1. Introduction
Networks are ubiquitous since their strong power to model real data, such as telecommunication networks, social networks, citation networks, and biological networks. There are many crucial networks analysis tasks to deal with, such as node classification, node clustering, link prediction, community detection, etc. Since a large portion of networks contain nodes with extra information, attributed networks are really worth to study. Earlier works suffer high computation and great space cost since they usually represent networks with adjacency matrix. Then, network embedding, also known as graph embedding or network representation learning is proposed, aiming to embed vertices into a low dimensional space where the network structural information and network properties are preserved, then it is easy to exploit off-the-shelf machine learning algorithms to analysis networks efficiently [
1].
First kind of network embedding models are methods such as DeepWalk [
2], node2vec [
3] and LINE [
4], which adopt language model word2vec [
5] in network representation. Second kind of models are based on matrix factorization, such as HOPE [
6], TADW [
7], and MNF-M [
8]. They construct a matrix first, then decompose it to get embeddings.
As for attribute network embedding, since Graph Neural Networks (GCNs) have achieved great success, many models combine the GCN to do network analysis tasks. VGAE [
9] uses the auto-encoder framework and combines the Variational auto-encoders (VAE) ([
10,
11]) and GCN to learn great node representation. Then ARVGA [
12] and CAN [
13] improve it by adding GAN or changing learning aim. GCN-based models inherit some common defects. VGAE and ARVGA use variational auto-encoders (VAE) to encode the data distribution of the latent codes in graphs while suffering from the drawback of the sampling process in VAE, which makes the latent representation not repeatable for the same sample and harms the performance in related tasks. These typical methods mostly ignore the global information between nodes.
To overcome the deficits mentioned before, we propose a framework (SAVGAE) to encode the graph structure and node attribute of attributed networks into compact representation. We use the higher proximity metric Rooted PageRank to include the local and global information between each pair of nodes. We design a delicate structure to employ the strong learning ability of VAE while circumventing the effect brought by the inner core sampling process through an adversarial training module. The delicate structure makes the representation exclusive for each node and the adversarial training module prevents the performance declining it may cause. We evaluate our model on three typical graph analysis tasks including link prediction, node clustering, and graph visualization meanwhile get competitive performance. In summary, our contributions are as follows:
We propose to use Rooted PageRank (RPR) rather than adjacency matrix as the reconstruction objective so as to preserve local and global proximity simultaneously in node representation.
We develop a novel graph autoencoder model combining generative adversarial networks and variational autoencoder, where we use the great learning ability of VAE and avoid the side effect it may cause on learning graph embedding.
We evaluate the proposed model over two tasks on three widely used graph benchmarks, and get competitive results.
We organize the rest of our paper as follows. In
Section 2, we introduce the work related to our model. In
Section 3, we detail our model SAVGAE. In
Section 4, we introduce the experiments setup and experiments results. Finally, in
Section 5, we draw our conclusions.
2. Related Work
In this section, we introduce the models related to our work. Firstly, we introduce the earlier network embedding methods, then we present attributed network embedding models. Finally, we introduce related models exploiting deep generative models.
2.1. Network Embedding
Network embedding assigns each node in the network a low dimensional representation while preserving the network information [
14]. Earlier works were related to dimension reduction techniques such as Isomap [
15], Locally Linear Embedding (LLE) [
16], and Laplacian Eigenmap (LE) [
17]. The time complexity of these methods is usually at least quadratic with respect to the number of nodes, thus suffering scalability issue [
1]. Recent models are directly designed for representing networks rather than reducing the high dimensionality of the non-relational data.
2.2. Attributed Network Embedding
Attributed Network Embedding exploits the topological structure and the node content simultaneously. Yang et al. prove that DeepWalk is equivalent to matrix factorization, and propose TADW to incorporate text features of nodes into network embedding [
7]. TriNDR [
18] is a coupled neural network model to exploit network structure, node content, and node labels to learn node representation. However, these models shallow models are either DeepWalk variants or based on the mechanism of matrix factorization. Recently Graph Neural Networks (GNNs) achieve excellent performance due to their strong representation power, and there are some works that exploit GNN in network embedding, such as VGAE, incorporating GCN into variational autoencoder framework.
2.3. Deep Generative Models
Generative models are an important class of models in probability statistics and machine learning, referring to a set of models used to generate observable data randomly. Generative models have a wide range of applications, and can be used to model different kinds of data, such as images, text, sound, etc. Thus, it is promising to use generative models in network representation learning. Deep generative models take advantage of the ability of deep neural networks to approximate arbitrary functions to model a complex distribution, Variational Autoencoder (VAE) and Generative Adversarial Networks (GANs) are two excellent classes.
AAE [
19] combines the GAN and autoencoder to perform variational inference. However, the model is not designed for networks. Recently, Dai et al. used the adversarial training in networks [
20]. However, the model cannot handle networks with node attributes. ARVGA also combines the GAN with the graph auto-encoder structure. However it only uses the GAN as a regularizer to enhance an intrinsic hypothesis of prior distribution in VAE. In contrast, our model uses the adversarial mechanism to improve the defect of VAE in attribute network embedding. Ref. [
21] also try to overcome the problem of VAE, but it aims at general data.
3. Model
In this section, we formalize the attribute network embedding task we focus on firstly. Then we present our Structural Adversarial Variational Graph Auto-Encoder (SAVGAE) model. Next we introduce the Structral Variational Graph Auto-Encoder(SVGA) module, which uses Rooted Pagerank to learn comprehensive topology structural information. Finally, we introduce the adversarial training module which overcomes the defects of the VAE.
3.1. Problem Definition
Definition 1 (Attibuted Network)
. A attributed network is defined as , where is the set of nodes, and denotes the sets of edges with . is the feature matrix with denoting the feature information associated with node . Meanwhile the topology information of the network can be represented by adjacency matrix . All notations are summarized in Table 1. Problem 1 (Attributed Network embedding)
. The goal is to project each node to a low-dimension vector :
where , such that embedding matrix Z should well preserve the topological structure and attribute information of the graph, given an attributed network denoted as . 3.2. Overall Architecture
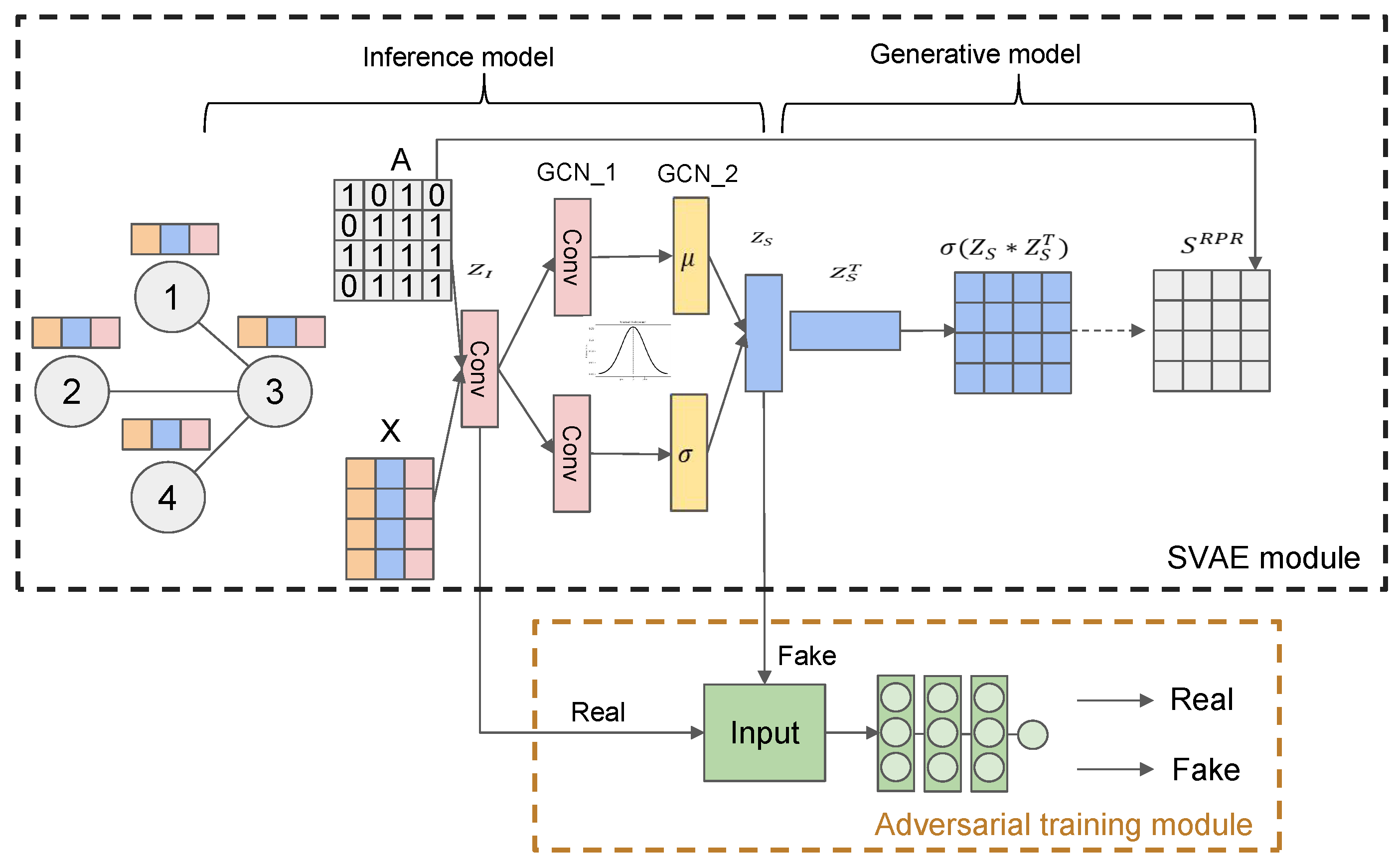
In this section, we introduce the overall architecture of our model. As
Figure 1 shows, our model consists of two modules. Firstly, the SVGAE module learns representations for nodes while preserving their overall structural information and attribute information. Then we propose adversarial training module to learn robust embeddings for nodes.
Structural Variational Graph Auto-encoder module. SVGAE takes in adjacency matrix and feature matrix and gets embeddings , then reconstructs the Rooted Pagerank matrix.
Adversarial training module. This module introduces the adversarial training based on the SVGAE to overcome the side effect of the VAE framework.
3.3. Structural Variational Graph Auto-Encoder Module
The Structural Variational Graph Auto-encoder consists of an inference model and a generative model. In our model, we adopt vanilla Graph Convolution Network layer [
22] in our graph auto-encoder model. The layerwise transformation is denoted as follows:
where
is the input layer,
is the adjacency matrix, and the
is the weights parameters to be learned in the training process. To be specific, our graph convolution here is defined as follows:
where
is the Relu activation function,
,
.
3.3.1. Rooted PageRank
Here we choose a high-order proximity Rooted PageRank (RPR)
[
23] as an alternative to
. As is defined in Equations (
4) and (
5), the
-th entry of
defines the probability that if there is a random walk starting from node
, it will stop at node
in the steady state.
is the parameter to control the probability of the current node walking to a neighbor node randomly rather than jumping back to the start node.
is transition matrix, and
. So we can use
to describe the node-to-node proximity. Specifically, the
i-th row of
, can be used to represent the global structural information of node
.
3.3.2. Inference Model
In the inference model, we get hidden variable
from the input adjacency matrix
and attribute matrix
.
The inference model can also be seen as an encoder.
which actually consists of two GCN layers.
and are matrix of mean vectors and matrix of log of the variance vectors respectively.
3.3.3. Generative Model
The generative model can also be seen as a decoder where traditional models aim at reconstructing the adjacency matrix
so the embedding matrix
can preserve the information of graph
. However,
just encode the one-order structural information of the graph which is not enough for learning comprehensive information of the graph structure. Therefore we adopt the
as the reconstruction aiming of the decoder. So the node embeddings preserve the comprehensive information of the network after loss function being optimized as it can reconstruct the Rooted Pagerank matrix to some extent.
3.3.4. Optimization
As a Variational graph auto-encoder model, we need optimize the variational lower bound
where the former indicates the reconstruction loss and the latter uses the Kullback-Leibler divergence as a regularizer of the variational auto-encoder.
3.4. Adversarial Training Module
In the previous section, we can find
is suitable for represent the nodes of the network as illustrated in
Figure 1. However, locating after the sampling process makes it not repeatable, so we decide to use
for its stability, and using adversarial training module to decrease the information loss between
and
to guarantee the latter is representative.
In our adversarial training module, the input of the discriminator is latent code either from
(real) or
(fake) and the output is a scalar deciding whether the input is true or not. The cost of the discriminator is
Then the objective to train the encoder of SVGAE module with discriminator of the adversarial training module is as follows:
3.5. Overall Algorithm
As in Algorithm 1, the SAVGAE model takes in the attributed network
, and generates
and
through Equations (
7) and (
10) in step 2. Then it takes m samples from
and
and trains the discriminator in step 6. Finally, it trains the SAVGAE through updating Equation (
12) in step 8 and gets the desired network embedding
.
| Algorithm 1 Algorithm for SAVGAE |
| Input:: Graph; T: the number of iterations of training process; K: the
number of steps for updating discriminator; d: the dimension of the embeddings |
| Output: |
| 1: for iterator = 1 to T do |
| 2: Generate matrix and by Equations (7) and (10); |
| 3: for iterator = 1 to K do |
| 4: Sample m samples from |
| 5: Sample m samples from |
| 6: Update the discriminator with its stochastic gradient |
| 7: end for |
| 8: Update the SVAE module by Equation (12) |
| 9: end for |
| 10: return |
4. Experiments
In this section, we evaluate the proposed model on two widely used tasks: link prediction and node clustering over 2 real-world datasets [
24]. Meanwhile, we compare our methods with baseline algorithms.
4.1. Datasets and Baselines
We report experimental results on two attributed graph datasets: Cora and Citeseer. They are citation networks with varying sizes where nodes and edges represent publications and citations. Node attributes of Cora and Citeseer are bag-of-words representing the content of the publication. The statistics of the datasets are summarized in
Table 2.
To validate and evaluate SAVGAE, we compare it with 7 baseline methods including DeepWalk, Spectral Clustering [
25], node2vec, GAE, VGAE, ARGA, ARVGA in link prediction task. In node clustering task, we include 6 methods K-means, Graph Encoder [
26], DNGR [
27], RTM [
28], RMSC [
29], TADW besides baselines in link prediction task.
4.2. Experimental Setup and Evaluation Methods
We detail the experimental setup and evaluation methods for link prediction and node clustering respectively. Link prediction task aims at inferring the missing edges from the network which is not complete. Following the experimental settings of VGAE, each dataset is partitioned into the training set, the validatoin set and the test set. We randomly remove 5% edges for validation and 10% edges for test which serve as positive samples. Then we sample the same number of non-existing links as negative samples from the pairs of unconnected nodes. The model is trained on the residual network where attributes of nodes are kept. The model can be evaluated by the ability to classify the positive samples and negative samples, and we employ the AUC(the area under a receiver operating characteristic curve) and AP(average precision) as the metrics for evaluation.
For the node clustering task, we employ the K-means algorithm based on the node embeddings learned from the training phase. Then we employ accuracy (Acc), normalized mutual information (NMI), F-score (F1), precision, and average rand index (ARI) as metrics, compared to the true class labels of nodes.
4.3. Main Results
In this section, we detail our model performance on link prediction and node clustering experiments. Then we validate the effectiveness of different parts of SAVGAE through ablation studies. Finally, we visualize the node embeddings.
4.3.1. Link Prediction
The experimental results on the link prediction task are reported in
Table 3. We can observe that SAVGAE achieves outstanding performance: consistently performs better than all baseline models. Compared with all the baselines, SAVGAE increased the AP score from around 2.9% compared with ARVGE, 3.9% compared with VGAE, 13.1% compared with VGAE without node features; 14.3% and 12.3% compared with Spectral Clustering and DeepWalk respectively.
4.3.2. Node Clustering
After getting node embeddings, we apply K-means algorithm to do the node clustering task. The results on Cora and Citeseer datasets are reported in
Table 4 and
Table 5. The results illustrate that SAVGA has achieved a great improvement compared with all the other baselines. For example, on Cora dataset, SAVGAE has increased the accuracy from 40.2% compared with K-means to 88.0% compared with Spectral Clustering; increased the F1 score from 44.5% compared with TADW to 102.8% compared with DeepWalk; and increased NMI from 55.0% compared with K-means to 10.4% compared with ARVGE. The results has demonstrated the superiority of our SAVGAE.
4.3.3. Graph Visualization
Visualization is another intuitive and effective way to assess the performance of network embedding methods. We get node representations through SAVGAE model, then map these points into 2D space by t-SNE [
30]. The color denotes the class label of the nodes in the dataset, which is not exploited in our unsupervised attributed network embedding. Therefore, nodes in the same color should locate closer to each other. As in
Figure 2, the meaningful clustering layout demonstrates the performance of SAVGAE.
4.4. Ablation Studies
To evaluate the performance of different parts of SAVGAE, we perform ablation studies on link prediction and node clustering task. “-RPR” means dicarding high-order proximity Rooted Pagerank and using adjacency matrix as learning objective. “-Adversarial training” means discarding adversarial training module. “-RPR and adversarial training” means discarding both Rooted Pagerank and adversarial training module. The results are illustrated in
Table 6,
Table 7 and
Table 8. We find that the hybrid model performs best.
5. Conclusions
In this paper, we propose SAVGAE, a novel attributed network embedding method, which preserves comprehensive topology structure and node content information of attributed network by modifying the VAE to handle graph data while integrating with generative adversarial networks. We use Rooted PageRank (RPR) rather than adjacency matrix as the reconstruction objective of the auto-encoder to preserve local and global proximity simultaneously in node representation while not increasing much model complexity. The sampling process is an indispensable part of VAE before getting a latent representation of the node, but it causes the problem that the node embeddings are not exclusive which may affect the downstream network analysis task. We modify the VAE structure and integrate with generative adversarial networks to learn an exclusive and robust embedding for each node to overcome the aforementioned problem. Empirically, we evaluate the generated network representations in a variety of network datasets and applications. The results demonstrate the superiority of our method compared with the state-of-the-art. Our future work will focus on proposing a generalized framework that can handle unseen nodes and networks rather than just learning embeddings on a single fixed network.
Author Contributions
Conceptualization, J.Z. and F.L.; methodology, J.Z., Y.W., D.L.; validation, J.Z.; formal analysis, J.Z.; writing—original draft preparation, J.Z.; writing—review and editing, F.L., Y.W., D.L.; supervision, G.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Strategic Priority Research Program of Chinese Academy of Sciences (grant number Y835120378).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
References
- Zhang, D.; Yin, J.; Zhu, X.; Zhang, C. Network representation learning: A survey. IEEE Trans. Big Data 2018, 6, 3–28. [Google Scholar] [CrossRef]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; ACM: New York, NY, USA, 2014; pp. 701–710. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 855–864. [Google Scholar]
- Tang, J.; Qu, M.; Wang, M.; Zhang, M.; Yan, J.; Mei, Q. Line: Large-scale information network embedding. In Proceedings of the 24th International Conference on World Wide Web, Florence, Italy, 18–22 May 2015; pp. 1067–1077. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2013; pp. 3111–3119. [Google Scholar]
- Ou, M.; Cui, P.; Pei, J.; Zhang, Z.; Zhu, W. Asymmetric transitivity preserving graph embedding. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 1105–1114. [Google Scholar]
- Yang, C.; Liu, Z.; Zhao, D.; Sun, M.; Chang, E.Y. Network representation learning with rich text information. IJCAI 2015, 2015, 2111–2117. [Google Scholar]
- Wang, X.; Cui, P.; Wang, J.; Pei, J.; Zhu, W.; Yang, S. Community preserving network embedding. Proc. AAAI Conf. Artif. Intell. 2017, 17, 3298239–3298270. [Google Scholar]
- Kipf, T.N.; Welling, M. Variational Graph Auto-Encoders. NIPS Workshop on Bayesian Deep Learning; Curran Associates: New York, NY, USA, 2016. [Google Scholar]
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Rezende, D.J.; Mohamed, S.; Wierstra, D. Stochastic backpropagation and approximate inference in deep generative models. arXiv 2014, arXiv:1401.4082. [Google Scholar]
- Pan, S.; Hu, R.; Long, G.; Jiang, J.; Yao, L.; Zhang, C. Adversarially Regularized Graph Autoencoder for Graph Embedding. arXiv 2018, arXiv:1802.04407. [Google Scholar]
- Meng, Z.; Liang, S.; Bao, H.; Zhang, X. Co-embedding attributed networks. In Proceedings of the Twelfth ACM International Conference on Web Search and Data Mining, Melbourne, VIC, Australia, 11–15 February 2019; ACM: New York, NY, USA, 2019; pp. 393–401. [Google Scholar]
- Cui, P.; Wang, X.; Pei, J.; Zhu, W. A survey on network embedding. IEEE Trans. Knowl. Data Eng. 2018, 31, 833–852. [Google Scholar] [CrossRef]
- Tenenbaum, J.B.; De Silva, V.; Langford, J.C. A global geometric framework for nonlinear dimensionality reduction. Science 2000, 290, 2319–2323. [Google Scholar] [CrossRef] [PubMed]
- Roweis, S.T.; Saul, L.K. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef] [PubMed]
- Belkin, M.; Niyogi, P. Laplacian eigenmaps and spectral techniques for embedding and clustering. Adv. Neural Inf. Proc. Syst. 2001, 14, 585–591. [Google Scholar]
- Pan, S.; Wu, J.; Zhu, X.; Zhang, C.; Wang, Y. Tri-party deep network representation. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence, New York, NY, USA, 9–15 July 2016; AAAI Press: Menlo Park, CA, USA, 2016; pp. 1895–1901. [Google Scholar]
- Makhzani, A.; Shlens, J.; Jaitly, N.; Goodfellow, I.; Frey, B. Adversarial autoencoders. arXiv 2015, arXiv:1511.05644. [Google Scholar]
- Dai, Q.; Li, Q.; Tang, J.; Wang, D. Adversarial network embedding. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; AAAI: Menlo Park, CA, USA, 2018. [Google Scholar]
- Zhang, X.; Yao, L.; Yuan, F. Adversarial variational embedding for robust semi-supervised learning. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; ACM: New York, NY, USA, 2019; pp. 139–147. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. In Proceedings of the 5th International Conference on Learning Representations, ICLR, Toulon, France, 24–26 April 2017. [Google Scholar]
- Liben-Nowell, D.; Kleinberg, J.M. The link-prediction problem for social networks. J. Assoc. Inf. Sci. Technol. 2007, 58, 1019–1031. [Google Scholar] [CrossRef]
- Sen, P.; Namata, G.; Bilgic, M.; Getoor, L.; Gallagher, B.; Eliassi-Rad, T. Collective Classification in Network Data. AI Mag. 2008, 29, 93–106. [Google Scholar] [CrossRef]
- Tang, L.; Liu, H. Leveraging social media networks for classification. Data Min. Knowl. Discov. 2011, 23, 447–478. [Google Scholar] [CrossRef]
- Tian, F.; Gao, B.; Cui, Q.; Chen, E.; Liu, T.Y. Learning deep representations for graph clustering. In Proceedings of the AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–13 February 2016; AAAI: Menlo Park, CA, USA, 2014; Volume 14, pp. 1293–1299. [Google Scholar]
- Cao, S.; Lu, W.; Xu, Q. Deep neural networks for learning graph representations. In Proceedings of the AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–13 February 2016; AAAI: Menlo Park, CA, USA, 2016; Volume 16, pp. 1145–1152. [Google Scholar]
- Chang, J.; Blei, D. Relational topic models for document networks. In Proceedings of the Artificial Intelligence and Statistics, Clearwater, FL, USA, 16–19 April 2009; pp. 81–88. [Google Scholar]
- Xia, R.; Pan, Y.; Du, L.; Yin, J. Robust multi-view spectral clustering via low-rank and sparse decomposition. In Proceedings of the Twenty-Eighth AAAI Conference on Artificial Intelligence, Québec City, QC, Canada, 27–31 July 2014; AAAI: Menlo Park, CA, USA, 2014; pp. 2149–2155. [Google Scholar]
- Maaten, L.V.D.; Hinton, G.E. Visualizing non-metric similarities in multiple maps. Mach. Learn. 2011, 87, 33–55. [Google Scholar] [CrossRef]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).