Abstract
The ultimate goal of most neuromusculoskeletal modeling research is to improve the treatment of movement impairments. However, even though neuromusculoskeletal models have become more realistic anatomically, physiologically, and neurologically over the past 25 years, they have yet to make a positive impact on the design of clinical treatments for movement impairments. Such impairments are caused by common conditions such as stroke, osteoarthritis, Parkinson’s disease, spinal cord injury, cerebral palsy, limb amputation, and even cancer. The lack of clinical impact is somewhat surprising given that comparable computational technology has transformed the design of airplanes, automobiles, and other commercial products over the same time period. This paper provides the author’s personal perspective for how neuromusculoskeletal models can become clinically useful. First, the paper motivates the potential value of neuromusculoskeletal models for clinical treatment design. Next, it highlights five challenges to achieving clinical utility and provides suggestions for how to overcome them. After that, it describes clinical, technical, collaboration, and practical needs that must be addressed for neuromusculoskeletal models to fulfill their clinical potential, along with recommendations for meeting them. Finally, it discusses how more complex modeling and experimental methods could enhance neuromusculoskeletal model fidelity, personalization, and utilization. The author hopes that these ideas will provide a conceptual blueprint that will help the neuromusculoskeletal modeling research community work toward clinical utility.
1. Introduction
Engineers love a challenging problem, and some of the most challenging problems that engineers seek to tackle involve human health. From an engineering perspective, the human neuromusculoskeletal system has the appearance of a well-designed mechanical system. It possesses rigid segments connected by extremely low-friction rotational joints. The joints are controlled by highly efficient linear actuators whose masses are strategically located as far inboard as possible. The control system for the actuators can stabilize a multi-segment inverted pendulum flawlessly under a wide variety of challenging movement conditions. Because of these similarities to an engineered system, the human neuromusculoskeletal system lends itself well to physics-based engineering analysis and simulation. This observation raises the possibility that the same computational technologies that have revolutionized the design of airplanes and automobiles over the past 25 years could also be used to revolutionize the design of surgical and rehabilitation treatments for movement impairments. Clinical conditions that could potentially benefit include stroke, osteoarthritis, Parkinson’s disease, spinal cord injury, cerebral palsy, limb amputation, and orthopedic cancer.
Though computational models representing different aspects of the human neuromusculoskeletal system have been developed for decades [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16], such models have yet to make a positive impact on the design of treatments for movement impairments. The neuromusculoskeletal modeling literature is full of journal articles with titles like “A model of x for doing y,” but actual clinical application of the model is rarely reported. Furthermore, few, if any, details are provided on how exactly the computational model would be used to improve clinical treatment design. Nonetheless, a few research groups have generated and validated initial clinical predictions using computational models, such as how an individual with medial knee osteoarthritis should walk differently to relieve pain and slow progression of the disease [17] and whether a child with an equinus deformity due to cerebral palsy should receive gastrocnemius lengthening surgery to improve walking function [18]. Unfortunately, such examples are the exception rather than the rule. As recently stated by the Stanford OpenSim team, “The potential to use subject-specific simulations to understand the causes of movement deviations and to assess treatment options is exciting, but has not been fully realized” [19].
Realization of this untapped potential could initiate a paradigm shift in the way treatments are designed for movement impairments (Figure 1). The current treatment design paradigm has not progressed substantially beyond generic interventions identified by costly and time-consuming clinical trials and selected using subjective clinical judgment. A clinical trial collects a large quantity of movement data from a large heterogeneous patient population and uses statistical models to test whether a particular intervention induces the desired outcome in general. In contrast, a future paradigm could use personalized interventions identified and selected using inexpensive and efficient objective computational models [20,21]. This alternate approach would collect a small quantity of movement data from a specific patient and use a patient-specific computational model to test how a wide variety of possible interventions would affect the patient’s functional outcome. In essence, this new approach would systematically apply virtual treatments to a virtual patient with the goal of identifying the most effective treatment for that patient. Computational treatment design using patient-specific neuromusculoskeletal models would be consistent with current clinical emphases on both personalized medicine and evidence-based medicine.
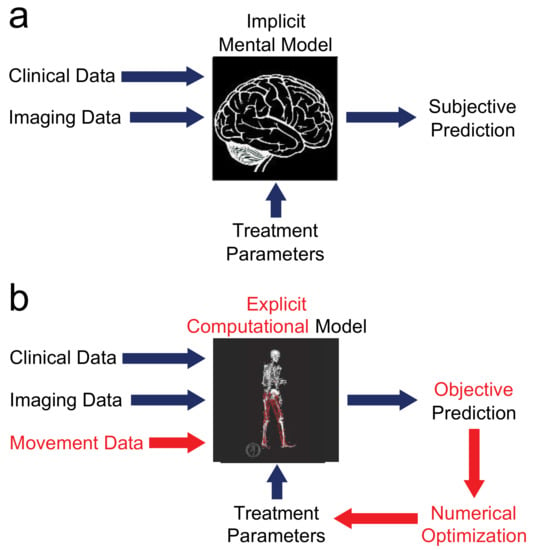
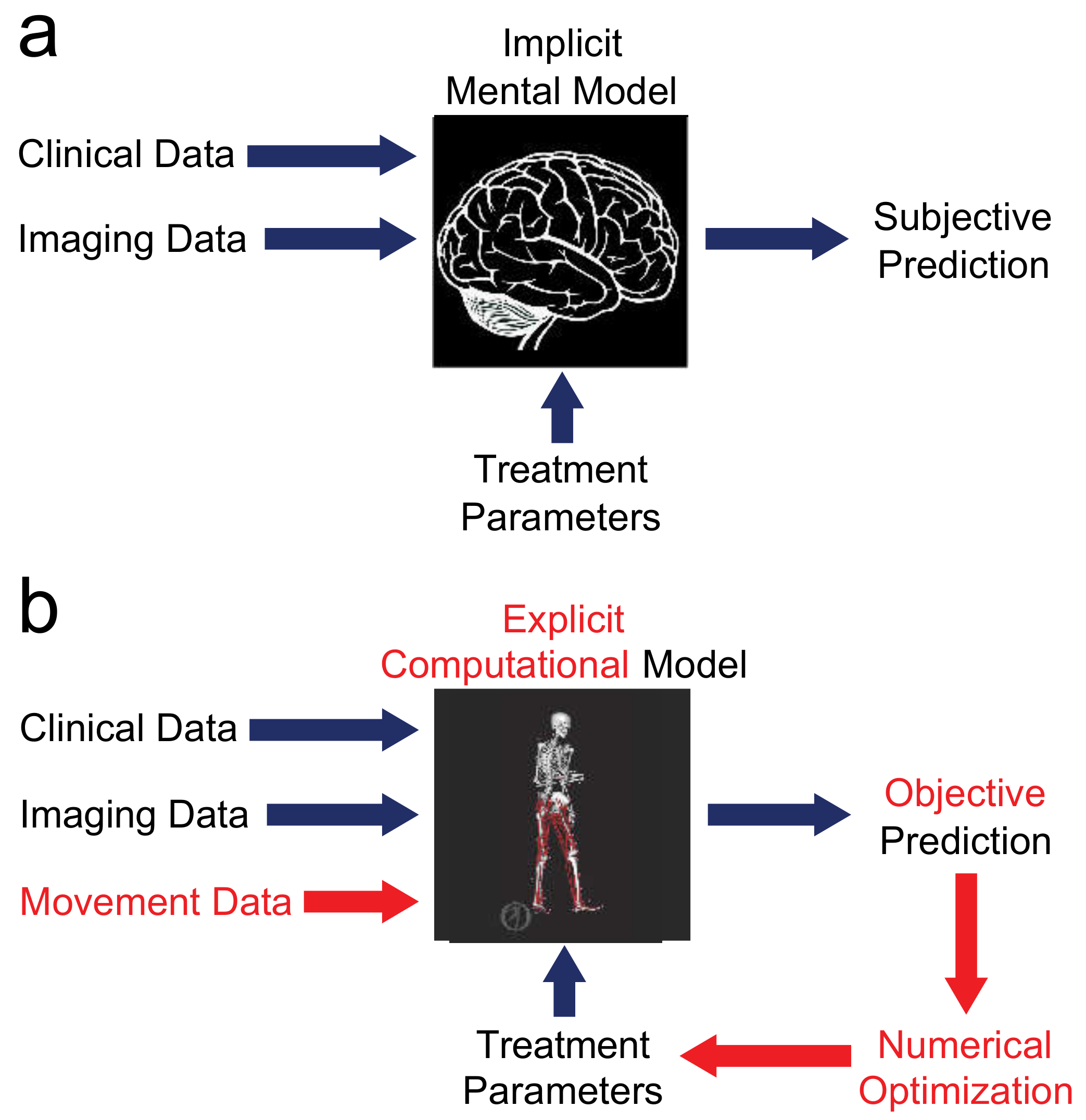
Figure 1.
Comparison of (a) current and (b) future treatment design paradigms. The current paradigm relies on an implicit mental model in the mind of the clinician. Given clinical and imaging data and proposed treatment parameters as inputs, the implicit mental model produces a subjective prediction of post-treatment function, so different clinicians may propose extremely different treatment designs. The future paradigm replaces the implicit mental model with an explicit computational model that obeys laws of physics and principles of physiology. With this approach, movement data are added to the inputs, the explicit computational model produces an objective prediction of post-treatment function, and the entire process is wrapped in numerical optimization to identify the treatment design that will maximize the patient’s functional outcome.
A computational approach to treatment design using personalized neuromusculoskeletal models possesses several advantages over the current treatment design paradigm. First, a computational approach would increase objectivity in treatment planning. Second, it would allow exploration of cause-and-effect relationships that map treatment decisions directly onto the patient’s functional outcome and do so without burning bridges. Third, it would facilitate identification of previously unknown treatments and sensitive treatment parameters. In addition, by using physics-based rather than machine learning models, computational treatment design would extrapolate well to new situations (e.g., post-treatment patient function) for which no experimental data are available. These advantages go beyond those provided by generic neuromusculoskeletal models, which can be used to elucidate factors that are, in general, likely to contribute to the development and progression of disease and thus may serve as targets for intervention (e.g., [22,23]).
This article provides the author’s personal perspective—a conceptual blueprint—for moving computational neuromusculoskeletal models to the point of clinical utility. The focus of the blueprint is on how the neuromusculoskeletal modeling research community can use commonly-employed modeling methods (i.e., rigid body dynamic, geometric, and lumped parameter) combined with commonly-available experimental data (i.e., video motion capture, ground reaction force, and electromyographic (EMG)) to achieve clinical utility. The article is structured as follows. In Section 2, motivation is provided for why computational neuromusculoskeletal models have strong potential to become clinically useful tools. Section 3 discusses challenges to progress that must be overcome if neuromusculoskeletal models are to become clinically useful and provides suggestions for how to overcome them. Section 4 provides a description of the clinical, technical, collaboration, and practical needs that must be satisfied for neuromusculoskeletal models to fulfill their clinical potential, along with recommendations for meeting them. Section 5 discusses opportunities involving more complex modeling and experimental methods that could enhance neuromusculoskeletal model fidelity, personalization, and utilization. Finally, Section 6 provides concluding thoughts on getting neuromusculoskeletal models across the threshold of clinical utility. While the emphasis of this article is on the computational design of treatments for movement impairments affecting gait (the author’s primary research focus), the concepts presented should apply equally well to research involving computational design of treatments for movement impairments affecting the upper extremities and spine.
2. Motivation for Modeling
For neuromusculoskeletal modeling researchers, the motivation for modeling is clear—there is significant potential for personalized models to improve the design of clinical treatments for movement impairments. However, clinical and basic science researchers often do not share the same perspective and are frequently skeptical about the potential benefits of computational modeling for this clinical area. This perspective is understandable given that computational treatment design for movement impairments has not yet produced numerous clinical “wins.” However, this perspective also introduces funding challenges that make rapid research progress difficult. For example, in the United States, the National Science Foundation primarily uses engineering reviewers, funds innovative modeling research, does not fund clinical research, and typically funds at a level that allows limited research progress. In contrast, the National Institutes of Health uses a significant number of clinical and basic science reviewers, primarily funds clinical and basic science research, is less likely to fund innovative modeling research, but funds at a level that allows rapid research progress. Thus, a potential funding mechanism gap exists for large innovative neuromusculoskeletal modeling research projects. If more rapid advances are to be made, modeling researchers need to provide clinical and basic science reviewers with strong, well-reasoned arguments for why model-based treatment design is worth pursuing. Below are several observations to help with building such arguments.
To begin, consider the following question: What would it take to make neuromusculoskeletal modeling a clinically useful tool for improving the design of treatments for movement impairments? To develop a computational process for treatment design, much less than one might expect. Over the past 25 years, neuromusculoskeletal modeling research has crept to the top of a precipice overlooking the “valley of death,” the proverbial chasm between fundamental research on one side and clinical applicability on the other [24]. With improvements in computational modeling capabilities and computer speed, the ability to personalize a neuromusculoskeletal model to represent a specific patient and then use that model to predict an optimal treatment for that patient is no longer science fiction [17,25,26,27]. “We have the technology.” [28] Instead, the main problem is making the computational technology readily accessible by neuromusculoskeletal modeling researchers working in collaboration with clinicians, which in turn is a problem tied to the funding mechanism gap noted above.
Is it worth trying to cross the “valley of death” in the hope that computational modeling can transform the design of treatments for movement impairments? The author argues that it is worth trying for at least four reasons. First, it is worth trying due to the significance of the clinical problems being addressed. Osteoarthritis, stroke, spinal cord injury, traumatic brain injury, and amputation affect roughly 19% of the U.S. adult population [29], with osteoarthritis and stroke being leading causes of serious long-term disability in adults worldwide [29,30,31]. Along with other conditions such as cerebral palsy, Parkinson’s disease, and orthopedic cancer, these conditions often significantly impair movement, resulting in substantial societal costs (e.g., health care, lost productivity), an increased risk of serious secondary health conditions (e.g., heart disease, diabetes), a reduction or even loss of independence, and a decreased quality of life [29,32]. Despite the significance of these clinical problems and the uniqueness of each patient’s clinical situation, treatment design has not progressed substantially beyond trial-and-error implementation of generic interventions selected through subjective clinical judgment. Even when the latest technology is used (e.g., rehabilitation robots, functional electrical stimulation), interventions are typically standardized rather than customized to the unique needs of the patient [33,34]. This state of affairs has contributed to suboptimal functional outcomes, with patients often recovering less function than desired. For example, for individuals with physician-diagnosed arthritis, approximately 44% experience significant functional limitations due to the disease [35]. Of those who receive total knee arthroplasty for knee osteoarthritis, as many as 17% are not satisfied with the outcome [36]. For individuals who suffer a stroke, only 65% regain ambulatory function, but their gait is typically slow, asymmetrical, and metabolically inefficient [37,38,39,40]. Furthermore, only 50% of individuals with walking dysfunction following stroke respond to intervention [41]. For individuals who suffer limb amputation, between 40% and 60% are not satisfied with their prosthesis [42]. For individuals who receive limb-sparing hemipelvectomy surgery for pelvic sarcoma, few are able to regain normal walking function [43,44]. We can and should do better. If individuals affected by impaired movement are to recover the most function possible, a new approach—one that is more effective, objective, and personalized—is needed for neurorehabilitation, surgical, and prosthetic treatment design.
Second, it is worth trying since the same computational design approach has revolutionized the design of products in the aerospace, automotive, heavy equipment, medical device, and numerous other industries [45,46,47,48,49,50]. In the early 1990s, Boeing was faced with the challenge of bringing a new long-range commercial airplane to market with significantly reduced development time and cost. That airplane was the Boeing 777, the most successful wide-body commercial airplane in aviation history [51,52]. To achieve its design goals, Boeing made the bold decision to reject the costly and time-consuming traditional design process involving trial-and-error design iterations performed experimentally on physical prototypes. Instead, it followed a cheaper and faster emerging design process involving systematic design iterations performed computationally on virtual prototypes. The Boeing 777 became the first airplane designed entirely on the computer [52,53], with the final design being developed in reduced time, at reduced cost, and with increased flight range, fuel efficiency, and mechanical reliability [52,54].
Twenty-five years later, it is impossible to fly in a new commercial aircraft or drive in a new car that was not designed using computational modeling. Why? Because computational models identify better final designs than can be found through physical intuition and manual iteration [45,55,56]. As noted in a recent BBC Future news story on airplane design, “By trawling through an exhaustive set of [design] options, computers typically find ones that a human would have missed” [45]. The Food and Drug Administration (FDA) in the United States has also acknowledged the potential benefits of computational modeling for the design of medical devices and interventions [57,58]. As recently stated by Dr. Tina Morrison, Deputy Director of the FDA’s Center for Devices and Radiological Health:
“FDA recognizes the public health benefits offered by modeling and simulation, including those in the area of in silico clinical trials (using individualized computer simulation in development and or regulatory evaluation of medical products, medical devices, or medical interventions).” [59]
Third, it is worth trying since the same computational design approach has already demonstrated success for designing personalized interventions for other clinical problems such as craniofacial bony defects [60,61,62] and occluded coronary arteries [57,63,64,65]. In both cases, the interventions are personalized using patient-specific physics-based computer models whose shapes are optimized. Unlike “black-box” machine learning models, physics-based models extrapolate and can accurately predict function for situations (such as post-treatment) outside the training boundaries. For craniofacial surgery, repeated finite element simulations allow clinician-engineer teams to design and implant personalized craniofacial prostheses that restore a patient’s jaw function and aesthetics [60,61,62]. For coronary artery stenting procedures, large numbers of computational fluid dynamics (CFD) simulations performed by the company HeartFlow allow clinician-engineer teams to “identify significant coronary artery disease and determine the optimal treatment pathway,” [63] reducing invasive coronary angiograms by 61%, unnecessary angiograms by 81%, and healthcare costs by 26%, all while improving patient quality of life and satisfaction [63,65,66]. Two related interventions designed using patient-specific physics-based models are not far behind—nasal airway obstruction surgery planned using CFD simulations [67] and atrial fibrillation ablation surgery planned using computational electrophysiological simulations [64,68].
Fourth, it is worth trying since the same computational approach has already demonstrated success for designing at least one personalized intervention for a movement impairment—a novel rehabilitation treatment for medial knee osteoarthritis [69]. The computational design process optimized the motion of a patient-specific full-body dynamic skeletal model to predict a personalized gait modification that would minimize an external indicator of medial knee contact force. The intervention was proven to work effectively on the patient for whom it was designed, providing benefits similar to those of high tibial osteotomy surgery simply by learning to walk differently [17]. It also reduced medial knee contact force significantly in a subject implanted with an instrumented tibial prosthesis [70]. This novel intervention, which is now being investigated by research teams around the world [71,72,73,74,75,76,77], was identified not by subjective clinical judgment but rather by an objective computational model.
These four reasons suggest that it is both worthwhile and strategic to “help [neuromusculoskeletal modeling] researchers engage in clinical research—and cross the valley of death” [24]. However, providing skeptical grant reviewers with a strong motivation is not enough. Significant challenges to progress and significant needs must be addressed to move neuromusculoskeletal modeling to the point of clinical utility.
3. Challenges to Progress
At least five challenges help explain why neuromusculoskeletal modeling has not yet progressed to the point of clinical utility.
3.1. Movement Data Alone Do Not Provide the Answer
Back in 1932, Schwartz and Heath stated in the Journal of Bone and Joint Surgery (American): “Empiricism fostered by trial and error, must continue to govern the therapy of abnormal function until measurement in some form improves the treatment of disabilities affecting the back and lower extremities” [78]. Restated more succinctly, their hypothesis was, “If we can measure it, we can fix it.” How has that hypothesis worked out over the past 90 years? We can now measure more movement-related quantities than ever before. Motion can be measured routinely using video-based motion capture technology, inertial measurement units, and even raw video images. Forces exerted on the ground and the environment can be measured using various types of load cells. Pressures under feet and within prosthetic sockets can be measured using various types of pressure sensors. Muscle electrical activity can be measured using surface and fine-wire EMG sensors. Highly accurate bone motion can be measured using single-plane and bi-plane dynamic x-ray techniques. Despite these advanced measurement capabilities, the resulting movement data have yet to produce transformative improvements in the treatment of movement impairments. The one exception is surgical treatment design for gait deficits caused by cerebral palsy, where gait lab measurements often change surgical decisions [79,80]. However, even for that clinical application, different clinicians will make different surgical recommendations given the same gait data for a specific patient [81]. Thus, as noted by clinician and biomechanics researcher Richard Brand, “Although locomotion analysis has always held great promise to aid the clinician, it has never lived up to its promise” [82].
Why has the ability to measure multiple aspects of human movement not lived up to its promise? The problem is that movement data tell us what a patient does but not necessarily how to fix the patient’s problem. Because the human neuromusculoskeletal system is a highly nonlinear dynamic system, it is difficult to predict intuitively how a change in system properties (e.g., changing the geometry of a bone, the length of a muscle, the attachment point of a muscle, the control strategy of the central nervous system, or the design of an assistive device) will change the patient’s movement function. The relationship between muscle excitation (as measured by EMG) and muscle force is described by nonlinear activation and contraction dynamics and nonlinear muscle force-length and force-velocity relationships [83]. The relationship between muscle force and muscle moments is described by nonlinear muscle moment arms [6]. Finally, the relationship between muscle moments and resulting body motion is described by nonlinear skeletal dynamics. Each of these nonlinearities contributes to the complexity of the human neuromusculoskeletal system, making intuition about how a particular treatment will affect the patient’s functional outcome unreliable. A more objective and reliable approach is needed to replace intuition.
3.2. Every Patient Is Unique
Each individual suffering from a movement impairment is unique neurologically, physiologically, and/or anatomically. Depending on the type and cause of movement impairment, individual differences can significantly impact the effectiveness of any proposed surgical or rehabilitation intervention. For example, “At present, the stroke rehabilitation field faces the challenge to tailor evidence-based treatment strategies to the needs of the individual stroke patient” [84]. For this reason, personalized rather than generic neuromusculoskeletal models are needed to support model-based intervention design for clinical conditions where patients exhibit significant heterogeneity [21,85]. Furthermore, without appropriate personalization, neuromusculoskeletal models do not reliably predict internal muscle and joint contact forces, body motion, or metabolic cost [25,86,87,88,89,90,91,92,93,94]. Unfortunately, the model personalization process is challenging, as some model parameter values cannot be measured directly and are only weakly observable. To make matters worse, individuals with movement impairments are likely to have important model parameter values that differ substantially from typical values obtained from healthy individuals. As stated in more general terms by the National Academy of Engineering, “Doctors have long known that people differ in susceptibility to disease and response to medicines. But with little guidance for understanding and adjusting to individual differences, treatments developed have generally been standardized for the many, rather than the few.” [95]
3.3. People Change over Time
By definition, “biomechanics” is the application of engineering mechanics to biological systems. One of the primary differences between a biological system and a man-made system is that biological systems have the capacity to adapt, either positively or negatively, over time. Such adaptation can influence both progression of and recovery from disease. However, how the human neuromusculoskeletal system will adapt over time to a specific clinical situation or intervention is difficult to simulate or predict. Apart from simulation studies of bone [96,97,98,99,100,101] and muscle [102] adaptation in response to mechanical loading, computational simulation of adaptation in the human neuromusculoskeletal system remains understudied. As noted by Richard Brand, “[D]espite powerful investigational tools, I would argue biomechanics has made a relatively minor impact in clinical practice primarily because most studies fail to account for the major distinction between living and nonliving systems: adaptability. While any study requires a clear question or hypothesis or goal, without accounting for adaptability, these studies might well be termed ‘necromechanical’” [103]. Although treatments for movement impairments by definition seek to elicit a change in function over time, most biomechanical research investigates only a snapshot in time and thus might be better labeled as necromechanical.
At least two methods exist for simulating neuromusculoskeletal adaptation. The first method is harder and involves simulating the process of adaptation. This method requires the development of fundamental adaptation principles that describe how the neuromusculoskeletal system changes gradually in response to an intervention, a change in mechanical environment, or simply the progression of time. For example, Reinkensmeyer and colleagues have suggested that the ability to model time-dependent mechanisms of neuroplasticity and motor learning, which reflect neural adaptation, could help predict a patient’s potential for recovery [104]. The second method is easier and involves predicting the outcome of adaptation. This method relies on identification of optimization principles that successfully predict a patient’s functional outcome once recovery has plateaued. For example, Meyer and colleagues have shown that minimization of changes in the modular control signals of an individual post-stroke can successfully predict how the individual will walk under new conditions [25]. For both methods, significant additional research is needed to identify appropriate methods for modeling and simulating adaptation so that functional outcome can be predicted reliably for individual patients.
3.4. Validation Is Often Weak
Although a computational neuromusculoskeletal model is only as useful as its ability to predict reality, the research community has made only limited progress at validating its predictions of human movement and the internal forces experienced by muscles, ligaments, and articular surfaces [105]. Since muscles are the actuators of human movement, unvalidated muscle force predictions have, in turn, limited progress in using computational models to predict post-treatment patient function for different treatment scenarios. Prediction of the internal forces experienced by muscles, ligaments, and articular surfaces is challenging due to the muscle redundancy problem [106,107,108,109]. Because the human body possesses approximately three times more muscles than degrees of freedom in the skeleton, no unique mathematical solution exists for the muscle forces generated during human movement.
The main reason for limited model validation is the lack of direct in vivo measurements of internal forces before treatment and of internal forces and patient function after treatment. In vivo measurement of hip and knee contact forces during walking and other activities has become possible through the use of instrumented implants [110,111,112,113,114,115,116,117,118,119,120,121,122]. Though limited to a small number of patients with implanted rather than healthy joints, these measurements provide valuable information about the in vivo loads experienced by articular contact surfaces during various functional activities. Several research groups have shared extensive human movement data sets that include in vivo hip or knee contact force measurements along with video motion capture, ground reaction force, EMG, single- or bi-plane fluoroscopy, and CT and/or MR data [121,122,123]. Some research groups are making extensive use of these unique shared data sets for model validation purposes. However, other research groups have not used them and consequently are now receiving negative grant proposal reviews in the United States from reviewers who are aware of these validation data sets. Direct measurement of muscle or tendon forces in vivo has historically been possible only under special circumstances [124,125,126,127,128,129]. However, researchers have recently developed a novel shear wave tensiometer to measure tendon forces in vivo during human movement [130,131]. These measurements are likely to become the gold standard for validating muscle force predictions generated by computational neuromusculoskeletal models.
An associated reason for limited model validation is that even when unique experimental data sets exist, some research groups are unwilling to share them with the research community. The main reason is the fear of generating research competition that could result in getting “scooped” on grant proposals or journal publications. To facilitate the “Grand Challenge Competition to Predict In Vivo Knee Loads,” the author and colleagues [121] freely disseminated the most widely utilized human movement data sets to date with over 18,000 downloads from SimTK [132]. Since the release of the first data set in 2011, the author’s research group has yet to be “scooped” in any grant proposal or journal publication, even though the data sets continue to be used extensively by the research community. While freely distributing these unique data sets has been nothing but beneficial for the author’s research program, taking the same step will likely require a “leap of faith” for some research groups. Regardless, funding agencies and publishers are providing a strong nudge by requiring that collected data sets used in publications be made freely available to the entire research community [133].
To date, no shared human movement data sets have been published that provide extensive measurements of patient function both before and after treatment, along with a quantitative and qualitative description of the treatment decisions implemented for the patient. Such data sets are needed so that model predictions of a patient’s post-treatment function can be validated given the patient’s pre-treatment movement data and the treatment plan implemented clinically. Since inaccurate musculoskeletal model predictions could lead to the design of ineffective or even harmful treatments, it is imperative that the neuromusculoskeletal modeling research community makes model validation efforts a higher priority.
3.5. Prediction of Post-Treatment Function Is Difficult
Even given a personalized neuromusculoskeletal model, it remains challenging to predict computationally how a particular patient will function following a planned intervention. This difficulty arises for at least two reasons. The first reason involves challenges in generating predictive simulations of human movement. In 2001, Anderson and Pandy [134] published a seminal paper on predicting human walking using a full-body neuromusculoskeletal model. The walking model was three-dimensional (3D), possessed 23 degrees of freedom controlled by 54 muscles, and included deformable foot-ground contact. 10,000 h of CPU time on a cluster was required to solve the nonlinear parameter optimization problem used to predict the walking motion. That study became the standard against which all other human walking predictions would be evaluated, and it inspired subsequent efforts to generate 3D walking predictions [17,25,26,135,136,137,138,139,140,141,142,143]. However, even today, few research groups possess the knowledge and technical expertise needed to generate complex muscle-actuated 3D predictions of human movement.
The second reason why prediction of patient function under novel conditions is difficult involves challenges in modeling a patient’s neural control strategy. To generate reliable predictions of a patient’s post-treatment movement function, researchers must be able to predict how a patient’s neural control strategy will change for the post-treatment conditions. When pain or fear of movement is a significant contributing factor to functional impairment, prediction of the patient’s post-treatment neural control strategy is especially challenging and will require further research. Recent studies have shown that to predict patient function under novel conditions, personalized neural control models are likely to be necessary [25], and furthermore, that pre-treatment muscle synergies may facilitate the prediction of post-treatment muscle excitations [144]. While numerous studies exist that describe complex, realistic sensorimotor control models incorporating elements such as supraspinal control, modularity, central pattern generators, and proprioceptive feedback [22,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159], these models are generic rather than personalized and are typically applied to simplified planar dynamic systems. More realistic musculoskeletal models have been coupled with simpler neural control models employing modular control [25,26,27,136,137,160,161,162,163] or proprioceptive feedback [164,165,166], but these models have yet to be evaluated in clinical treatment design scenarios. Since human movement impairments often involve a significant neurological component (e.g., stroke, spinal cord injury, cerebral palsy, even osteoarthritis), modeling approaches that can reliably predict a patient’s neural control strategy following a planned intervention are important to develop.
4. Description of Needs
To address these five challenges and turn neuromusculoskeletal modeling into a clinically useful tool, the neuromusculoskeletal modeling research community must address at least four categories of needs: (1) clinical, (2) technical, (3) collaboration, and (4) practical. Clinical needs address the scope of clinical problems that can potentially be tackled with computational models and the paradigm needed for using models to design personalized interventions. Technical needs address the scope of technical developments required for model-based treatment design to become broadly feasible for the neuromusculoskeletal modeling research community. Collaboration needs address the scope of collaboration between clinicians and neuromusculoskeletal modeling researchers needed to move model-designed treatments from benchtop to bedside. Finally, practical needs address the scope of additional issues that must be addressed for neuromusculoskeletal modeling to become clinically useful.
4.1. Clinical Needs
Only certain clinical problems involving impaired movement are likely to benefit from a treatment design approach based on computational modeling. For example, there would be little benefit to performing computational modeling for clinical problems where “cookie-cutter” treatment plans already work well. At the other extreme, there would also be little benefit for clinical problems where genetic and molecular factors play a predominant role. Clinical problems involving orthopedic surgery, physical rehabilitation, or neurorehabilitation are likely to benefit the most from a computational approach to treatment design.
Clinical situations where a patient could benefit from computational treatment design can be identified by the presence of three elements. The first element is a “clinical measure” [82]. This element is logical since computational models produce quantitative predictions. As stated by Richard Brand, “The ‘acid tests’ of clinical usefulness of any measure ... are whether that measure predicts a different outcome than would be predicted without the measure, or whether the measure suggests a different treatment (or a different implementation of the same treatment) than would be recommended without the measure” [82]. In essence, an appropriate clinical measure will define a quantifiable “target” that the clinician wants to “hit” through treatment. Clinical measures that meet these criteria will be highly correlated with some direct measure of clinical outcome, such as slowing disease progression, decreasing pain, or increasing function. Examples of clinical measures include medial knee contact force or the peak knee adduction moment for individuals with medial knee osteoarthritis or walking speed and bilateral symmetry for individuals with stroke or Parkinson’s disease. Identification of the best clinical measure for any particular patient and movement impairment will require the combined input of clinicians, the patient, and researchers.
The second element is “clinical requirements.” For computational models to be used for orthopedic surgery, physical rehabilitation, or neurorehabilitation treatment design, five clinical requirements should be met. First, the movement impairment should be influenced heavily by patient-specific factors. Such factors include unique neurological, physiological, or anatomical characteristics, as noted above. Second, standard treatment approaches should be either ineffective or inconsistent at restoring movement function. If standard treatment approaches already work well, then a computational approach to treatment design is unnecessary. Third, a clinical measure can be identified that can quantify initial clinical state and final treatment outcome. Without such a measure, the computational model would have no quantitative “target” to be “hit” through treatment. Fourth, clinical treatment parameters can be identified that can be changed by the clinician, can be modified in the model, and can affect the clinical measure. Identification of appropriate clinical treatment parameters requires close collaboration between clinician and researcher. Fifth, the clinical measure can be measured in the lab prior to treatment, predicted with a computational model to design the treatment, and measured again in the lab following the treatment. This process of “closing the loop” is essential for evaluating whether a computational model can predict functional outcome initially for treatments not designed with the model and subsequently for treatments designed using the model.
The third element is a “clinical model.” Three model requirements should be met for a computational model to be used for clinical treatment design. First, a clinical model should be predictive, capable of predicting how the clinical measure will change in response to a proposed treatment implementation. Second, a clinical model should be minimally complex, containing only those model elements necessary to predict the clinical measure reliably for an individual patient. Third, a clinical model should be patient-specific, capable of having relevant model parameter values calibrated to the patient’s pre-treatment movement and other data.
A real-life example involving computational design of a personalized rehabilitation treatment for medial knee osteoarthritis demonstrates how these three elements work together [17]. The patient studied had knee pain due to medial compartment knee osteoarthritis and was seeking an effective non-surgical treatment option. The goal became to design a modified gait pattern that reduced the patient’s knee pain. The selected clinical measure was the peak knee adduction moment. This quantity is highly correlated with the rate of medial knee osteoarthritis progression [167], and a low value following high tibial osteotomy surgery is associated with the best long-term functional outcome [168]. For clinical requirements, knee loading is heavily influenced by patient-specific leg alignment and walking pattern, standard non-surgical treatment approaches are ineffective at relieving pain and restoring walking function, the peak knee adduction moment can quantify initial clinical state and final treatment outcome, the subject’s walking motion can be modified and will change the peak knee adduction moment, and finally the peak knee adduction moment can be measured in the lab prior to treatment, predicted with a computational walking model to design treatment, and measured again in the lab following treatment. The clinical model predicted the patient’s peak knee adduction moment for different modified walking motions, was minimally complex by modeling only the skeleton (i.e., no muscle or neural control models) and omitting foot-ground contact models, and utilized a patient-specific kinematic structure and mass distribution. An optimization problem was formulated to predict the patient-specific gait modifications that would minimize the patient’s knee adduction moment while still producing a normal-looking walking motion. The optimization predicted that a modified gait pattern involving knee medialization during stance phase could produce a 34% reduction in the patient’s peak knee adduction moment. After learning to perform the predicted gait modification, the patient was able to achieve a 37% reduction when retested in a gait lab [17]. This reduction was comparable to that expected from high tibial osteotomy surgery [169] and decreased the patient’s risk of osteoarthritis progression by a factor of 10 [167]. The patient incorporated the modified gait pattern into his daily life, experienced significant pain reduction, and was able to return to running and recreational sports activities.
4.2. Technical Needs
While the potential is great for using computational models to design more effective treatments for movement impairments, realization of this potential is currently limited by two technical challenges. The first technical challenge involves model personalization. As noted above, for clinical conditions where every patient is unique, personalized models are needed to support model-based intervention design. For movement impairments in particular, it can be beneficial to personalize four aspects of a patient’s neuromusculoskeletal model using the patient’s pre-treatment movement and neurophysiological data [25,26], where the model elements requiring personalization will depend on the clinical problem at hand. These four aspects are: (1) joint model personalization, where parameters defining patient-specific joint centers and functional axes are calibrated [170,171,172,173,174,175,176], (2) muscle-tendon model personalization, where parameters defining patient-specific Hill-type muscle force generation [83,88,90,177,178,179,180,181,182,183,184] and (if desired) surrogate musculoskeletal geometry [185,186] are calibrated, (3) foot-ground contact model personalization (for walking impairments), where parameters defining patient-specific deformable ground contact force generation are calibrated [187], and (4) neural control model personalization, where parameters defining a patient-specific neural control structure are calibrated [25].
While computational methods already exist for personalizing each of these four aspects of a neuromusculoskeletal model (Table 1, 2nd through 5th columns), few research groups are personalizing their models. Furthermore, those that do typically personalize only one or two aspects of their models (review Table 1). The main reason is simple—model personalization is hard, requiring development of specialized optimization approaches that identify model parameter values that fit a patient’s movement and neurophysiological data as closely as possible. Even within the author’s research group where all four aspects are personalized [25,26], it takes a new Ph.D. student six months or longer to learn how to use the lab’s existing model personalization processes. While musculoskeletal modeling software such as OpenSim [19,188] and AnyBody [189] facilitate the process of constructing models and performing standard analyses (e.g., model scaling, inverse kinematics, inverse dynamics, forward dynamics, static optimization), these tools do not currently provide standard methods for personalizing these four model aspects. Realizing this gap, the Stanford OpenSim team recently noted, “More development is needed to streamline the process of creating and validating simulations of individuals with impairments” [19].

Table 1.
Overview of neuromusculoskeletal modeling research involving model personalization and motion prediction using optimization methods.
The second technical challenge involves treatment optimization. Treatment optimization adjusts neural control signals along with surgical, neural control, internal implant, and/or external device parameters in the patient’s personalized model to achieve a specified treatment goal (e.g., maximize walking speed and symmetry). As recently stated by the OpenSim team, “The prediction of outcome due to treatment or intervention (surgery, physical training, biofeedback, etc.) remains the ultimate goal of musculoskeletal modeling and simulation” [19]. However, even if an appropriately personalized neuromusculoskeletal model is available, using that model to predict how the patient will function following a specific type of intervention or a specific implementation of the intervention remains challenging. Since Anderson and Pandy’s foundational predictive walking simulation [134], a number of research groups have developed the capability to predict human movement under novel conditions (Table 1, last column). Despite these advances, most predictive simulations of human movement use scaled generic two-dimensional models rather than personalized 3D models and are rarely performed for treatment optimization purposes. Furthermore, most predictive movement simulations solve a tracking optimization problem, where the goal is to reproduce an experimentally measured motion rather than predict a new motion for a post-treatment situation. Similar to model personalization, the main reason why treatment optimization has not gained widespread use is that it is hard, again requiring specialized optimization approaches, in this case to predict how the patient will function after a planned intervention is applied to the patient’s model.
Realizing this challenge, neuromusculoskeletal modeling researchers have explored different nonlinear optimization methods derived from the field of optimal control for performing predictive simulations of human movement. Early efforts employed a direct shooting method [8,246]. For this method, the optimization design variables are model controls (e.g., muscle excitations), repeated forward dynamic simulations are performed explicitly through numerical integration, and individual time frames are solved sequentially by time marching. The disadvantages of this approach are that repeated forward dynamic simulations are susceptible to numerical integration drift and other integration problems, especially when intermittent foot-ground contact is involved, plus unstable movements such as walking cannot be predicted without some form of stabilizing feedback control. To circumvent these problems, researchers have recently converged on direct collocation as the preferred method for predicting human movement [25,26,190,232,242,243,245,247,248]. For this method, the optimization design variables are model controls as well as states, repeated forward dynamic simulations are performed implicitly as part of the optimization problem formulation, and all time frames are solved simultaneously, thereby eliminating time marching. While this approach produces a larger nonlinear optimization problem, it eliminates numerical integration drift and stability problems, and it is generally more reliable for predicting new motions. While some researchers have implemented their own direct collocation optimal control methods [190,191,242,243,245,247,249,250,251], implementation of these methods is non-trivial, leading other researchers to use academic direct collocation software such as GPOPS-II [25,26], CasADi [135,136], and OpenSim Moco [248]. Despite the availability of these programs, incorporating a generic or personalized neuromusculoskeletal model into one of these packages remains challenging, limiting the use of this approach by the neuromusculoskeletal modeling research community.
The common thread in these two technical challenges is ease of use, which includes ease of implementation. Ideally in a matter of hours, researchers would be able to personalize a neuromusculoskeletal model to a patient’s movement and neurophysiological data and then use the model to predict the optimal treatment for the patient. Given the computational technologies and methods that exist today, the main reason this ideal situation is not already a reality (at least for some clinical problems) is the lack of the software tools necessary to make the model personalization and treatment optimization processes fast and easy to set up and perform. As stated in a recently review article on the use of musculoskeletal models in clinical practice, “A specific focus should be increasing the access and usability for non-expert users” [252]. For model personalization and treatment optimization to become widespread, the barriers to entry need to be lowered substantially and the entire process needs to be made easily accessible to the broader neuromusculoskeletal modeling research community.
4.3. Collaboration Needs
Even if the technical needs described above were satisfied, a significant collaboration challenge would still exist. This challenge involves finding clinical and neuromusculoskeletal modeling researchers who have the mindsets needed to work effectively together.
An illustration is helpful for understanding the joint mindset needed for effective collaboration. The illustration involves two children playing in the same sandbox and highlights the need for a concept called “shared intellectual investment.” One child represents a clinician, the other child represents a modeler, and the sandbox represents the clinical problem at hand. In the least productive scenario, the clinician says to the modeler, “I don’t need modeling capabilities. Why are you in my sandbox?” Similarly, the modeler says to the clinician, “I don’t need clinical perspective. Why are you in my sandbox?” While rarely stated explicitly, this scenario is probably the most common, contributing greatly to the lack of progress. In a slightly more productive scenario, the clinician says to the modeler, “Come work on my clinical problem. Come build in my sandbox.” In this case, intellectual investment in solving the clinical problem comes primarily from the clinical side. Similarly, the modeler says to the clinician, “Come use my models. Come build in my sandbox.” Here, intellectual investment comes primarily from the modeling side. While this scenario is better than the first one, it is still far from ideal, as each side sees itself as independent from the other. In the most productive scenario, the clinician and modeler say to each other, “How could modeling help clinical problems? Let’s build in the sandbox together.” For this scenario, the sandbox becomes a shared space of shared intellectual investment, where the clinician wants to work closely with the modeler and vice versa. Both sides enter into the sandbox together with humility and appreciation for the capabilities and perspective that the other side brings to the problem. Though such collaborations exist today, they are far less common than they need to be for computational models to make a significant positive impact on treatment design for movement impairments.
Creation of a collaboration marked by “shared intellectual investment” requires a commitment from both the clinician and the modeler. The clinician needs to be willing to teach the modeler about the details of the clinical problem and to document and measure the treatment decisions implemented in the patient’s treatment plan. That information is critical to allow the modeler to evaluate post-treatment predictions of patient function given a model of the patient constructed from pre-treatment data and the treatment decisions implemented by the clinician. On the other side, the modeler needs to be willing to teach the clinician about the quantities that models can reliably predict and to learn as much as possible about the issues present in the clinical problem. While the urgency of daily required tasks (e.g., surgery, clinical care, teaching, research, administration) makes this additional effort inconvenient for both sides, it is a long-term investment with the potential to pay high dividends.
4.4. Practical Needs
Beyond the previous three categories of needs, a fourth category involves practical considerations that are not rocket science but rather strategic decisions. One practical consideration is the focus of neuromusculoskeletal modeling research. Engineering researchers love technology development. It can be difficult for us to know when to stop developing technology and start applying the technology we have been developing to important problems. It is the author’s belief that a subset of movement impairment clinical problems exists today that could benefit greatly from existing computational modeling methods. A greater focus on identifying those problems, rather than on developing more technology, could lead to the initial “clinical wins” that would help propel model-based treatment design forward.
Another practical consideration is the need for extensive experimental data sets that would allow neuromusculoskeletal modeling researchers to develop and evaluate model-based treatment design methods. Such data sets would be collected from specific patients and include pre- and post-treatment (following plateau in recovery) movement and neurophysiological data, along with documented and measured treatment decisions implemented by the clinician. The specific experimental data needed would depend on the clinical problem but could include video motion capture, EMG, and (for walking) ground reaction data, along with medical imaging data (e.g., CT and/or MR). Pre- and post-treatment imaging data could be valuable for improving personalization of the patient’s pre-treatment model and measuring certain treatment decisions made by the clinician (e.g., locations of bone cuts). Having pre- and post-treatment data and treatment decisions available for specific patients would allow researchers to apply the actual treatment decisions to the patient’s pre-treatment model and predict the patient’s post-treatment function. Differences between measured and predicted post-treatment function would then be used to improve and refine the modeling methods without the risk of negatively impacting the patient’s clinical care. Once retrospective prediction of post-treatment patient function can be performed reliably, prospective prediction of post-treatment function could begin to be utilized clinically as long as any necessary regulatory approvals had been addressed. The author is aware of only one study to date that has “closed the loop” by validating a prospective patient-specific model-based prediction of post-treatment function using post-treatment movement data collected from the patient [17].
Since few neuromusculoskeletal modeling research groups possess the financial, experimental, and clinical resources needed to generate pre- and post-treatment movement and neurophysiological data, the research community will need well-curated data sets to begin learning how to “close the loop” for model-based treatment design. One way to accelerate the generation and distribution of such data sets would be to organize international competitions similar to the “Knee Grand Challenge Competition” organized previously by the author and colleagues [121]. For example, modelers and clinicians together could develop a “Cerebral Palsy Grand Challenge Competition,” “Stroke Grand Challenge Competition,” or “Pelvic Sarcoma Grand Challenge Competition.” Competition organizers would provide pre-treatment movement and neurophysiological data collected from a specific patient, along with details of the treatment plan implemented by the clinician. Competing research teams would create a personalized neuromusculoskeletal model of the patient using the pre-treatment data, implement the patient’s actual treatment plan in the patient’s model, and then generate a blinded post-treatment prediction of the specified clinical measure. The blinded prediction would be submitted to the competition organizers, and only then would competitors be given the patient’s post-treatment movement and neurophysiological data, including the specified clinical measure, to evaluate their predictions and identify the weak links in their modeling methods. The best blinded predictions, and the subsequent modeling modifications that improved the predictions, would be presented in a special session of an annual conference and published in a special issue of a relevant journal, similar to the approach used successfully for the “Knee Grand Challenge Competition.” Following several competitions, designing and evaluating treatments for movement impairments would become common practice for at least a segment of the neuromusculoskeletal modeling research community.
5. Opportunities for Enhancement
Thus far, this article has focused on how to get neuromusculoskeletal modeling to the point of clinical utility using commonly-employed modeling methods (i.e., rigid body dynamic, geometric, and lumped parameter) and commonly-available experimental data (i.e., video motion capture, ground reaction force, and EMG). This focus is consistent with the well-known principle of minimum model complexity–engineers should use the minimum complexity model that can reliably predict the quantities of interest. In the author’s opinion, significant clinical utility can be achieved in the near future without moving beyond commonly-used and minimally-complex modeling and experimental approaches. Furthermore, personalized modeling and predictive simulation capabilities that are based on commonly-used approaches are the most likely to gain widespread use by the neuromusculoskeletal modeling research community. At the same time, if researchers are willing to go beyond common approaches, then related research areas employing more complex modeling and experimental methods could enhance model fidelity, personalization, and utilization.
5.1. Enhanced Model Fidelity
Opportunities for enhancement exist for generating higher fidelity neuromusculo- skeletal models that capture short- and long-term tissue-level behavior. For some clinical problems where functional outcome depends on tissue-level stresses and strains, rigid body skeletal models controlled by lumped-parameter Hill-type muscle-tendon models will not be sufficient. To capture tissue-level effects in muscles, ligaments, bones, and/or articular cartilage, finite element (FE) models with deformable tissue properties are the logical choice [102,253,254,255,256,257,258,259,260,261,262,263]. In muscles, for example, non-uniform tissue-level behavior could be important for predicting muscle hypertrophy or injury in response to exercise, the effects of aging and disuse, the progression of muscular dystrophy, and muscle function in microgravity. In ligaments, it could be important for predicting failure mechanisms and the best way to perform reconstructions. In bones, it could be important for predicting the risk of fracture due to unfavorable loading conditions, osteoporosis, or joint replacement. In articular cartilage, it could be important for predicting conditions that initiate, accelerate, or slow the development of osteoarthritis.
For more complex clinical problems where functional outcome depends on how muscle, bone, and/or cartilage tissue changes over time, FE models can be combined with either empirical or agent-based adaptation models. As noted earlier, bone adaptation to an altered loading environment is the classic historical example of adaptive simulations. However, recent work has extended adaptive FE simulation methods to muscle and articular cartilage as well. For example, 3D FE models of muscle have been combined with agent-based adaptation models to simulate disease-related changes in muscle function caused by muscular dystrophy [255] and to propose improved tissue regeneration approaches to treat severe muscle injuries [102]. For articular cartilage, a specimen-specific 3D FE model of a patellofemoral joint has been coupled with an empirical adaptation model to predict articular cartilage wear over time during in vitro testing [264]. The challenge with adaptive simulations is that a new FE analysis must be performed each time the adaptation model updates the FE model’s tissue properties. Furthermore, since it is unrealistic to simulate thousands or millions of motion cycles, the number of cycles for which the most recent FE simulation results can be extrapolated must be determined (see [265,266] for further details).
5.2. Enhanced Model Personalization
Opportunities for enhancement also exist for personalizing important properties of musculoskeletal models. Personalization of musculoskeletal geometry can be enhanced using imaging data and recently published software tools. These tools, which include NMSBuilder [267], the MAP Client [268], and STAPLE [269], can turn patient imaging (e.g., CT, MR) and/or surface marker data into personalized geometric OpenSim models (for a comparison of the capabilities of these three tools, see [270]). Together, such tools allow for generation of personalized bone geometries, muscle lines of action, and/or joint definitions, significantly improving the speed and accuracy with which personalized geometric musculoskeletal models can be created. A recent study has also provided a detailed step-by-step procedure for creating subject-specific lower limb musculoskeletal geometric models from MR imaging data [271].
Personalization of model parameter values can be enhanced using probabilistic modeling methods. Probabilistic modeling samples combinations of model inputs to characterize the distribution of corresponding model outputs. This approach can help identify model parameter values to which the outputs of interest are sensitive, thereby revealing which parameter values are worth personalizing and which are not [272,273]. For example, one probabilistic modeling study showed that when calculating lower extremity inverse dynamic joint moments during walking, errors in body segment mass properties have little influence, while errors in joint positions and orientations can have a significant influence [91]. Other probabilistic modeling studies have investigated which parameters in Hill-type muscle-tendon models and geometric musculoskeletal models are the most important to personalize when estimating lower extremity muscle forces during walking [274,275,276,277]. These studies have reported that optimal muscle fiber length, tendon slack length, and peak isometric force in Hill-type muscle-tendon models [274,275,276] and the attachment points of the iliacus and psoas muscles, but not those of other muscles [277], are the most critical to calibrate to subject data when estimating muscle forces.
Personalization of model parameter values can also be enhanced using various imaging methods. In some cases, imaging methods permit direct measurement of model parameter values. For example, magnetic resonance imaging (MRI) can be used to measure in vivo muscle volumes [278], which can then be converted to muscle peak isometric forces. Diffusion-tensor MRI can be used to perform in vivo measurements of muscle pennation angle and fiber type distributions [279,280]. Ultrasonography can be used to identify subject-specific force-strain parameters for tendons and optimal fiber length and pennation angle for muscles [281,282,283]. In other cases, imaging methods provide novel in vivo measurements that could improve estimation of model parameter values using optimization methods. For example, tendon forces measured in vivo by shear wave tensiometry [130,131] could be employed as additional constraints when personalizing muscle-tendon model parameter values using EMG, kinematic, and kinetic data.
5.3. Enhanced Model Utilization
Finally, opportunities for enhancement exist for making utilization of complex models faster and easier through surrogate modeling methods. Surrogate modeling is a special case of supervised machine learning that fits a computationally “fast” model to the input-output characteristics of a computationally “slow” model, simulation, or optimization (for a broader perspective on the use of machine learning methods for personalizing neuromusculoskeletal models, see [284]). The surrogate model essentially becomes a fast black-box model that replaces the slow original model, simulation, or optimization.
The surrogate modeling process generally involves a sequence of six steps [285]. First, identify one or more outputs of interest from the model, simulation, or optimization. Second, identify the inputs that affect the outputs of interest, where the number of inputs will ideally be small (e.g., <10) so that the surrogate model fitting process will not become overly complex. Third, sample combinations of inputs (called “sample points”) within pre-defined bounds using design of experiments (e.g., Latin hypercube) to spread out the sample points evenly throughout the (possibly high-dimensional) design space. Fourth, perform an analysis, simulation, or optimization for each sample point to generate the corresponding outputs of interest. Fifth, fit each output of interest as a function of the sample point inputs using a black box fitting method (e.g., polynomial response surfaces, Kriging, support vector machines, artificial neural networks). Sixth, evaluate the accuracy of the resulting surrogate models using sample points not included in the fitting process. While it can take hours or days of computation time to evaluate all of the input combinations, this computational cost is paid only once upfront, and the final surrogate models will be orders of magnitude faster.
For neuromusculoskeletal modeling applications, surrogate models have two primary benefits. First, surrogate models can make “slow” computational models “fast” for use within other “fast” models. In the context of neuromusculoskeletal modeling, the “fast” model is typically a rigid body dynamic skeletal model controlled by lumped-parameter muscle-tendon models, while the “slow” model is typically a geometric musculoskeletal model or a tissue-level elastic continuum model. Since the mechanics of the slow and fast models interact, the two models should ideally be simulated simultaneously rather than sequentially. Surrogate models have been developed to calculate muscle-tendon lengths and moment arms produced by geometric musculoskeletal models [185,186], stresses in foot tissues produced by FE models [192], strain fields in long bones produced by FE models [286], and contact forces, stresses, and/or wear in natural and artificial knees produced by FE or elastic foundation models [220,287,288,289,290,291,292].
Second, surrogate models can make “slow” computational simulations or optimizations “fast” for clinical implementation. Performing treatment design iterations with a physics-based model can require extensive computation time. If the clinician is satisfied with the final predicted treatment design, then long computation time may not be a problem. However, if the clinician would like to interact with the model and observe in real time the effect of different treatment decisions, then long computation time will be unacceptable. To circumvent this problem, one can fit a surrogate model to the outputs (e.g., clinical measures) of repeated simulations or optimizations [173,293]. The resulting surrogate models would allow the clinician to evaluate different treatment scenarios in real time. This approach has been used successfully with FDA approval by HeartFlow [63], where the barrier to clinical implementation was excessive computation time for thousands of CFD simulations. The same surrogate models can also be used to perform ultra-fast optimizations [285]. Clinicians and engineers working together could rapidly investigate tradeoffs in functional outcomes for different decisions about the relative importance of different simulation outputs included in the cost function.
6. Conclusions
In this article, the author has provided a conceptual blueprint for how neuromusculoskeletal models can become clinically useful. At the core of this blueprint is the need to make model personalization and treatment optimization easy to implement and easy to use by the broad neuromusculoskeletal modeling research community. In addition to motivating the use of neuromusculoskeletal models to improve treatment design for movement impairments, the author has highlighted challenges to progress along with suggestions for overcoming them, a description of needs with recommendations for addressing them, and opportunities for enhancement involving more complex modeling and experimental methods. In the short term, if a subset of movement impairment problems can be identified to which existing neuromusculoskeletal modeling methods could be applied effectively, the field could generate some initial clinical “wins” that would help propel it across the threshold of clinical utility and open up a new paradigm for treatment design. In the long term, model-based treatment design for movement impairments will need the endorsement of regulatory (e.g., the FDA in the United States, the European Medicines Agency in Europe) and health technology assessment (e.g., the Health Technology Assessment Programme in the United Kingdom) agencies, which would certify the clinical utility of the approach and facilitate crossing the “valley of death.” If computational modeling and simulation can do for the design of movement impairment treatments even a fraction of what they have done for the design of airplanes and automobiles, their clinical impact will be transformative.
Funding
The author would like to thank the Cancer Prevention and Research Institute of Texas (grant RR170026) for supporting this work.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The author thanks his clinical collaborators Carolynn Patten, Valerae Lewis, Gerard Francisco, and James Chang for valuable discussions that were instrumental in forming many of the ideas in this article. He would also like to thank Richard Brand for inspiring the author’s search for a formal model-based treatment design paradigm.
Conflicts of Interest
The author declares no conflict of interest.
References
- Hatze, H. A comprehensive model for human motion simulation and its application to the take-off phase of the long jump. J. Biomech. 1981, 14, 135–142. [Google Scholar] [CrossRef]
- Mena, D.; Mansour, J.M.; Simon, S.R. Analysis and synthesis of human swing leg motion during gait and its clinical applications. J. Biomech. 1981, 14, 823–832. [Google Scholar] [CrossRef]
- Zajac, F.E.; Wicke, R.W.; Levine, W.S. Dependence of jumping performance on muscle properties when humans use only calf muscles for propulsion. J. Biomech. 1984, 17, 513–523. [Google Scholar] [CrossRef]
- Delp, S.L.; Loan, J.P.; Hoy, M.G.; Zajac, F.E.; Topp, E.L.; Rosen, J.M. An interactive graphics-based model of the lower extremity to study orthopaedic surgical procedures. IEEE Trans. Biomed. Eng. 1990, 37, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Audu, M.L.; Davy, D.T. The influence of muscle model complexity in musculoskeletal motion modeling. J. Biomech. Eng. 1985, 107, 147–157. [Google Scholar] [CrossRef]
- Hoy, M.G.; Zajac, F.E.; Gordon, M.E. A musculoskeletal model of the human lower extremity: The effect of muscle, tendon, and moment arm on the moment-angle relationship of musculotendon actuators at the hip, knee, and ankle. J. Biomech. 1990, 23, 157–169. [Google Scholar] [CrossRef]
- Yamaguchi, G.T.; Zajac, F.E. A planar model of the knee joint to characterize the knee extensor mechanism. J. Biomech. 1989, 22, 1–10. [Google Scholar] [CrossRef]
- Pandy, M.G.; Zajac, F.E.; Sim, E.; Levine, W.S. An optimal control model for maximum-height human jumping. J. Biomech. 1990, 23, 1185–1198. [Google Scholar] [CrossRef]
- Gerritsen, K.G.; van den Bogert, A.J.; Hulliger, M.; Zernicke, R.F. Intrinsic muscle properties facilitate locomotor control—A computer simulation study. Mot. Control 1998, 2, 206–220. [Google Scholar] [CrossRef]
- Blankevoort, L.; Kuiper, J.H.; Huiskes, R.; Grootenboer, H.J. Articular contact in a three-dimensional model of the knee. J. Biomech. 1991, 24, 1019–1031. [Google Scholar] [CrossRef]
- Fregly, B.J.; Zajac, F.E. A state-space analysis of mechanical energy generation, absorption, and transfer during pedaling. J. Biomech. 1996, 29, 81–90. [Google Scholar] [CrossRef]
- Neptune, R.R.; Hull, M.L. A theoretical analysis of preferred pedaling rate selection in endurance cycling. J. Biomech. 1999, 32, 409–415. [Google Scholar] [CrossRef]
- Raasch, C.C.; Zajac, F.E.; Ma, B.; Levine, W.S. Muscle coordination of maximum-speed pedaling. J. Biomech. 1997, 30, 595–602. [Google Scholar] [CrossRef]
- Hubbard, M. Computer simulation in sport and industry. J. Biomech. 1993, 26 (Suppl. S1), 53–61. [Google Scholar] [CrossRef]
- Van Soest, A.J.; Huijing, P.A.; Solomonow, M. The effect of tendon on muscle force in dynamic isometric contractions: A simulation study. J. Biomech. 1995, 28, 801–807. [Google Scholar] [CrossRef]
- Yeadon, M.R.; Atha, J.; Hales, F.D. The simulation of aerial movement--IV A computer simulation model. J. Biomech. 1990, 23, 85–89. [Google Scholar] [CrossRef]
- Fregly, B.J.; Reinbolt, J.A.; Rooney, K.L.; Mitchell, K.H.; Chmielewski, T.L. Design of patient-specific gait modifications for knee osteoarthritis rehabilitation. IEEE Trans. Biomed. Eng. 2007, 54, 1687–1695. [Google Scholar] [CrossRef]
- Rajagopal, A.; Kidziński, Ł.; McGlaughlin, A.; Hicks, J.; Delp, S.; Schwartz, M. Pre-operative gastrocnemius lengths in gait predict outcomes following gastrocnemius lengthening surgery in children with cerebral palsy. PLoS ONE 2020, 15, e0233706. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Hicks, J.L.; Uchida, T.K.; Habib, A.; Dembia, C.L.; Dunne, J.J.; Ong, C.F.; DeMers, M.S.; Rajagopal, A.; Millard, M.; et al. OpenSim: Simulating musculoskeletal dynamics and neuromuscular control to study human and animal movement. PLoS Comput. Biol. 2018, 14, e1006223. [Google Scholar] [CrossRef] [PubMed]
- Fregly, B.J.; Boninger, M.L.; Reinkensmeyer, D.J. Personalized neuromusculoskeletal modeling to improve treatment of mobility impairments: A perspective from European research sites. J. Neuroeng. Rehabil. 2012, 9, 18. [Google Scholar] [CrossRef]
- McMorland, A.J.C.; Runnalls, K.D.; Byblow, W.D. A neuroanatomical framework for upper limb synergies after stroke. Front. Hum. Neurosci. 2015, 9, 82. [Google Scholar] [CrossRef]
- Song, S.; Geyer, H. Predictive neuromechanical simulations indicate why walking performance declines with ageing. J. Physiol. 2018, 596, 1199–1210. [Google Scholar] [CrossRef]
- Thelen, D.G.; Chumanov, E.S.; Sherry, M.A.; Heiderscheit, B.C. Neuromusculoskeletal models provide insights into the mechanisms and rehabilitation of hamstring strains. Exerc. Sport Sci. Rev. 2006, 34, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Butler, D. Crossing the valley of death. Nature 2008, 453, 840–842. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J.; Eskinazi, I.; Jackson, J.N.; Rao, A.V.; Patten, C.; Fregly, B.J. Muscle synergies facilitate computational prediction of subject-specific walking motions. Front. Bioeng. Biotechnol. 2016, 4, 77. [Google Scholar] [CrossRef]
- Sauder, N.R.; Meyer, A.J.; Allen, J.L.; Ting, L.H.; Kesar, T.M.; Fregly, B.J. Computational design of FastFES treatment to improve propulsive force symmetry during post-stroke gait: A feasibility study. Front. Neurorobotics 2019, 13, 80. [Google Scholar] [CrossRef]
- Pitto, L.; Kainz, H.; Falisse, A.; Wesseling, M.; van Rossom, S.; Hoang, H.; Papageorgiou, E.; Hallemans, A.; Desloovere, K.; Molenaers, G.; et al. SimCP: A simulation platform to predict gait performance following orthopedic intervention in children with cerebral palsy. Front. Neurorobotics 2019, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Harve Bennett Productions, Silverton Productions, and Universal Television. The Six Million Dollar Man; Harve Bennett Productions, Silverton Productions, and Universal Television: Los Angeles, CA, USA, 1974. [Google Scholar]
- Ma, V.Y.; Chan, L.; Carruthers, K.J. Incidence, prevalence, costs, and impact on disability of common conditions requiring rehabilitation in the United States: Stroke, spinal cord injury, traumatic brain injury, multiple sclerosis, osteoarthritis, rheumatoid arthritis, limb loss, and back pa. Arch. Phys. Med. Rehabil. 2014, 95, 986–995. [Google Scholar] [CrossRef]
- Ovbiagele, B.; Goldstein, L.B.; Higashida, R.T.; Howard, V.J.; Johnston, S.C.; Khavjou, O.A.; Lackland, D.T.; Lichtman, J.H.; Mohl, S.; Sacco, R.L.; et al. Forecasting the future of stroke in the United States: A policy statement from the American Heart Association and American Stroke Association. Stroke 2013, 44, 2361–2375. [Google Scholar] [CrossRef]
- Theis, K.A.; Roblin, D.W.; Helmick, C.G.; Luo, R. Prevalence and causes of work disability among working-age US adults, 2011-2013, NHIS. Disabil. Health J. 2018, 11, 108–115. [Google Scholar] [CrossRef]
- Praemer, A.; Furner, S.; Rice, D.P. Musculoskeletal Conditions in the United States; American Academy of Orthopaedic Surgeons: Rosemont, IL, USA, 1999. [Google Scholar]
- Proietti, T.; Crocher, V.; Roby-Brami, A.; Jarrasse, N. Upper-limb robotic exoskeletons for neurorehabilitation: A review on control strategies. IEEE Rev. Biomed. Eng. 2016, 9, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.L.; Ting, L.H.; Kesar, T.M. Gait rehabilitation using functional electrical stimulation induces changes in ankle muscle coordination in stroke survivors: A preliminary study. Front. Neurol. 2018, 9, 1127. [Google Scholar] [CrossRef] [PubMed]
- Barbour, K.E.; Helmick, C.G.; Boring, M.; Brady, T.J. Vital signs: Prevalence of doctor-diagnosed arthritis and arthritis-attributable activity limitation-United States, 2013-2015. Mmwr Morb. Mortal. Wkly. Rep. 2017, 66, 246–253. [Google Scholar] [CrossRef]
- Dunbar, M.J.; Richardson, G.; Robertsson, O. I can’t get no satisfaction after my total knee replacement: Rhymes and reasons. Bone Joint J. 2013, 95-B, 148–152. [Google Scholar] [CrossRef]
- Brandstater, M.E.; de Bruin, H.; Gowland, C.; Clark, B.M. Hemiplegic gait: Analysis of temporal variables. Arch. Phys. Med. Rehabil. 1983, 64, 583–587. [Google Scholar]
- Chen, G.; Patten, C.; Kothari, D.H.; Zajac, F.E. Gait differences between individuals with post-stroke hemiparesis and non-disabled controls at matched speeds. Gait Posture 2005, 22, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Patten, C.; Kothari, D.H.; Zajac, F.E. Gait deviations associated with post-stroke hemiparesis: Improvement during treadmill walking using weight support, speed, support stiffness, and handrail hold. Gait Posture 2005, 22, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Olney, S.J.; Monga, T.N.; Costigan, P.A. Mechanical energy of walking of stroke patients. Arch. Phys. Med. Rehabil. 1986, 67, 92–98. [Google Scholar] [CrossRef]
- Duncan, P.W.; Sullivan, K.J.; Behrman, A.L.; Azen, S.P.; Wu, S.S.; Nadeau, S.E.; Dobkin, B.H.; Rose, D.K.; Tilson, J.K.; Cen, S.; et al. Body-weight–supported treadmill rehabilitation after stroke. N. Engl. J. Med. 2011, 364, 2026–2036. [Google Scholar] [CrossRef] [PubMed]
- Baars, E.C.; Schrier, E.; Dijkstra, P.U.; Geertzen, J.H.B. Prosthesis satisfaction in lower limb amputees: A systematic review of associated factors and questionnaires. Medicine 2018, 97, e12296. [Google Scholar] [CrossRef] [PubMed]
- Gerrand, C.H.; Bell, R.S.; Griffin, A.M.; Wunder, J.S. Instability after major tumor resection: Prevention and treatment. Orthop. Clin. N. Am. 2001, 32, 697–710. [Google Scholar] [CrossRef]
- Lewis, V.O. Internal hemipelvectomies: Resection and reconstructive options. Oper. Tech. Orthop. 2014, 24, 74–78. [Google Scholar] [CrossRef]
- Heaven, D. The Designer Changing the Way Aircraft are Built. BBC Future. 2018. Available online: https://www.bbc.com/future/article/20181129-the-ai-transforming-the-way-aircraft-are-built (accessed on 29 December 2020).
- Koebler, J. SpaceX is Using These Simulations to Design the Rocket That’ll Take us to Mars. Vice. 2015. Available online: https://www.vice.com/en_us/article/ezv79w/spacex-i (accessed on 29 December 2020).
- Alexander, C. A Brief History of Computing in Formula 1. McLaren Website. 2016. Available online: https://www.mclaren.com/racing/team/a-brief-history-of-computing-in-F1-1052199/ (accessed on 29 December 2020).
- Niccolai, J. The America’s Cup: Nerves, Skill, and Computer Design. PCWorld. 2013. Available online: https://www.pcworld.com/article/2049820/the-americas-cup-nerves-skill-and-a-lot-of-computers.html (accessed on 29 December 2020).
- Dandy, G.; Daniell, T.; Foley, B.; Warner, R. Planning and Design of Engineering Systems, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Gualdrón, C.-I.L.; Ibarra, E.-R.B.; Bohórquez, A.-P.; Bohórquez, M.I.G. Present and future for technologies to develop patient-specific medical devices: A systematic review approach. Med. Devices Evid. Res. 2019, 12, 253–273. [Google Scholar] [CrossRef]
- Sharma, K.J.; Bowonder, B. The making of the Boeing 777: A case study in concurrent engineering. Int. J. Manuf. Technol. Manag. 2004, 6, 254–264. [Google Scholar] [CrossRef]
- Zhang, B. The glorious history of the best plane Boeing has ever built. Business Insider. 2018. Available online: https://www.businessinsider.nl/boeing-777-history-2017-6/ (accessed on 29 December 2020).
- Weiss, S.I.; Amir, A.R. Aerospace Industry-Design Methods. In Encyclopædia Britannica; Encyclopædia Britannica Inc.: Chicago, IL, USA, 2019. [Google Scholar]
- Petroski, H. Engineering: The Boeing 777. Am. Sci. 1995, 83, 519–522. [Google Scholar]
- Vishwas, M.; Vinyas, M.; Puneeth, K. Application of computer aided engineering in reducing the product development life cycle. Int. J. Innov. Res. Sci. Technol. 2016, 2, 19–22. [Google Scholar]
- Miller, L. Product innovation through computational prototypes and supercomputing. IEEE Comput. Sci. Eng. 2017, 19, 9–17. [Google Scholar] [CrossRef]
- Morrison, T.M.; Pathmanathan, P.; Adwan, M.; Margerrison, E. Advancing regulatory science with computational modeling for medical devices at the FDA’s Office of Science and Engineering Laboratories. Front. Med. 2018, 5, 241. [Google Scholar] [CrossRef]
- James, G. Using Simulation in Medical Device Design-Image-Based Modeling and Simulation are Becoming Increasingly Common for Device Design. Orthopedic Design & Technology. 2015. Available online: https://www.odtmag.com/issues/2015-11-01/view_features/using-simulation-in-medical-device-design (accessed on 29 December 2020).
- Morrison, T. How Simulation Can Transform Regulatory Pathways. In Food and Drug Administration; U.S. Food & Drug Administration; 2018. Available online: https://www.fda.gov/science-research/about-science-research-fda/how-simulation-can-transform-regulatory-pathways (accessed on 29 December 2020).
- Mehrotra, D.; Howlader, D.; Singh, P.K.; Gupta, S. Patient specific three-dimensional implant for reconstruction of complex mandibular defect. J. Craniofacial Surg. 2019, 30, e308–e311. [Google Scholar]
- Sutradhar, A.; Park, J.; Carrau, D.; Miller, M.J. Experimental validation of 3D printed patient-specific implants using digital image correlation and finite element analysis. Comput. Biol. Med. 2014, 52, 8–17. [Google Scholar] [CrossRef]
- Pinheiro, M.; Alves, J.L. The feasibility of a custom-made endoprosthesis in mandibular reconstruction: Implant design and finite element analysis. J. Cranio Maxillo Facial Surg. 2015, 43, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. HeartFlow. HeartFlow Website. 2020. Available online: https://www.heartflow.com/ (accessed on 29 December 2020).
- Gray, R.A.; Pathmanathan, P. Patient-specific cardiovascular computational modeling: Diversity of personalization and challenges. J. Cardiovasc. Transl. Res. 2018, 11, 80–88. [Google Scholar] [CrossRef]
- Hlatky, M.A.; de Bruyne, B.; Pontone, G.; Patel, M.R.; Norgaard, B.L.; Byrne, R.A.; Curzen, N.; Purcell, I.; Gutberlet, M.; Rioufol, G.; et al. Quality-of-life and economic outcomes of assessing fractional flow reserve with computed tomography angiography: PLATFORM. J. Am. Coll. Cardiol. 2015, 66, 2315–2323. [Google Scholar] [CrossRef]
- Douglas, P.S.; Pontone, G.; Hlatky, M.A.; Patel, M.R.; Norgaard, B.L.; Byrne, R.A.; Curzen, N.; Purcell, I.; Gutberlet, M.; Rioufol, G.; et al. Clinical outcomes of fractional flow reserve by computed tomographic angiography-guided diagnostic strategies vs usual care in patients with suspected coronary artery disease: The prospective longitudinal trial of FFR(CT): Outcome and resource impacts st. Eur. Heart J. 2015, 36, 3359–3367. [Google Scholar] [CrossRef]
- Lee, T.S.; Goyal, P.; Li, C.; Zhao, K. Computational fluid dynamics to evaluate the effectiveness of inferior turbinate reduction techniques to improve nasal airflow. JAMA Facial Plast. Surg. 2018, 20, 263–270. [Google Scholar] [CrossRef]
- Boyle, P.M.; Zghaib, T.; Zahid, S.; Ali, R.L.; Deng, D.; Franceschi, W.H.; Hakim, J.B.; Murphy, M.J.; Prakosa, A.; Zimmerman, S.L.; et al. Computationally guided personalized targeted ablation of persistent atrial fibrillation. Nat. Biomed. Eng. 2019, 3, 870–879. [Google Scholar] [CrossRef]
- Uchida, T.; Delp, S. Biomechanics of Movement: Science of Sports, Robotics, and Rehabiliation; MIT Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Fregly, B.J.; D’Lima, D.D.; Colwell, C.W. Effective gait patterns for offloading the medial compartment of the knee. J. Orthop. Res. 2009, 27, 1016–1021. [Google Scholar] [CrossRef]
- Gerbrands, T.A.; Pisters, M.F.; Theeven, P.J.R.; Verschueren, S.; Vanwanseele, B. Lateral trunk lean and medializing the knee as gait strategies for knee osteoarthritis. Gait Posture 2017, 51, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Ferrigno, C.; Wimmer, M.A.; Trombley, R.M.; Lundberg, H.J.; Shakoor, N.; Thorp, L.E. A reduction in the knee adduction moment with medial thrust gait is associated with a medial shift in center of plantar pressure. Med. Eng. Phys. 2016, 38, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Gerbrands, T.A.; Pisters, M.F.; Vanwanseele, B. Individual selection of gait retraining strategies is essential to optimally reduce medial knee load during gait. Clin. Biomech. 2014, 29, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, L.K.; Laubach, L.L.; Barrios, J.A. Effect of specific gait modifications on medial knee loading, metabolic cost and perception of task difficulty. Clin. Biomech. 2013, 28, 649–654. [Google Scholar] [CrossRef]
- Shull, P.B.; Lurie, K.L.; Cutkosky, M.R.; Besier, T.F. Training multi-parameter gaits to reduce the knee adduction moment with data-driven models and haptic feedback. J. Biomech. 2011, 44, 1605–1609. [Google Scholar] [CrossRef]
- Barrios, J.A.; Crossley, K.M.; Davis, I.S. Gait retraining to reduce the knee adduction moment through real-time visual feedback of dynamic knee alignment. J. Biomech. 2010, 43, 2208–2213. [Google Scholar] [CrossRef]
- Dowling, A.V.; Fisher, D.S.; Andriacchi, T.P. Gait modification via verbal instruction and an active feedback system to reduce peak knee adduction moment. J. Biomech. Eng. 2010, 132, 071007. [Google Scholar] [CrossRef]
- Schwartz, R.P.; Heath, A.L. The pneumographic method of recording gait. J. Bone Jt. Surg. (Am.) 1932, 14, 783–794. [Google Scholar]
- Lofterød, B.; Terjesen, T.; Skaaret, I.; Huse, A.; Jahnsen, R. Preoperative gait analysis has a substantial effect on orthopedic decision making in children with cerebral palsy: Comparison between clinical evaluation and gait analysis in 60 patients. Acta Orthop. 2007, 78, 74–80. [Google Scholar] [CrossRef]
- Lofterød, B.; Terjesen, T. Results of treatment when orthopaedic surgeons follow gait-analysis recommendations in children with CP. Dev. Med. Child Neurol. 2008, 50, 503–509. [Google Scholar] [CrossRef]
- Ferrari, A.; Brunner, R.; Faccioli, S.; Reverberi, S.; Benedetti, M. Gait analysis contribution to problems identification and surgical planning in CP patients: An agreement study. Eur. J. Phys. Rehabil. Med. 2015, 51, 39–48. [Google Scholar] [PubMed]
- Brand, R.A. Locomotion analysis. In Oxford Textbook of Orthopaedics and Trauma, 1st ed.; Bulstrode, C., Buckwalter, J.J., Carr, A., Marsh, L., Fairbank, J., Wilson-MacDonald, J., Bowden, G., Eds.; Oxford University Press: New York, NY, USA, 2002; pp. 439–454. [Google Scholar]
- Zajac, F.E. Muscle and tendon: Properties, models, scaling, and application to biomechanics and motor control. Crit. Rev. Biomed. Eng. 1989, 17, 359–411. [Google Scholar]
- Hatem, S.M.; Saussez, G.; della Faille, M.; Prist, V.; Zhang, X.; Dispa, D.; Bleyenheuft, Y. Rehabilitation of motor function after stroke: A multiple systematic review focused on techniques to stimulate upper extremity recovery. Front. Hum. Neurosci. 2016, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Akhras, M.A.; Bortoletto, R.; Madehkhaksar, F.; Tagliapietra, L. Neural and musculoskeletal modeling: Its role in neurorehabilitation. In Emerging Therapies in Neurorehabilitation II (Biosystems & Biorobotics), 1st ed.; Pons, J.L., Raya, R., González, J., Eds.; Springer: New York, NY, USA, 2016; pp. 109–144. [Google Scholar]
- Serrancoli, G.; Kinney, A.L.; Fregly, B.J.; Font-Llagunes, J.M. Neuromusculoskeletal model calibration significantly affects predicted knee contact forces for walking. J. Biomech. Eng. 2016, 138, 081001. [Google Scholar] [CrossRef] [PubMed]
- Żuk, M.; Syczewska, M.; Pezowicz, C. Influence of uncertainty in selected musculoskeletal model parameters on muscle forces estimated in inverse dynamics-based static optimization and hybrid approach. J. Biomech. Eng. 2018, 140, 121001. [Google Scholar] [CrossRef]
- Lloyd, D.G.; Besier, T.F. An EMG-driven musculoskeletal model to estimate muscle forces and knee joint moments in vivo. J. Biomech. 2003, 36, 765–776. [Google Scholar] [CrossRef]
- Manal, K.; Buchanan, T.S. Use of an EMG-driven biomechanical model to study virtual injuries. Med. Sci. Sports Exerc. 2005, 37, 1917–1923. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J.; Patten, C.; Fregly, B.J. Lower extremity EMG-driven modeling of walking with automated adjustment of musculoskeletal geometry. PLoS ONE 2017, 12, e0179698. [Google Scholar] [CrossRef] [PubMed]
- Reinbolt, J.A.; Haftka, R.T.; Chmielewski, T.L.; Fregly, B.J. Are patient-specific joint and inertial parameters necessary for accurate inverse dynamics analyses of gait? IEEE Trans. Biomed. Eng. 2007, 54, 782–793. [Google Scholar] [CrossRef]
- Moissenet, F.; Modenese, L.; Dumas, R. Alterations of musculoskeletal models for a more accurate estimation of lower limb joint contact forces during normal gait: A systematic review. J. Biomech. 2017, 63, 8–20. [Google Scholar] [CrossRef]
- Arones, M.; Shourijeh, M.S.; Patten, C.; Fregly, B.J. Musculoskeletal model personalization affects metabolic cost estimates for walking. Front. Bioeng. Biotechnol. 2020, in press. [Google Scholar] [CrossRef]
- Gerus, P.; Sartori, M.; Besier, T.F.; Fregly, B.J.; Delp, S.L.; Banks, S.A.; Pandy, M.G.; D’Lima, D.D.; Lloyd, D.G. Subject-specific knee joint geometry improves predictions of medial tibiofemoral contact forces. J. Biomech. 2013, 46, 2778–2786. [Google Scholar] [CrossRef]
- National Academy of Engineering, Engineering Better Medicines. 2010. Available online: http://www.engineeringchallenges.org/challenges/medicines.aspx (accessed on 29 December 2020).
- Carter, D.R.; Vasu, R.; Harris, W.H. Stress changes in the femoral head due to porous ingrowth surface replacement arthroplasty. J. Biomech. 1984, 17, 737–747. [Google Scholar] [CrossRef]
- Fyhrie, D.P.; Schaffler, M.B. The adaptation of bone apparent density to applied load. J. Biomech. 1995, 28, 135–146. [Google Scholar] [CrossRef]
- Beaupré, G.S.; Orr, T.E.; Carter, D.R. An approach for time-dependent bone modeling and remodeling-application: A preliminary remodeling simulation. J. Orthop. Res. 1990, 8, 662–670. [Google Scholar] [CrossRef]
- Ruimerman, R.; Hilbers, P.; van Rietbergen, B.; Huiskes, R. A theoretical framework for strain-related trabecular bone maintenance and adaptation. J. Biomech. 2005, 38, 931–941. [Google Scholar] [CrossRef]
- Jacobs, C.R.; Levenston, M.E.; Beaupré, G.S.; Simo, J.C.; Carter, D.R. Numerical instabilities in bone remodeling simulations: The advantages of a node-based finite element approach. J. Biomech. 1995, 28, 449–459. [Google Scholar] [CrossRef]
- Siffert, R.S.; Luo, G.M.; Cowin, S.C.; Kaufman, J.J. Dynamic relationships of trabecular bone density, architecture, and strength in a computational model of osteopenia. Bone 1996, 18, 197–206. [Google Scholar] [CrossRef]
- Martin, K.S.; Virgilio, K.M.; Peirce, S.M.; Blemker, S.S. Computational modeling of muscle regeneration and adaptation to advance muscle tissue regeneration strategies. Cells Tissues Organs 2016, 202, 250–266. [Google Scholar] [CrossRef]
- Brand, R.A. Biomechanics or necromechanics? Or how to interpret biomechanical studies. Iowa Orthop. J. 2002, 22, 110–115. [Google Scholar] [PubMed]
- Reinkensmeyer, D.J.; Burdet, E.; Casadio, M.; Krakauer, J.W.; Kwakkel, G.; Lang, C.E.; Swinnen, S.P.; Ward, N.S.; Schweighofer, N. Computational neurorehabilitation: Modeling plasticity and learning to predict recovery. J. Neuroeng. Rehabil. 2016, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.L.; Uchida, T.K.; Seth, A.; Rajagopal, A.; Delp, S.L. Is my model good enough? Best practices for verification and validation of musculoskeletal models and simulations of movement. J. Biomech. Eng. 2015, 137, 020905. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, D.R.; Brand, R.A.; Cheng, C.; Arora, J.S. Direct comparison of muscle force predictions using linear and nonlinear programming. J. Biomech. Eng. 1987, 109, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, K.R.; An, K.W.; Litchy, W.J.; Chao, E.Y. Physiological prediction of muscle forces--I Theoretical formulation. Neuroscience 1991, 40, 781–792. [Google Scholar] [CrossRef]
- Li, G.; Kaufman, K.R.; Chao, E.Y.; Rubash, H.E. Prediction of antagonistic muscle forces using inverse dynamic optimization during flexion/extension of the knee. J. Biomech. Eng. 1999, 121, 316–322. [Google Scholar] [CrossRef]
- Jinha, A.; Ait-Haddou, R.; Binding, P.; Herzog, W. Antagonistic activity of one-joint muscles in three-dimensions using non-linear optimisation. Math. Biosci. 2006, 202, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.R.; Heller, M.O.; Bergmann, G.; Duda, G.N. Tibio-femoral loading during human gait and stair climbing. J. Orthop. Res. 2004, 22, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, G.; Deuretzbacher, G.; Heller, M.; Graichen, F.; Rohlmann, A.; Strauss, J.; Duda, G.N. Hip contact forces and gait patterns from routine activities. J. Biomech. 2001, 34, 859–871. [Google Scholar] [CrossRef]
- Brand, R.A.; Pedersen, D.R.; Davy, D.T.; Kotzar, G.M.; Heiple, K.G.; Goldberg, V.M. Comparison of hip force calculations and measurements in the same patient. J. Arthroplast. 1994, 9, 45–51. [Google Scholar] [CrossRef]
- Graichen, F.; Bergmann, G. Four-channel telemetry system for in vivo measurement of hip joint forces. J. Biomed. Eng. 1991, 13, 370–374. [Google Scholar] [CrossRef]
- Heller, M.O.; Bergmann, G.; Deuretzbacher, G.; Dürselen, L.; Pohl, M.; Claes, L.; Haas, N.P.; Duda, G.N. Musculo-skeletal loading conditions at the hip during walking and stair climbing. J. Biomech. 2001, 34, 883–893. [Google Scholar] [CrossRef]
- Rohlmann, A.; Graichen, F.; Bender, A.; Kayser, R.; Bergmann, G. Loads on a telemeterized vertebral body replacement measured in three patients within the first postoperative month. Clin. Biomech. 2008, 23, 147–158. [Google Scholar] [CrossRef]
- Stansfield, B.W.; Nicol, A.C.; Paul, J.P.; Kelly, I.G.; Graichen, F.; Bergmann, G. Direct comparison of calculated hip joint contact forces with those measured using instrumented implants An evaluation of a three-dimensional mathematical model of the lower limb. J. Biomech. 2003, 36, 929–936. [Google Scholar] [CrossRef]
- Kaufman, K.R.; Kovacevic, N.; Irby, S.E.; Colwell, C.W. Instrumented implant for measuring tibiofemoral forces. J. Biomech. 1996, 29, 667–671. [Google Scholar] [CrossRef]
- D’Lima, D.D.; Patil, S.; Steklov, N.; Slamin, J.E.; Colwell, C.W. The Chitranjan Ranawat Award: In vivo knee forces after total knee arthroplasty. Clin. Orthop. Relat. Res. 2005, 440, 45–49. [Google Scholar] [CrossRef]
- D’Lima, D.D.; Steklov, N.; Patil, S.; Colwell, C.W. The Mark Coventry Award: In vivo knee forces during recreation and exercise after knee arthroplasty. Clin. Orthop. Relat. Res. 2008, 466, 2605–2611. [Google Scholar] [CrossRef] [PubMed]
- D’Lima, D.D.; Townsend, C.P.; Arms, S.W.; Morris, B.A.; Colwell, C.W. An implantable telemetry device to measure intra-articular tibial forces. J. Biomech. 2005, 38, 299–304. [Google Scholar] [CrossRef]
- Fregly, B.J.; Besier, T.F.; Lloyd, D.G.; Delp, S.L.; Banks, S.A.; Pandy, M.G.; D’Lima, D.D. Grand challenge competition to predict in vivo knee loads. J. Orthop. Res. 2012, 30, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.; Schütz, P.; Bergmann, G.; List, R.; Postolka, B.; Hitz, M.; Dymke, J.; Damm, P.; Duda, G.; Gerber, H.; et al. A comprehensive assessment of the musculoskeletal system: The CAMS-Knee data set. J. Biomech. 2017, 65, 32–39. [Google Scholar] [CrossRef]
- Anonymous. Orthoload. OrthoLoad—Loading of Orthopaedic Implants Website. 2020. Available online: https://orthoload.com/ (accessed on 29 December 2020).
- Finni, T.; Komi, P.V.; Lukkariniemi, J. Achilles tendon loading during walking: Application of a novel optic fiber technique. Eur. J. Appl. Physiol. Occup. Physiol. 1998, 77, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Komi, P.V.; Fukashiro, S.; Järvinen, M. Biomechanical loading of Achilles tendon during normal locomotion. Clin. Sports Med. 1992, 11, 521–531. [Google Scholar] [CrossRef]
- Ishikawa, M.; Komi, P.V.; Grey, M.J.; Lepola, V.; Bruggemann, G.P. Muscle-tendon interaction and elastic energy usage in human walking. J. Appl. Physiol. 2005, 99, 603–608. [Google Scholar] [CrossRef]
- Dennerlein, J.T.; Diao, E.; Mote, C.D.; Rempel, D.M. Tensions of the flexor digitorum superficialis are higher than a current model predicts. J. Biomech. 1998, 31, 295–301. [Google Scholar] [CrossRef]
- Schuind, F.; Garcia-Elias, M.; Cooney, W.P.; An, K.N. Flexor tendon forces: In vivo measurements. J. Hand Surg. 1992, 17, 291–298. [Google Scholar] [CrossRef]
- Gregor, R.J.; Komi, P.V.; Järvinen, M. Achilles tendon forces during cycling. Int. J. Sports Med. 1987, 8 (Suppl. S1), 9–14. [Google Scholar] [CrossRef]
- Martin, J.A.; Brandon, S.C.E.; Keuler, E.M.; Hermus, J.R.; Ehlers, A.C.; Segalman, D.J.; Allen, M.S.; Thelen, D.G. Gauging force by tapping tendons. Nat. Commun. 2018, 9, 1592. [Google Scholar] [CrossRef] [PubMed]
- Acuña, S.A.; Ebrahimi, A.; Pomeroy, R.L.; Martin, J.A.; Thelen, D.G. Achilles tendon shear wave speed tracks the dynamic modulation of standing balance. Physiol. Rep. 2019, 7, e14298. [Google Scholar] [CrossRef]
- Fregly, B.J.; D’Lima, D.D.; Besier, T.F. SimTK: Grand Challenge Competition to Predict In Vivo Knee Loads: Project Home. SimTK.org Project. 2021. Available online: https://simtk.org/projects/kneeloads (accessed on 8 February 2021).
- Erdemir, A.; Hunter, P.J.; Holzapfel, G.A.; Loew, L.M.; Middleton, J.; Jacobs, C.R.; Nithiarasu, P.; Löhner, R.; Wei, G.; Winkelstein, B.A.; et al. Perspectives on sharing models and related resources in computational biomechanics research. J. Biomech. Eng. 2018, 140, 0247011. [Google Scholar] [CrossRef]
- Anderson, F.C.; Pandy, M.G. Dynamic optimization of human walking. J. Biomech. Eng. 2001, 123, 381–390. [Google Scholar] [CrossRef]
- Falisse, A.; Serrancolí, G.; Dembia, C.L.; Gillis, J.; Jonkers, I.; de Groote, F. Rapid predictive simulations with complex musculoskeletal models suggest that diverse healthy and pathological human gaits can emerge from similar control strategies. J. R. Soc. Interface 2019, 16, 20190402. [Google Scholar] [CrossRef] [PubMed]
- Falisse, A.; Pitto, L.; Kainz, H.; Hoang, H.; Wesseling, M.; van Rossom, S.; Papageorgiou, E.; Bar-On, L.; Hallemans, A.; Desloovere, K.; et al. Physics-based simulations to predict the differential effects of motor control and musculoskeletal deficits on gait dysfunction in cerebral palsy: A retrospective case study. Front. Hum. Neurosci. 2020, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.L.; Kautz, S.A.; Neptune, R.R. The influence of merged muscle excitation modules on post-stroke hemiparetic walking performance. Clin. Biomech. 2013, 28, 697–704. [Google Scholar] [CrossRef]
- Miller, R.H. A comparison of muscle energy models for simulating human walking in three dimensions. J. Biomech. 2014, 47, 1373–1381. [Google Scholar] [CrossRef]
- Miller, R.H.; Esterson, A.Y.; Shim, J.K. Joint contact forces when minimizing the external knee adduction moment by gait modification: A computer simulation study. Knee 2015, 22, 481–489. [Google Scholar] [CrossRef]
- John, C.T.; Anderson, F.C.; Higginson, J.S.; Delp, S.L. Stabilisation of walking by intrinsic muscle properties revealed in a three-dimensional muscle-driven simulation. Comput. Methods Biomech. Biomed. Eng. 2013, 16, 452–462. [Google Scholar] [CrossRef]
- Steele, K.M.; van der Krogt, M.M.; Schwartz, M.H.; Delp, S.L. How much muscle strength is required to walk in a crouch gait? J. Biomech. 2012, 45, 2564–2569. [Google Scholar] [CrossRef]
- Hamner, S.R.; Seth, A.; Delp, S.L. Muscle contributions to propulsion and support during running. J. Biomech. 2010, 43, 2709–2716. [Google Scholar] [CrossRef]
- Knarr, B.A.; Reisman, D.S.; Binder-Macleod, S.A.; Higginson, J.S. Understanding compensatory strategies for muscle weakness during gait by simulating activation deficits seen post-stroke. Gait Posture 2013, 38, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Pitto, L.; van Rossom, S.; Desloovere, K.; Molenaers, G.; Huenaerts, C.; de Groote, F.; Jonkers, I. Pre-treatment EMG can be used to model post-treatment muscle coordination during walking in children with cerebral palsy. PLoS ONE 2020, 15, e0228851. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, N.; Yamazaki, N. Generation of human bipedal locomotion by a bio-mimetic neuro-musculo-skeletal model. Biol. Cybern. 2001, 84, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, A.J.; Haenen, W.P.; Rozendaal, L.A. Stability of bipedal stance: The contribution of cocontraction and spindle feedback. Biol. Cybern. 2003, 88, 293–301. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jo, S.; Massaquoi, S.G. A model of cerebrocerebello-spinomuscular interaction in the sagittal control of human walking. Biol. Cybern. 2007, 96, 279–307. [Google Scholar] [CrossRef]
- Jo, S. A neurobiological model of the recovery strategies from perturbed walking. Biosystems 2007, 90, 750–768. [Google Scholar] [CrossRef]
- Geyer, H.; Herr, H. A muscle-reflex model that encodes principles of legged mechanics produces human walking dynamics and muscle activities. IEEE Trans. Neural Syst. Rehabil. Eng. 2010, 18, 263–273. [Google Scholar] [CrossRef]
- Kistemaker, D.A.; van Soest, A.J.K.; Wong, J.D.; Kurtzer, I.; Gribble, P.L. Control of position and movement is simplified by combined muscle spindle and Golgi tendon organ feedback. J. Neurophysiol. 2013, 109, 1126–1139. [Google Scholar] [CrossRef]
- Tsianos, G.A.; Goodner, J.; Loeb, G.E. Useful properties of spinal circuits for learning and performing planar reaches. J. Neural Eng. 2014, 11, 056006. [Google Scholar] [CrossRef] [PubMed]
- Byadarhaly, K.V.; Perdoor, M.C.; Minai, A.A. A modular neural model of motor synergies. Neural Netw. 2012, 32, 96–108. [Google Scholar] [CrossRef]
- Sreenivasa, M.; Ayusawa, K.; Nakamura, Y. Modeling and identification of a realistic spiking neural network and musculoskeletal model of the human arm, and an application to the stretch reflex. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 24, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Hardesty, R.L.; Boots, M.T.; Yakovenko, S.; Gritsenko, V. Computational evidence for nonlinear feedforward modulation of fusimotor drive to antagonistic co-contracting muscles. Sci. Rep. 2020, 10, 10625. [Google Scholar] [CrossRef] [PubMed]
- Aoi, S.; Ohashi, T.; Bamba, R.; Fujiki, S.; Tamura, D.; Funato, T.; Senda, K.; Ivanenko, Y.; Tsuchiya, K. Neuromusculoskeletal model that walks and runs across a speed range with a few motor control parameter changes based on the muscle synergy hypothesis. Sci. Rep. 2019, 9, 369. [Google Scholar] [CrossRef]
- Dzeladini, F.; van den Kieboom, J.; Ijspeert, A. The contribution of a central pattern generator in a reflex-based neuromuscular model. Front. Hum. Neurosci. 2014, 8, 371. [Google Scholar] [CrossRef]
- Markin, S.N.; Klishko, A.N.; Shevtsova, N.A.; Lemay, M.A.; Prilutsky, B.I.; Rybak, I.A. Afferent control of locomotor CPG: Insights from a simple neuromechanical model. Ann. N. Y. Acad. Sci. 2010, 1198, 21–34. [Google Scholar] [CrossRef]
- Endo, K.; Herr, H. A model of muscle-tendon function in human walking at self-selected speed. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 352–362. [Google Scholar] [CrossRef]
- Franklin, D.W.; Burdet, E.; Tee, K.P.; Osu, R.; Chew, C.-M.; Milner, T.E.; Kawato, M. CNS learns stable, accurate, and efficient movements using a simple algorithm. J. Neurosci. 2008, 28, 11165–11173. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.L.; Ting, L.H. Optimization of muscle activity for task-level goals predicts complex changes in limb forces across biomechanical contexts. PLoS Comput. Biol. 2012, 8, e1002465. [Google Scholar] [CrossRef] [PubMed]
- Sartori, M.; Gizzi, L.; Lloyd, D.G.; Farina, D. A musculoskeletal model of human locomotion driven by a low dimensional set of impulsive excitation primitives. Front. Comput. Neurosci. 2013, 7, 79. [Google Scholar] [CrossRef]
- Razavian, R.S.; Ghannadi, B.; McPhee, J. A synergy-based motor control framework for the fast feedback control of musculoskeletal systems. J. Biomech. Eng. 2018, 141, 031009. [Google Scholar] [CrossRef]
- Al Borno, M.; Hicks, J.L.; Delp, S.L. The effects of motor modularity on performance, learning and generalizability in upper-extremity reaching: A computational analysis. J. R. Soc. Interface 2020, 17, 20200011. [Google Scholar] [CrossRef]
- Williams, I.; Constandinou, T.G. Computationally efficient modeling of proprioceptive signals in the upper limb for prostheses: A simulation study. Front. Neurosci. 2014, 8, 181. [Google Scholar] [CrossRef]
- Falisse, A.; Bar-On, L.; Desloovere, K.; Jonkers, I.; de Groote, F. A spasticity model based on feedback from muscle force explains muscle activity during passive stretches and gait in children with cerebral palsy. PLoS ONE 2018, 13, e0208811. [Google Scholar] [CrossRef] [PubMed]
- De Groote, F.; Blum, K.P.; Horslen, B.C.; Ting, L.H. Interaction between muscle tone, short-range stiffness and increased sensory feedback gains explains key kinematic features of the pendulum test in spastic cerebral palsy: A simulation study. PLoS ONE 2018, 13, e0205763. [Google Scholar] [CrossRef]
- Miyazaki, T.; Wada, M.; Kawahara, H.; Sato, M.; Baba, H.; Shimada, S. Dynamic load at baseline can predict radiographic disease progression in medial compartment knee osteoarthritis. Ann. Rheum. Dis. 2002, 61, 617–622. [Google Scholar] [CrossRef]
- Prodromos, C.C.; Andriacchi, T.P.; Galante, J.O. A relationship between gait and clinical changes following high tibial osteotomy. J. Bone Jt. Surg. Am. Vol. 1985, 67, 1188–1194. [Google Scholar] [CrossRef]
- Wada, M.; Imura, S.; Nagatani, K.; Baba, H.; Shimada, S.; Sasaki, S. Relationship between gait and clinical results after high tibial osteotomy. Clin. Orthop. Relat. Res. 1998, 354, 180–188. [Google Scholar] [CrossRef]
- Van den Bogert, A.J.; Smith, G.D.; Nigg, B.M. In vivo determination of the anatomical axes of the ankle joint complex: An optimization approach. J. Biomech. 1994, 27, 1477–1488. [Google Scholar] [CrossRef]
- Charlton, I.W.; Tate, P.; Smyth, P.; Roren, L. Repeatability of an optimised lower body model. Gait Posture 2004, 20, 213–221. [Google Scholar] [CrossRef]
- Reinbolt, J.A.; Schutte, J.F.; Fregly, B.J.; Koh, B.I.; Haftka, R.T.; George, A.D.; Mitchell, K.H. Determination of patient-specific multi-joint kinematic models through two-level optimization. J. Biomech. 2005, 38, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Reinbolt, J.A.; Haftka, R.T.; Chmielewski, T.L.; Fregly, B.J. A computational framework to predict post-treatment outcome for gait-related disorders. Med. Eng. Phys. 2008, 30, 434–443. [Google Scholar] [CrossRef]
- Piazza, S.J.; Okita, N.; Cavanagh, P.R. Accuracy of the functional method of hip joint center location: Effects of limited motion and varied implementation. J. Biomech. 2001, 34, 967–973. [Google Scholar] [CrossRef]
- Piazza, S.J.; Erdemir, A.; Okita, N.; Cavanagh, P.R. Assessment of the functional method of hip joint center location subject to reduced range of hip motion. J. Biomech. 2004, 37, 349–356. [Google Scholar] [CrossRef]
- Besier, T.F.; Sturnieks, D.L.; Alderson, J.A.; Lloyd, D.G. Repeatability of gait data using a functional hip joint centre and a mean helical knee axis. J. Biomech. 2003, 36, 1159–1168. [Google Scholar] [CrossRef]
- Millard, M.; Uchida, T.; Seth, A.; Delp, S.L. Flexing computational muscle: Modeling and simulation of musculotendon dynamics. J. Biomech. Eng. 2013, 135, 021005. [Google Scholar] [CrossRef] [PubMed]
- Bogey, R.A.; Perry, J.; Gitter, A.J. An EMG-to-force processing approach for determining ankle muscle forces during normal human gait. IEEE Trans. Neural Syst. Rehabil. Eng. 2005, 13, 302–310. [Google Scholar] [CrossRef]
- Buchanan, T.S.; Lloyd, D.G.; Manal, K.; Besier, T.F. Estimation of muscle forces and joint moments using a forward-inverse dynamics model. Med. Sci. Sports Exerc. 2005, 37, 1911–1916. [Google Scholar] [CrossRef]
- Winby, C.R.; Lloyd, D.G.; Besier, T.F.; Kirk, T.B. Muscle and external load contribution to knee joint contact loads during normal gait. J. Biomech. 2009, 42, 2294–2300. [Google Scholar] [CrossRef]
- Gerus, P.; Rao, G.; Buchanan, T.S.; Berton, E. A clinically applicable model to estimate the opposing muscle groups contributions to isometric and dynamic tasks. Ann. Biomed. Eng. 2010, 38, 2406–2417. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Rudolph, K.S.; Manal, K.T. EMG-driven modeling approach to muscle force and joint load estimations: Case study in knee osteoarthritis. J. Orthop. Res. 2012, 30, 377–383. [Google Scholar] [CrossRef]
- Sartori, M.; Reggiani, M.; Farina, D.; Lloyd, D.G. EMG-driven forward-dynamic estimation of muscle force and joint moment about multiple degrees of freedom in the human lower extremity. PLoS ONE 2012, 7, e52618. [Google Scholar] [CrossRef] [PubMed]
- Gardinier, E.S.; Manal, K.; Buchanan, T.S.; Snyder-Mackler, L. Minimum detectable change for knee joint contact force estimates using an EMG-driven model. Gait Posture 2013, 38, 1051–1053. [Google Scholar] [CrossRef]
- Menegaldo, L.L.; Fleury, A.d.; Weber, H.I. Moment arms and musculotendon lengths estimation for a three-dimensional lower-limb model. J. Biomech. 2004, 37, 1447–1453. [Google Scholar] [CrossRef] [PubMed]
- Sartori, M.; Reggiani, M.; van den Bogert, A.J.; Lloyd, D.G. Estimation of musculotendon kinematics in large musculoskeletal models using multidimensional B-splines. J. Biomech. 2012, 45, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.N.; Hass, C.J.; Fregly, B.J. Development of a subject-specific foot-ground contact model for walking. J. Biomech. Eng. 2016, 138, 091002. [Google Scholar] [CrossRef]
- Delp, S.L.; Anderson, F.C.; Arnold, A.S.; Loan, P.; Habib, A.; John, C.T.; Guendelman, E.; Thelen, D.G. OpenSim: Open-source software to create and analyze dynamic simulations of movement. IEEE Trans. Biomed. Eng. 2007, 54, 1940–1950. [Google Scholar] [CrossRef]
- Damsgaard, M.; Rasmussen, J.; Christensen, S.T.; Surma, E.; de Zee, M. Analysis of musculoskeletal systems in the AnyBody Modeling System. Simul. Model. Pract. Theory 2006, 14, 1100–1111. [Google Scholar] [CrossRef]
- Van den Bogert, A.J.; Blana, D.; Heinrich, D. Implicit methods for efficient musculoskeletal simulation and optimal control. Procedia Iutam 2011, 2, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; van den Bogert, A.J. Optimality principles for model-based prediction of human gait. J. Biomech. 2010, 43, 1055–1060. [Google Scholar] [CrossRef]
- Halloran, J.P.; Erdemir, A.; van den Bogert, A.J. Adaptive surrogate modeling for efficient coupling of musculoskeletal control and tissue deformation models. J. Biomech. Eng. 2009, 131, 011014. [Google Scholar] [CrossRef]
- Ackermann, M.; van den Bogert, A.J. Predictive simulation of gait at low gravity reveals skipping as the preferred locomotion strategy. J. Biomech. 2012, 45, 1293–1298. [Google Scholar] [CrossRef]
- McLean, S.G.; Su, A.; van den Bogert, A.J. Development and validation of a 3-D model to predict knee joint loading during dynamic movement. J. Biomech. Eng. 2003, 125, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Koelewijn, A.D.; Dorschky, E.; van den Bogert, A.J. A metabolic energy expenditure model with a continuous first derivative and its application to predictive simulations of gait. Comput. Methods Biomech. Biomed. Eng. 2018, 21, 521–531. [Google Scholar] [CrossRef]
- Van den Bogert, A.J.; Samorezov, S.; Davis, B.L.; Smith, W.A. Modeling and optimal control of an energy-storing prosthetic knee. J. Biomech. Eng. 2012, 134, 051007. [Google Scholar] [CrossRef]
- Koelewijn, A.D.; van den Bogert, A.J. Joint contact forces can be reduced by improving joint moment symmetry in below-knee amputee gait simulations. Gait Posture 2016, 49, 219–225. [Google Scholar] [CrossRef]
- Buchanan, T.S.; Lloyd, D.G.; Manal, K.; Besier, T.F. Neuromusculoskeletal modeling: Estimation of muscle forces and joint moments and movements from measurements of neural command. J. Appl. Biomech. 2004, 20, 367–395. [Google Scholar] [CrossRef]
- Rao, G.; Berton, E.; Amarantini, D.; Vigouroux, L.; Buchanan, T.S. An EMG-driven biomechanical model that accounts for the decrease in moment generation capacity during a dynamic fatigued condition. J. Biomech. Eng. 2010, 132, 071003. [Google Scholar] [CrossRef]
- Shao, Q.; Bassett, D.N.; Manal, K.; Buchanan, T.S. An EMG-driven model to estimate muscle forces and joint moments in stroke patients. Comput. Biol. Med. 2009, 39, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Manal, K.; Buchanan, T.S. An electromyogram-driven musculoskeletal model of the knee to predict in vivo joint contact forces during normal and novel gait patterns. J. Biomech. Eng. 2013, 135, 021014. [Google Scholar] [CrossRef]
- Manal, K.; Gravare-Silbernagel, K.; Buchanan, T.S. A real-time EMG-driven musculoskeletal model of the ankle. Multibody Syst. Dyn. 2012, 28, 169–180. [Google Scholar] [CrossRef][Green Version]
- Pàmies-Vilà, R.; Font-Llagunes, J.M.; Lugris, U.; Cuadrado, J. Parameter identification method for a three-dimensional foot-ground contact model. Mech. Mach. Theory 2014, 75, 107–116. [Google Scholar] [CrossRef]
- Higginson, J.S.; Zajac, F.E.; Neptune, R.R.; Kautz, S.A.; Delp, S.L. Muscle contributions to support during gait in an individual with post-stroke hemiparesis. J. Biomech. 2006, 39, 1769–1777. [Google Scholar] [CrossRef]
- Liu, M.Q.; Anderson, F.C.; Pandy, M.G.; Delp, S.L. Muscles that support the body also modulate forward progression during walking. J. Biomech. 2006, 39, 2623–2630. [Google Scholar] [CrossRef]
- Arnold, A.S.; Anderson, F.C.; Pandy, M.G.; Delp, S.L. Muscular contributions to hip and knee extension during the single limb stance phase of normal gait: A framework for investigating the causes of crouch gait. J. Biomech. 2005, 38, 2181–2189. [Google Scholar] [CrossRef]
- Goldberg, S.R.; Anderson, F.C.; Pandy, M.G.; Delp, S.L. Muscles that influence knee flexion velocity in double support: Implications for stiff-knee gait. J. Biomech. 2004, 37, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Piazza, S.J.; Delp, S.L. Three-dimensional dynamic simulation of total knee replacement motion during a step-up task. J. Biomech. Eng. 2001, 123, 599–606. [Google Scholar] [CrossRef]
- Ong, C.F.; Hicks, J.L.; Delp, S.L. Simulation-based design for wearable robotic systems: An optimization framework for enhancing a standing long jump. IEEE Trans. Biomed. Eng. 2016, 63, 894–903. [Google Scholar] [CrossRef]
- Sartori, M.; Llyod, D.G.; Farina, D. Neural data-driven musculoskeletal modeling for personalized neurorehabilitation technologies. IEEE Trans. Biomed. Eng. 2016, 63, 879–893. [Google Scholar] [CrossRef]
- Sartori, M.; Farina, D.; Lloyd, D.G. Hybrid neuromusculoskeletal modeling to best track joint moments using a balance between muscle excitations derived from electromyograms and optimization. J. Biomech. 2014, 47, 3613–3621. [Google Scholar] [CrossRef]
- Gonzalez-Vargas, J.; Sartori, M.; Dosen, S.; Torricelli, D.; Pons, J.L.; Farina, D. A predictive model of muscle excitations based on muscle modularity for a large repertoire of human locomotion conditions. Front. Comput. Neurosci. 2015, 9, 114. [Google Scholar] [CrossRef]
- Febrer-Nafría, M.; Pallarès-López, R.; Fregly, B.J.; Font-Llagunes, J.M. Comparison of different optimal control formulations for generating dynamically consistent orthosis-assisted crutch walking simulations. Mech. Mach. Theory 2020, 154, 104031. [Google Scholar] [CrossRef]
- Febrer-Nafría, M.; Pallarès-López, R.; Fregly, B.J.; Font-Llagunes, J.M. Prediction of three-dimensional crutch walking patterns using a torque-driven model. Multibody Syst. Dyn. 2021, 51, 1–19. [Google Scholar] [CrossRef]
- Ao, D.; Shourijeh, M.S.; Patten, C.; Fregly, B.J. Evaluation of synergy extrapolation for predicting unmeasured muscle excitations from measured muscle synergies. Front. Comput. Neurosci. 2020, 14, 588943. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.P.; Kinney, A.L.; Banks, S.A.; D’Lima, D.D.; Besier, T.F.; Lloyd, D.G.; Fregly, B.J. Muscle synergies may improve optimization prediction of knee contact forces during walking. J. Biomech. Eng. 2014, 136, 021031. [Google Scholar] [CrossRef] [PubMed]
- Fregly, B.J.; Reinbolt, J.A.; Chmielewski, T.L. Evaluation of a patient-specific cost function to predict the influence of foot path on the knee adduction torque during gait. Comput. Methods Biomech. Biomed. Eng. 2008, 11, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Fregly, B.J. Computational assessment of combinations of gait modifications for knee osteoarthritis rehabilitation. IEEE Trans. Biomed. Eng. 2008, 55, 2104–2106. [Google Scholar] [CrossRef]
- Koh, B.-I.; Reinbolt, J.A.; George, A.D.; Haftka, R.T.; Fregly, B.J. Limitations of parallel global optimization for large-scale human movement problems. Med. Eng. Phys. 2009, 31, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Eskinazi, I.; Fregly, B.J. A computational framework for simultaneous estimation of muscle and joint contact forces and body motion using optimization and surrogate modeling. Med. Eng. Phys. 2018, 54, 56–64. [Google Scholar] [CrossRef]
- Falisse, A.; van Rossom, S.; Jonkers, I.; de Groote, F. EMG-driven optimal estimation of subject-specific Hill model muscle-tendon parameters of the knee joint actuators. IEEE Trans. Biomed. Eng. 2017, 64, 2253–2262. [Google Scholar]
- Jansen, K.; de Groote, F.; Aerts, W.; de Schutter, J.; Duysens, J.; Jonkers, I. Altering length and velocity feedback during a neuro-musculoskeletal simulation of normal gait contributes to hemiparetic gait characteristics. J. Neuroeng. Rehabil. 2014, 11, 78. [Google Scholar] [CrossRef]
- Jonkers, I.; Spaepen, A.; Papaioannou, G.; Stewart, C. An EMG-based, muscle driven forward simulation of single support phase of gait. J. Biomech. 2002, 35, 609–619. [Google Scholar] [CrossRef]
- Hoang, H.X.; Pizzolato, C.; Diamond, L.E.; Lloyd, D.G. Subject-specific calibration of neuromuscular parameters enables neuromusculoskeletal models to estimate physiologically plausible hip joint contact forces in healthy adults. J. Biomech. 2018, 80, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Millard, M.; McPhee, J.; Kubica, E. Multi-step forward dynamic gait simulation. In Multibody Dynamics: Computational Methods and Applications; Bottasso, C., Ed.; Springer: Amsterdam, The Netherlands, 2009; pp. 25–43. [Google Scholar]
- Shourijeh, M.S.; McPhee, J. Foot–ground contact modeling within human gait simulations: From Kelvin–Voigt to hyper-volumetric models. Multibody Syst. Dyn. 2015, 35, 393–407. [Google Scholar] [CrossRef]
- Brown, P.; McPhee, J. A 3D ellipsoidal volumetric foot-ground contact model for forward dynamics. Multibody Syst. Dyn. 2018, 42, 447–467. [Google Scholar] [CrossRef]
- Razavian, R.S.; Mehrabi, N.; McPhee, J. A model-based approach to predict muscle synergies using optimization: Application to feedback control. Front. Comput. Neurosci. 2015, 9, 121. [Google Scholar]
- Shourijeh, M.S.; McPhee, J. Forward dynamic optimization of human gait simulations: A global parameterization approach. J. Comput. Nonlinear Dyn. 2014, 9, 31018. [Google Scholar] [CrossRef]
- Mehrabi, N.; Razavian, R.S.; McPhee, J. A physics-based musculoskeletal driver model to study steering tasks. J. Comput. Nonlinear Dyn. 2015, 10, 21012. [Google Scholar] [CrossRef]
- Mehrabi, N.; Razavian, R.S.; Ghannadi, B.; McPhee, J. Predictive simulation of reaching moving targets using nonlinear model predictive control. Front. Comput. Neurosci. 2017, 10, 143. [Google Scholar] [CrossRef]
- Umberger, B.R.; Miller, R.H. Optimal control modeling of human movement. In Handbook of Human Motion; Müller, B., Ed.; Springer: New York, NY, USA, 2018; pp. 1–22. [Google Scholar]
- Miller, R.H.; Hamill, J. Optimal footfall patterns for cost minimization in running. J. Biomech. 2015, 48, 2858–2864. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.L.; Neptune, R.R. Three-dimensional modular control of human walking. J. Biomech. 2012, 45, 2157–2163. [Google Scholar] [CrossRef]
- McGowan, C.P.; Neptune, R.R.; Clark, D.J.; Kautz, S.A. Modular control of human walking: Adaptations to altered mechanical demands. J. Biomech. 2010, 43, 412–419. [Google Scholar] [CrossRef]
- Neptune, R.R.; Clark, D.J.; Kautz, S.A. Modular control of human walking: A simulation study. J. Biomech. 2009, 42, 1282–1287. [Google Scholar] [CrossRef]
- Sasaki, K.; Neptune, R.R. Differences in muscle function during walking and running at the same speed. J. Biomech. 2006, 39, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- Neptune, R.R. Optimization algorithm performance in determining optimal controls in human movement analyses. J. Biomech. Eng. 1999, 121, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Pandy, M.G.; Anderson, F.C.; Hull, D.G. A parameter optimization approach for the optimal control of large-scale musculoskeletal systems. J. Biomech. Eng. 1992, 114, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Anderson, F.C.; Pandy, M.G. A dynamic optimization solution for vertical jumping in three dimensions. Comput. Methods Biomech. Biomed. Eng. 1999, 2, 201–231. [Google Scholar] [CrossRef]
- Dorn, T.W.; Lin, Y.-C.; Pandy, M.G. Estimates of muscle function in human gait depend on how foot-ground contact is modelled. Comput. Methods Biomech. Biomed. Eng. 2012, 15, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Porsa, S.; Lin, Y.-C.; Pandy, M.G. Direct methods for predicting movement biomechanics based upon optimal control theory with implementation in OpenSim. Ann. Biomed. Eng. 2016, 44, 2542–2557. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.-F.; Umberger, B.R. Generating optimal control simulations of musculoskeletal movement using OpenSim and MATLAB. PeerJ 2016, 4, e1638. [Google Scholar] [CrossRef] [PubMed]
- Gidley, A.D.; Marsh, A.P.; Umberger, B.R. Performance criteria for generating predictive optimal control simulations of bicycle pedaling. Comput. Methods Biomech. Biomed. Eng. 2019, 22, 11–20. [Google Scholar] [CrossRef]
- Nguyen, V.Q.; Johnson, R.T.; Sup, F.C.; Umberger, B.R. Bilevel optimization for cost function determination in dynamic simulation of human gait. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Pandy, M.G.; Zajac, F.E. Optimal muscular coordination strategies for jumping. J. Biomech. 1991, 24, 1–10. [Google Scholar] [CrossRef]
- Rohani, F.; Richter, H.; van den Bogert, A.J. Optimal design and control of an electromechanical transfemoral prosthesis with energy regeneration. PLoS ONE 2017, 12, e0188266. [Google Scholar]
- Dembia, C.L.; Bianco, N.A.; Falisse, A.; Hicks, J.L.; Delp, S.L. OpenSim Moco: Musculoskeletal optimal control. PLoS Comput. Biol. 2020, 16, e1008493. [Google Scholar] [CrossRef]
- Wang, H.; van den Bogert, A. Identification of postural controllers in human standing balance. J. Biomech. Eng. 2021, 143, 041001. [Google Scholar] [CrossRef]
- Lin, Y.C.; Pandy, M.G. Three-dimensional data-tracking dynamic optimization simulations of human locomotion generated by direct collocation. J. Biomech. 2017, 59, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Walter, J.P.; Pandy, M.G. Predictive simulations of neuromuscular coordination and joint-contact loading in human gait. Ann. Biomed. Eng. 2018, 46, 1216–1227. [Google Scholar] [CrossRef] [PubMed]
- Killen, B.A.; Falisse, A.; de Groote, F.; Jonkers, I. In silico-enhanced treatment and rehabilitation planning for patients with musculoskeletal disorders: Can musculoskeletalm and dynamic simulations really impact current clinical practice? Appl. Sci. 2020, 10, 7255. [Google Scholar] [CrossRef]
- Blemker, S.S.; Pinsky, P.M.; Delp, S.L. A 3D model of muscle reveals the causes of nonuniform strains in the biceps brachii. J. Biomech. 2005, 38, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, N.M.; Blemker, S.S. Musculotendon variability influences tissue strains experienced by the biceps femoris long head muscle during high-speed running. J. Biomech. 2014, 47, 3325–3333. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, K.M.; Martin, K.S.; Peirce, S.M.; Blemker, S.S. Multiscale models of skeletal muscle reveal the complex effects of muscular dystrophy on tissue mechanics and damage susceptibility. Interface Focus 2015, 5, 20140080. [Google Scholar] [CrossRef]
- Navacchia, A.; Hume, D.R.; Rullkoetter, P.J.; Shelburne, K.B. A computationally efficient strategy to estimate muscle forces in a finite element musculoskeletal model of the lower limb. J. Biomech. 2019, 84, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Hume, D.R.; Rullkoetter, P.J.; Shelburne, K.B. ReadySim: A computational framework for building explicit finite element musculoskeletal simulations directly from motion laboratory data. Int. J. Numer. Methods Biomed. Eng. 2020, 36, e3396. [Google Scholar] [CrossRef] [PubMed]
- Perillo-Marcone, A.; Taylor, M. Effect of varus/valgus malalignment on bone strains in the proximal tibia after TKR: An explicit finite element study. J. Biomech. Eng. 2007, 129, 1–11. [Google Scholar] [CrossRef]
- Maas, S.A.; Ateshian, G.A.; Weiss, J.A. FEBio: History and advances. Annu. Rev. Biomed. Eng. 2017, 19, 279–299. [Google Scholar] [CrossRef]
- Anderson, A.E.; Peters, C.L.; Tuttle, B.D.; Weiss, J.A. Subject-specific finite element model of the pelvis: Development, validation and sensitivity studies. J. Biomech. Eng. 2005, 127, 364–373. [Google Scholar] [CrossRef]
- Pegg, E.C.; Walter, J.; Mellon, S.J.; Pandit, H.G.; Murray, D.W.; D’Lima, D.D.; Fregly, B.J.; Gill, H.S. Evaluation of factors affecting tibial bone strain after unicompartmental knee replacement. J. Orthop. Res. 2013, 31, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Erdemir, A. Open Knee: Open source modeling and simulation in knee biomechanics. J. Knee Surg. 2016, 29, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Pandy, M. Direct validation of human knee-joint contact mechanics derived from subject-specific finite-element models of the tibiofemoral and patellofemoral joints. J. Biomech. Eng. 2020, 142, 071001. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Patil, S.; Steklov, N.; Bae, W.; D’Lima, D.D.; Sah, R.L.; Fregly, B.J. Finite element modelling of in vitro articular cartilage wear in the patellofemoral joint. In Proceedings of the 2012 ASME Summer Bioengineering Conference, Fajardo, PR, USA, 20–23 June 2012; pp. 163–164. [Google Scholar]
- Knight, L.A.; Pal, S.; Coleman, J.C.; Bronson, F.; Haider, H.; Levine, D.L.; Taylor, M.; Rullkoetter, P.J. Comparison of long-term numerical and experimental total knee replacement wear during simulated gait loading. J. Biomech. 2007, 40, 1550–1558. [Google Scholar] [CrossRef]
- Zhao, D.; Sakoda, H.; Sawyer, W.G.; Banks, S.A.; Fregly, B.J. Predicting knee replacement damage in a simulator machine using a computational model with a consistent wear factor. J. Biomech. Eng. 2008, 130, 011004. [Google Scholar] [CrossRef]
- Valente, G.; Crimi, G.; Vanella, N.; Schileo, E.; Taddei, F. nmsBuilder: Freeware to create subject-specific musculoskeletal models for OpenSim. Comput. Methods Programs Biomed. 2017, 152, 85–92. [Google Scholar] [CrossRef]
- Killen, B.A.; da Luz, S.B.; Lloyd, D.G.; Carleton, A.D.; Zhang, J.; Besier, T.F.; Saxby, D.J. Automated creation and tuning of personalised muscle paths for OpenSim musculoskeletal models of the knee joint. Biomech. Model. Mechanobiol. 2021. Online ahead of print.. [Google Scholar]
- Modenese, L.; Renault, J.-B. Automatic generation of personalised skeletal models of the lower limb from three-dimensional bone geometries. J. Biomech. 2021, 116, 110186. [Google Scholar] [CrossRef] [PubMed]
- Modenese, L.; Renault, J.-B. What are the differences between the STAPLE toolbox, NMSBuilder and the MAP Client for generating OpenSim models? 2021. Available online: https://github.com/modenaxe/msk-STAPLE (accessed on 14 February 2021).
- Modenese, L.; Montefiori, E.; Wang, A.; Wesarg, S.; Viceconti, M.; Mazzà, C. Investigation of the dependence of joint contact forces on musculotendon parameters using a codified workflow for image-based modelling. J. Biomech. 2018, 73, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Myers, C.A.; Laz, P.J.; Shelburne, K.B.; Davidson, B.S. A probabilistic approach to quantify the impact of uncertainty propagation in musculoskeletal simulations. Ann. Biomed. Eng. 2015, 43, 1098–1111. [Google Scholar] [CrossRef]
- Valente, G.; Pitto, L.; Testi, D.; Seth, A.; Delp, S.L.; Stagni, R.; Viceconti, M.; Taddei, F. Are subject-specific musculoskeletal models robust to the uncertainties in parameter identification? PLoS ONE 2014, 9, e112625. [Google Scholar] [CrossRef]
- Ackland, D.C.; Lin, Y.-C.; Pandy, M.G. Sensitivity of model predictions of muscle function to changes in moment arms and muscle-tendon properties: A Monte-Carlo analysis. J. Biomech. 2012, 45, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Navacchia, A.; Myers, C.A.; Rullkoetter, P.J.; Shelburne, K.B. Prediction of in vivo knee joint loads using a global probabilistic analysis. J. Biomech. Eng. 2016, 138, 4032379. [Google Scholar] [CrossRef]
- Bujalski, P.; Martins, J.; Stirling, L. A Monte Carlo analysis of muscle force estimation sensitivity to muscle-tendon properties using a Hill-based muscle model. J. Biomech. 2018, 79, 67–77. [Google Scholar] [CrossRef]
- Bosmans, L.; Valente, G.; Wesseling, M.; van Campen, A.; de Groote, F.; de Schutter, J.; Jonkers, I. Sensitivity of predicted muscle forces during gait to anatomical variability in musculotendon geometry. J. Biomech. 2015, 48, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Handsfield, G.G.; Meyer, C.H.; Hart, J.M.; Abel, M.F.; Blemker, S.S. Relationships of 35 lower limb muscles to height and body mass quantified using MRI. J. Biomech. 2014, 47, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Lansdown, D.A.; Ding, Z.; Wadington, M.; Hornberger, J.L.; Damon, B.M. Quantitative diffusion tensor MRI-based fiber tracking of human skeletal muscle. J. Appl. Physiol. 2007, 103, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Scheel, M.; von Roth, P.; Winkler, T.; Arampatzis, A.; Prokscha, T.; Hamm, B.; Diederichs, G. Fiber type characterization in skeletal muscle by diffusion tensor imaging. Nmr Biomed. 2013, 26, 1220–1224. [Google Scholar] [CrossRef]
- Gerus, P.; Rao, G.; Berton, E. Ultrasound-based subject-specific parameters improve fascicle behaviour estimation in Hill-type muscle model. Comput. Methods Biomech. Biomed. Eng. 2015, 18, 116–123. [Google Scholar] [CrossRef]
- Li, L.; Tong, K.Y.; Hu, X.L.; Hung, L.K.; Koo, T.K.K. Incorporating ultrasound-measured musculotendon parameters to subject-specific EMG-driven model to simulate voluntary elbow flexion for persons after stroke. Clin. Biomech. 2009, 24, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Bohm, S.; Marzilger, R.; Mersmann, F.; Santuz, A.; Arampatzis, A. Operating length and velocity of human vastus lateralis muscle during walking and running. Sci. Rep. 2018, 8, 5066. [Google Scholar] [CrossRef]
- Saxby, D.J.; Killen, B.A.; Pizzolato, C.; Carty, C.P.; Diamond, L.E.; Modenese, L.; Fernandez, J.; Davico, G.; Barzan, M.; Lenton, G.; et al. Machine learning methods to support personalized neuromusculoskeletal modelling. Biomech. Model. Mechanobiol. 2020, 19, 1169–1185. [Google Scholar] [CrossRef]
- Queipo, N.V.; Haftka, R.T.; Shyy, W.; Goel, T.; Vaidyanathana, R.; Tuckerb, P.K. Surrogate-based analysis and optimization. Prog. Aerosp. Sci. 2005, 41, 1–28. [Google Scholar] [CrossRef]
- Ziaeipoor, H.; Martelli, S.; Pandy, M.; Taylor, M. Efficacy and efficiency of multivariate linear regression for rapid prediction of femoral strain fields during activity. Med. Eng. Phys. 2019, 63, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Farr, J.; Carter, K.; Fregly, B.J. Response surface optimization for joint contact model evaluation. J. Appl. Biomech. 2006, 22, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Haftka, R.T.; Queipo, N.V.; Fregly, B.J. Two-dimensional surrogate contact modeling for computationally efficient dynamic simulation of total knee replacements. J. Biomech. Eng. 2009, 131, 041010. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Haftka, R.T.; Queipo, N.V.; Fregly, B.J. Surrogate articular contact models for computationally efficient multibody dynamic simulations. Med. Eng. Phys. 2010, 32, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Eskinazi, I.; Fregly, B.J. Surrogate modeling of deformable joint contact using artificial neural networks. Med. Eng. Phys. 2015, 37, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Eskinazi, I.; Fregly, B.J. An open-source toolbox for surrogate modeling of joint contact mechanics. IEEE Trans. Biomed. Eng. 2016, 63, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Marra, M.A.; Andersen, M.S.; Damsgaard, M.; Koopman, B.F.J.M.; Janssen, D.; Verdonschot, N. Evaluation of a surrogate contact model in force-dependent kinematic simulations of total knee replacement. J. Biomech. Eng. 2017, 139, 081001. [Google Scholar] [CrossRef]
- O’Rourke, D.; Martelli, S.; Bottema, M.; Taylor, M. A computational efficient method to assess the sensitivity of finite-element models: An illustration with the hemipelvis. J. Biomech. Eng. 2016, 138, 121008. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).