233, 404–414. [Google Scholar] [CrossRef] [Green Version]
- Bayley, V.L.; Fansler, S.L.; Smith, J.L.; Bolton, H. Reconciling apparent variability in effects of biochar amendment on soil enzyme activities by assay optimization. Soil Biol. Biochem. 2011, 43, 296–301. [Google Scholar] [CrossRef]
- Frene, J.P.; Frazier, M.; Liu, S.; Clark, B.; Parker, M.; Gardner, T.G. Early response of organic matter dynamics to pine-biochar in sandy soil under peach trees. Agrosyst. Geosci. Environ. 2020, 3, e200094. [Google Scholar] [CrossRef]
- Culman, S.W.; Snapp, S.S.; Freeman, M.A.; Schipanski, M.E.; Beniston, J.; Lal, R. Permanganate oxidizable carbon reflects a processed soil fraction that is sensitive to management. Soil Sci. Soc. Am. J. 2012, 76, 494–504. [Google Scholar] [CrossRef] [Green Version]
- Weil, R.R.; Islam, K.R.; Stine, M.A.; Gruver, J.B.; Samson-Liebig, S.E. Estimating active carbon for soil quality assessment: A simplified method for laboratory and field use. Am. J. Altern. Agric. 2003, 18, 3–17. [Google Scholar]
- Elliott, E.T. Aggregate structure and carbon, nitrogen, and phosphorus in native and cultivated soils. Soil Sci. Soc. Am. J. 1986, 50, 627–633. [Google Scholar] [CrossRef]
- van Bavel, C.H.M. Mean Weight-Diameter of Soil Aggregates as a Statistical Index of Aggregation. Soil Sci. Soc. Am. J. 1950, 14, 20–23. [Google Scholar] [CrossRef] [Green Version]
- Schutter, M.E.; Dick, R.P. Comparison of fatty acid methyl ester (FAME) methods for characterizing microbial communities. Soil Sci. Soc. Am. J. 2000, 64, 1659–1668. [Google Scholar] [CrossRef]
- Zelles, L. Fatty Acid Patterns of Microbial Phospholipids and Lipopolysaccharides; Schinner, F., Öhlinger, R., Kandeler, E., Margesin, R., Eds.; Methods in Soil Biology; Springer: Berlin, Germany, 1996; pp. 80–93. [Google Scholar]
- Zelles, L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere 1997, 35, 275–294. [Google Scholar] [CrossRef]
- Zelles, L. Fatty acid patterns of microbial phospholipids and lipopolysaccharides in the characterization of microbial communities in soil: A review. Biol. Fertil. Soils 1999, 29, 111–129. [Google Scholar] [CrossRef]
- Frostegard, A.; Baath, E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soil 1996, 22, 59–65. [Google Scholar] [CrossRef]
- Li, C.; Fultz, L.; Moore-Kucera, J.; Acosta-Martinez, V.; Karkala, M.; Weindorf, D.C. Soil microbial community restoration in Conservation Reserve Program semi-arid grasslands. Soil Biol. Biochem. 2018, 118, 166–177. [Google Scholar] [CrossRef]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis, Part 2. Microbiological and Biochemical Properties; Bottomley, P.S., Angle, J.S., Weaver, R.W., Eds.; SSSA Book Series; Soil Science Society of America: Madison, WI, USA, 1994; pp. 775–833. [Google Scholar]
- Parham, J.A.; Deng, S.P. Detection, quantification and characterization of β-glucosaminidase activity in soil. Soil Biol. Biochem. 2000, 32, 1183–1190. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: http://www.R-project.org/ (accessed on 12/9/2019).
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Simpson, G.L.; Stevens, M.H.H. Vegan: Community Ecology Package. Version 1 11 14. 2008. Available online: http://vegan.r-forge.r-project.org (accessed on 12/9/2019).
- Kindler, R.; Siemen, J.; Kaiser, K. Dissolved carbon leaching from soil is a crucial component of the net ecosystem carbon balance. Glob. Chang. Biol. 2011, 17, 1167–1185. [Google Scholar] [CrossRef] [Green Version]
- Spokas, K.A.; Baker, J.M.; Reicosky, D.C. Ethylene: Potential key for biochar amendment impacts. Plant Soil 2010, 1, 443–452. [Google Scholar] [CrossRef]
- Santos, F.; Torn, M.S.; Bird, J.A. Biological degradation of pyrogenic organic matter in temperate forest soils. Soil Biol. Biochem. 2012, 51, 115–124. [Google Scholar] [CrossRef]
- Luo, Y.; Dungait, J.A.; Zhao, X.; Brookes, P.; Durenkamp, M.; Li, G.; Lin, Q. Pyrolysis temperature during biochar production alters its subsequent utilization by microorganisms in an acid arable soil. Land Degrad. Dev. 2018, 29, 2183–2188. [Google Scholar] [CrossRef]
- Jiang, X.; Denef, K.; Stewart, C.; Cotrufo, M.F. Controls and dynamics of biochar decomposition and soil microbial abundance, composition, and carbon use efficiency during long-term biochar-amended soil incubations. Biol. Fertil. Soils 2016, 52, 1–14. [Google Scholar] [CrossRef]
- Costa, O.; Raaijmakers, J.M.; Kuramae, E.E. Microbial Extracellular Polymeric Substances: Ecological Function and Impact on Soil Aggregation. Front. Microbial. 2018, 9, 1636. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Wang, S.; Tian, L.; Li, S.; Shen, Y.; Tian, C. Long-term biochar application influences soil microbial community and its potential roles in semi-arid farmland. Appl. Soil Ecol. 2017, 117, 10–15. [Google Scholar] [CrossRef]
- Schimel, J.; Balser, T.C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef]
- Fanin, N.; Kardol, P.; Farrell, M.; Nilsson, M.C.; Gundale, M.J.; Wardle, D.A. The ratio of Gram-positive to Gram-negative bacterial PLFA markers as an indicator of carbon availability in organic soils. Soil Biol. Biochem. 2019, 128, 111–114. [Google Scholar] [CrossRef]
- Marschner, P. Soil Microbial Community Structure and Function Assessed by FAME, PLFA and DGGE—Advantages and Limitations. In Advanced Techniques in Soil Microbiology; Varma, A., Oelmüller, R., Eds.; Springer: Berlin, Germany, 2007; pp. 181–200. ISBN 978-3-540-70864-3. [Google Scholar]
- Bossio, D.A.; Scow, K.M.; Gunapala, N.; Graham, K.J. Determinants of soil microbial communities: Effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb. Ecol. 1998, 36, 1–12. [Google Scholar] [CrossRef]
- Wang, X.; Song, S.; Liang, G.; Zhang, Q.; Ai, C.; Zhou, W. Maize biochar addition rate influence soil enzyme activity and microbial community composition in fluvo-aqui soil. Appl. Soil Ecol. 2015, 96, 265–272. [Google Scholar] [CrossRef]
- Crim, P.M.; Cumming, R.J. Extracellular Soil Enzyme Activities in High-Elevation Mixed Red Spruce Forests in Central Appalachia, USA. Forests 2020, 11, 468. [Google Scholar] [CrossRef] [Green Version]
- Nannipieri, P.; Giagnoni, L.; Renella, G.; Puglisi, E.; Ceccanti, B.; Masciandaro, G.; Fornasier, F.; Moscatelli, M.; Marinari, S. Soil enzymology: Classical and molecular approachs. Soil Fertil. Biol. 2012, 48, 743–762. [Google Scholar] [CrossRef]
- Brookes, P.C.; Cayuela, M.L.; Contin, M.; Nobili, M.D.; Kemmitt, S.J.; Mondini, C. The mineralization of fresh and humified soil organic matter by the soil microbial biomass. Waste Manag. 2008, 28, 716–722. [Google Scholar] [CrossRef]
- Paz-Ferreiro, J.; Gascó, G; Gutiérrez, B.; Méndez, A. Soil biochemical activities and the geometric mean of enzyme activities after application of sewage sludge and sewage sludge biochar to soil. Biol. Fertil. Soils 2012, 48, 511–517. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Lauber, C.L.; Weintraub, M.N.; Ahmed, B.; Allison, S.D.; Crenshaw, C.; Contosta, A.R.; Cusack, D.; Frey, S.; Gallo, M.E.; et al. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Burns, R.G. Enzyme activity in soil: Locations and a possible role in microbial ecology. Soil Biol. Biochem. 1982, 14, 423–427. [Google Scholar] [CrossRef]
- Bowles, T.M.; Acosta-Martinez, V.; Calderon, F.; Jackson, L.E. Soil enzymatic activities, microbial communities, and carbon and nitrogen availability in organic agroecosystem across an intensively managed agricultural landscape. Soil Biol. Biochem. 2014, 68, 252–262. [Google Scholar] [CrossRef]
- Brantley, K.Y.; Savin, M.C.; Brye, K.R.; Longer, D.E. Pine woodchip biochar impact on soil nutrient concentrations and corn yield in a silt loam in the mid-southern U.S. Agriculture 2015, 5, 30–47. [Google Scholar] [CrossRef] [Green Version]
- Demisie, W.; Lio, Z.; Zhang, M. Effect of biochar on carbon fractions and enzyme activity of red soil. Catena 2014, 121, 214–221. [Google Scholar] [CrossRef]


| Year | Treatments | Total FAME | Total Bacteria | Total Fungi | Gram Positive | Gram Negative | Actinomycetes | Saprophytic Fungi | AMF |
|---|---|---|---|---|---|---|---|---|---|
| 0–15 cm | |||||||||
| 2018 | Control | 94.04 (13.3) | 23.25 (3.1) | 14.88 (3.8) | 13.44 (1.6) | 3.63 (0.6) | 6.17 (0.9) | 13.18 (3.1) | 1.92 (0.9) |
| 5%-Sur | 104.15 (4.7) | 24.33 (2.1) | 16.04 (2.6) | 14.57 (1.1) | 3.59 (0.3) | 6.16 (0.7) | 13.92 (2.4) | 2.12 (0.2) | |
| 5%-Inc | 90.38 (11.9) | 22.45 (2.9) | 12.15 (2.0) | 13.25 (1.6) | 3.25 (0.5) | 5.94 (0.8) | 10.44 (1.6) | 1.71 (0.9) | |
| 10%-Inc | 107.44 (12.2) | 27.79 (3.5) | 17.54 (3.0) | 16.30 (2.0) | 4.19 (0.5) | 7.29 (1.0) | 15.11 (2.7) | 2.43 (0.4) | |
| 2019 | Control | 96.14 (20.0) | 23.59 (3.5) | 18.73 (3.0) | 13.48 (2.2) | 3.60 (0.6) | 6.50 (0.6) | 16.45 (3.2) | 2.27 (0.4) |
| 5%-Sur | 139.52 (10.4) | 33.47 (2.9) | 26.24 (2.2) | 18.97 (1.9) | 5.11 (0.4) | 9.39 (0.6) | 22.45 (2.4) | 3.79 (0.6) | |
| 5%-Inc | 109.24 (12.9) | 27.39 (2.8) | 20.02 (2.0) | 15.56 (1.7) | 3.96 (0.5) | 7.87 (0.6) | 16.57 (2.2) | 3.45 (0.5) | |
| 10%-Inc | 115.69 (12.8) | 28.39 (3.2) | 18.98 (1.8) | 16.10 (1.9) | 4.43 (0.6) | 7.85 (0.7) | 15.76 (1.8) | 3.22 (0.9) | |
| ANOVA | Treat | n.s. | n.s. | n.s. | 0.0338 | n.s. | n.s. | n.s. | n.s. |
| Year | 0.0333 | 0.0222 | 0.00248 | n.s. | 0.0473 | 0.0017 | 0.000013 | 0.0073 | |
| TxY | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | |
| 15–30 cm | |||||||||
| 2018 | Control | 38.08 (22.6) | 8.26 (2.8) | 6.56 (3.1) | 5.01 (3.3) | 1.48 (1.0) | 1.76 (0.6) | 5.73 (3.6) | 0.83 (0.1) |
| 5%-Sur | 46.87 (35.1) | 10.29 (4.5) | 6.68 (2.5) | 5.50 (1.1) | 1.70 (0.5) | 3.07 (0.8) | 5.75 (3.1) | 0.93 (0.3) | |
| 5%-Inc | 52.09 (33.5) | 9.02 (8.8) | 6.12 (3.5) | 5.44 (2.6) | 1.45 (0.6) | 2.16 (0.9) | 5.48 (2.5) | 0.64 (0.2) | |
| 10%-Inc | 33.39 (35.7) | 8.23 (4.2) | 5.12 (4.0) | 4.68 (2.1) | 1.41 (0.3) | 2.13 (2.9) | 4.60 (2.6) | 0.51 (0.4) | |
| Control | 56.88 (25.1) | 10.73 (4.4) | 11.99 (2.4) | 6.07 (2.1) | 1.80 (1.0) | 2.85 (1.3) | 10.58 (3.0) | 1.40 (0.8) | |
| 5%-Sur | 57.12 (27.3) | 13.83 (5.1) | 9.83 (1.3) | 7.78 (2.7) | 2.24 (1.1) | 3.80 (1.6) | 7.97 (2.4) | 1.85 (0.2) | |
| 2019 | 5%-Inc | 58.18 (24.1) | 13.60 (4.1) | 10.62 (1.1) | 7.63 (1.9) | 2.16 (1.0) | 3.80 (1.3) | 9.03 (3.0) | 1.58 (0.2) |
| 10%-Inc | 63.91 (21.9) | 15.31 (5.9) | 11.68 (3.3) | 8.59 (3.0) | 2.50 (1.2) | 4.20 (1.8) | 9.72 (3.9) | 1.96 (0.7) | |
| ANOVA | Treat. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. |
| Year | 0.0115 | 0.0004 | 0.0005 | 0.0015 | 0.0004 | 0.0001 | 0.0073 | 0.0001 | |
| TxY | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | |
| Year | Treatment | F:B Ratio | G+/G− Ratio | AMF/SF Ratio | AMF/TB Ratio | SF/TB Ratio | cy/pre Ratio | Shannon Index |
|---|---|---|---|---|---|---|---|---|
| 0–15 cm | ||||||||
| 2018 | Control | 0.64 (0.07) | 3.73 (0.11) | 0.15 (0.03) | 0.08 (0.01) | 0.57 (0.07) | 0.58 (0.01)a | 131.54 (35.45) |
| 5%-Sur | 0.66 (0.1) | 4.21 (0.27) | 0.17 (0.03) | 0.09 (0.01) | 0.58 (0.1) | 0.49 (0.04)b | 150.99 (27.06) | |
| 5%-Inc | 0.54 (0.04) | 4.07 (0.02) | 0.17 (0.01) | 0.08 (0.01) | 0.46 (0.03) | 0.47 (0.01)b | 110.41 (85.86) | |
| 10%-Inc | 0.64 (0.04) | 3.9 (0.09) | 0.17 (0.02) | 0.09 (0.01) | 0.55 (0.05) | 0.47 (0.01)b | 154.20 (31.00) | |
| 2019 | Control | 0.76 (0.1) | 3.84 (0.17) | 0.15 (0.01) | 0.09 (0.01) | 0.66 (0.1) | 0.48 (0.02)b | 134.01 (68.39) |
| 5%-Sur | 0.78 (0.03) | 3.77 (0.21) | 0.18 (0.02) | 0.11 (0.01) | 0.66 (0.04) | 0.41 (0.02)c | 229.88 (43.62) | |
| 5%-Inc | 0.74 (0.05) | 4.07 (0.29) | 0.21 (0.02) | 0.13 (0.01) | 0.61 (0.05) | 0.41 (0.01)c | 154.95 (55.31) | |
| 10%-Inc | 0.67 (0.07) | 3.69 (0.12) | 0.21 (0.04) | 0.11 (0.02) | 0.56 (0.06) | 0.41 (0.01)c | 175.93 (81.82) | |
| ANOVA | Treat | n.s. | n.s. | n.s. | n.s. | n.s. | 0.0001 | n.s. |
| Year | 0.0232 | n.s. | n.s. | 0.0002 | n.s. | 0.0001 | 0.001 | |
| TxY | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | |
| 15–30 cm | ||||||||
| 2018 | Control | 0.77 (0.05) b | 3.35 (0.09) | 0.15 (0.02) | 0.1 (0.01) | 0.67 (0.05) b | 0.52 (0.03) | 24.40 (6.48) |
| 5%-Sur | 0.72 (0.04) b | 3.5 (0.22) | 0.23 (0.05) | 0.13 (0.02) | 0.59 (0.04) b | 0.44 (0.01) | 92.48 (55.66) | |
| 5%-Inc | 0.66 (0.05) b | 3.68 (0.37) | 0.13 (0.02) | 0.07 (0.01) | 0.58 (0.05) b | 0.42 (0.08) | 47.73 (26.18) | |
| 10%-Inc | 0.62 (0.03) b | 3.33 (0.1) | 0.11 (0.01) | 0.06 (0.01) | 0.56 (0.03) b | 0.51 (0.03) | 8.08 (4.04) | |
| Control | 1.09 (0.23) a | 3.36 (0.14) | 0.19 (0.06) | 0.14 (0.02) | 0.96 (0.24) a | 0.38 (0.03) | 40.82 (16.64) | |
| 5%-Sur | 0.65 (0.06) b | 3.33 (0.37) | 0.16 (0.02) | 0.09 (0.01) | 0.56 (0.05) b | 0.46 (0.04) | 47.75 (34.40) | |
| 2019 | 5%-Inc | 0.77 (0.05) b | 3.5 (0.12) | 0.18 (0.01) | 0.12 (0.01) | 0.66 (0.04) b | 0.4 (0.02) | 42.08 (18.68) |
| 10%-Inc | 0.72 (0.05) b | 3.33 (0.09) | 0.19 (0.01) | 0.12 (0.01) | 0.6 (0.04) b | 0.42 (0.02) | 64.00 (24.74) | |
| ANOVA | Treat. | 0.0282 | n.s. | n.s. | n.s. | 0.0429 | n.s. | n.s. |
| Year | 0.0298 | n.s. | 0.0066 | 0.0002 | n.s. | 0.0182 | n.s. | |
| TxY | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | n.s. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frene, J.P.; Frazier, M.; Liu, S.; Clark, B.; Parker, M.; Gardner, T. Early Effect of Pine Biochar on Peach-Tree Planting on Microbial Community Composition and Enzymatic Activity. Appl. Sci. 2021, 11, 1473. https://doi.org/10.3390/app11041473
Frene JP, Frazier M, Liu S, Clark B, Parker M, Gardner T. Early Effect of Pine Biochar on Peach-Tree Planting on Microbial Community Composition and Enzymatic Activity. Applied Sciences. 2021; 11(4):1473. https://doi.org/10.3390/app11041473
Chicago/Turabian StyleFrene, Juan Pablo, Mattie Frazier, Shuang Liu, Bernadette Clark, Michael Parker, and Terrence Gardner. 2021. "Early Effect of Pine Biochar on Peach-Tree Planting on Microbial Community Composition and Enzymatic Activity" Applied Sciences 11, no. 4: 1473. https://doi.org/10.3390/app11041473






