An RNA Triangle with Six Ribozyme Units Can Promote a Trans-Splicing Reaction through Trimerization of Unit Ribozyme Dimers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Design
2.2. Plasmid Construction and RNA Preparation
2.3. Preparation of L7Ae Protein and L7Ae–EGFP Fusion Protein
2.4. Electrophoretic Mobility Shift Assay (EMSA) of kUrd Oligomers
2.5. Atomic Force Microscopy
2.6. Electrophoretic Mobility Shift Assay (EMSA) of the kUrd Trimer with L7Ae Protein
2.7. Substrate Cleavage Reaction
2.8. Trans-Splicing Monitored by Fluorescence of Spinach RNA–DFHBI
3. Results
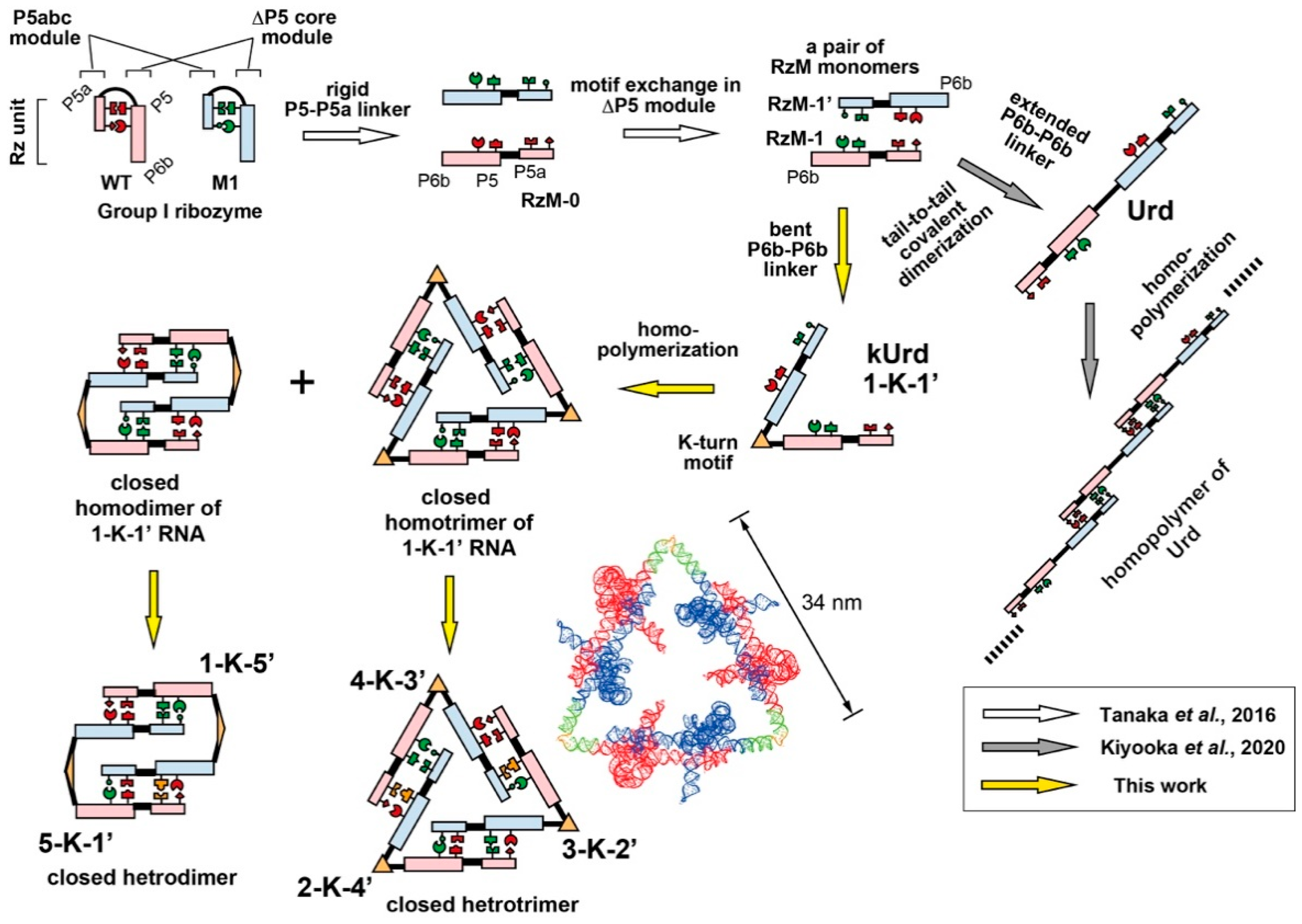
3.1. Design of Unit Ribozyme Dimers (Urds) with a Bent Linker
3.2. Homooligomerization of kUrds
3.3. Block Copolymerization of kUrds
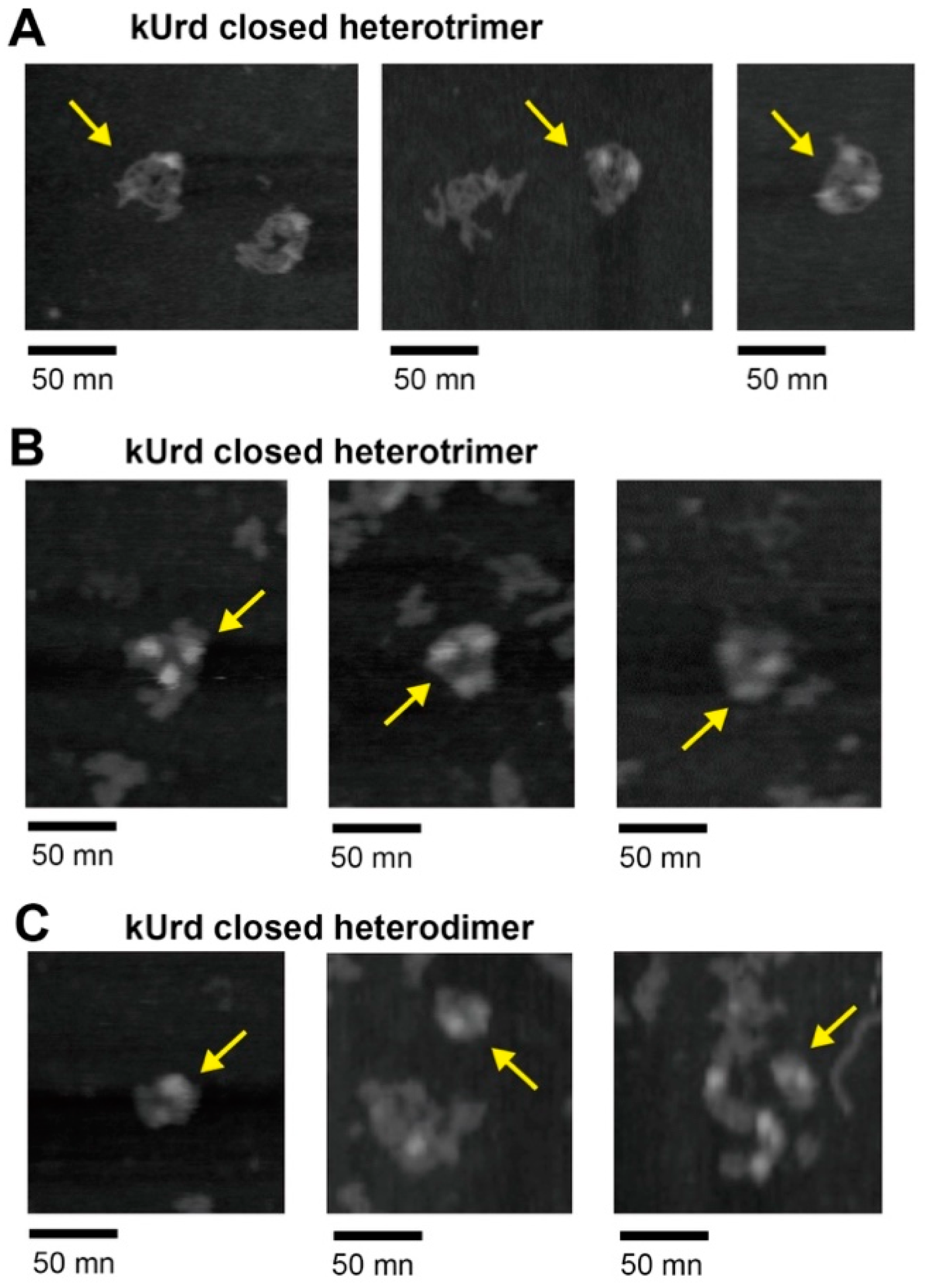
3.4. Atomic Force Microscopy Analysis of kUrd Heterotrimer and Heterodimer
3.5. Decoration of the kUrd Closed Trimer with Kink-Turn Binding Protein L7Ae
3.6. Catalytic Activities of kUrds and Their Oligomers
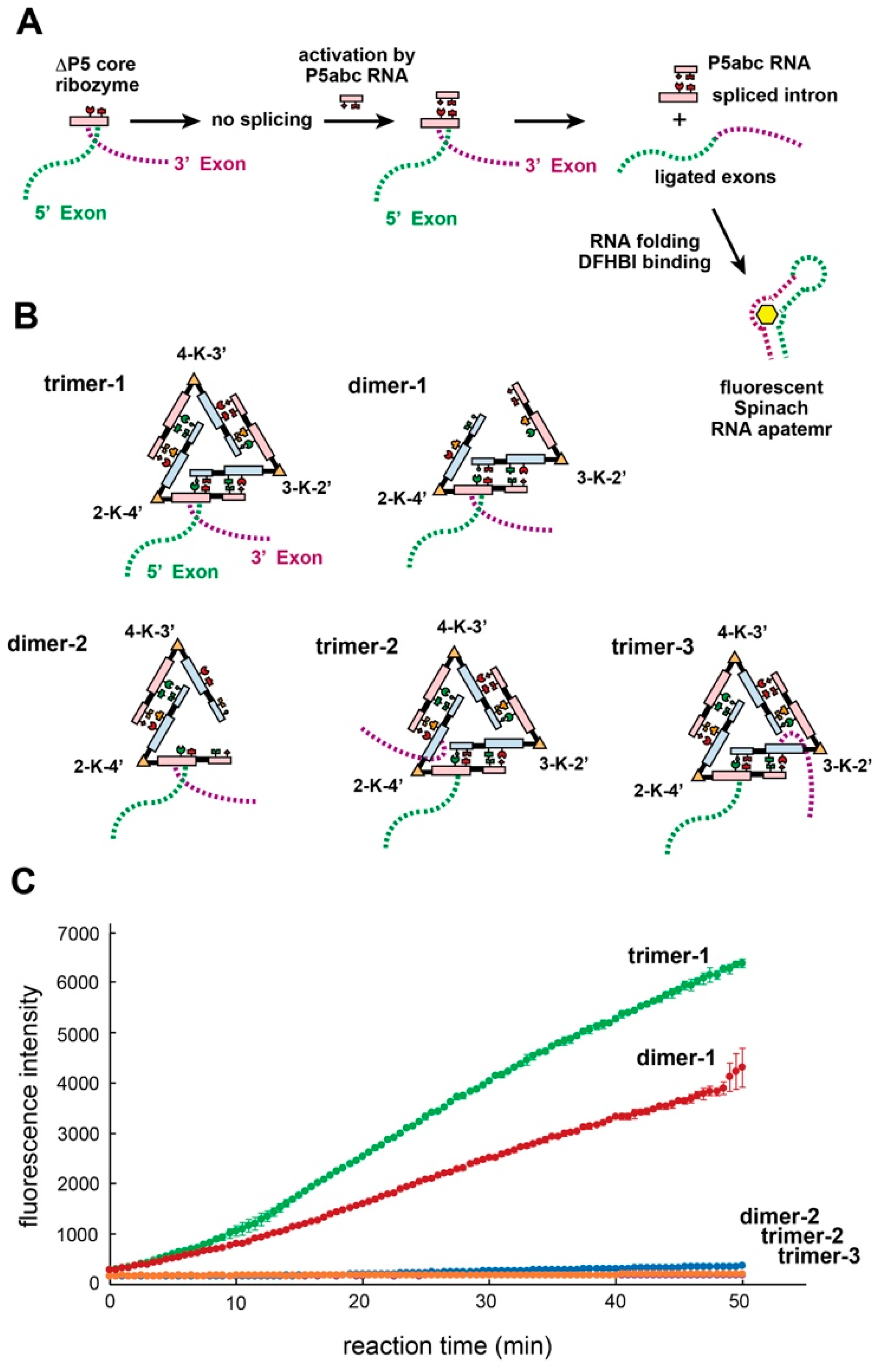
3.7. Trans-Splicing Reaction Promoted by kUrd Oligomer Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goodsell, D.S.; Olson, A.J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 105–153. [Google Scholar] [CrossRef]
- Lesieur, C. The assembly of protein oligomers: Old stories and new perspectives with graph theory. In Oligomerization of Chemical and Biological Compounds; Lesieur, C., Ed.; IntecOpen: London, UK, 2014. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.; Thornton, J.M. Protein-protein interactions: A review of protein dimer structures. Prog. Biophys. Mol. Biol. 1995, 63, 31–65. [Google Scholar] [CrossRef]
- Glover, D.J.; Clark, D.S. Protein calligraphy: A new concept begins to take shape. ACS Cent. Sci. 2016, 2, 438–444. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Luo, Q.; Hou, C.; Liu, J. Nanostructures based on protein self-assembly: From hierarchical construction to bioinspired materials. Nano Today 2017, 14, 16–41. [Google Scholar] [CrossRef]
- Kuan, S.L.; Bergamini, F.R.G.; Weil, T. Functional protein nanostructures: A chemical toolbox. Chem. Soc. Rev. 2018, 47, 9069–9105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyken, S.E.; Chen, Z.; Groves, B.; Langan, R.A.; Oberdorfer, G.; Ford, A.; Gilmore, J.M.; Xu, C.; DiMaio, F.; Pereira, J.H.; et al. De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science 2016, 352, 680–687. [Google Scholar] [CrossRef] [Green Version]
- Fallas, J.A.; Ueda, G.; Sheffler, W.; Nguyen, V.; McNamara, D.E.; Sankaran, B.; Pereira, J.H.; Parmeggiani, F.; Brunette, T.J.; Cascio, D.; et al. Computational design of self-assembling cyclic protein homo-oligomers. Nat. Chem. 2017, 9, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Kuhlman, B.; O’Neill, J.W.; Kim, D.E.; Zhang, K.Y.; Baker, D. Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc. Natl. Acad. Sci. USA 2001, 98, 10687–10691. [Google Scholar] [CrossRef] [Green Version]
- Stranges, P.B.; Machius, M.; Miley, M.J.; Tripathy, A.; Kuhlman, B. Computational design of a symmetric homodimer using beta-strand assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 20562–20567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, Y.; Huang, P.S.; Hsu, F.C.; Huang, S.J.; Mayo, S.L. Computational design and experimental verification of a symmetric protein homodimer. Proc. Natl. Acad. Sci. USA 2015, 112, 10714–10719. [Google Scholar] [CrossRef] [Green Version]
- Fedor, M.J.; Williamson, J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005, 6, 399–412. [Google Scholar] [CrossRef]
- Weinberg, C.E.; Weinberg, Z.; Hammann, C. Novel ribozymes: Discovery, catalytic mechanisms, and the quest to understand biological function. Nucleic Acids Res. 2019, 47, 9480–9494. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R. Prospects for riboswitch discovery and analysis. Mol. Cell 2011, 4, 867–879. [Google Scholar] [CrossRef] [Green Version]
- Butcher, S.E.; Pyle, A.M. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011, 44, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, J.; Fujita, Y.; Maeda, Y.; Furuta, H.; Ikawa, Y. GNRA/receptor interacting modules: Versatile modular units for natural and artificial RNA architectures. Methods 2011, 54, 226–238. [Google Scholar] [CrossRef]
- Jarrell, K.A.; Dietrich, R.C.; Perlman, P.S. Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell. Biol. 1988, 8, 2361–2366. [Google Scholar] [CrossRef] [Green Version]
- van der Horst, G.; Christian, A.; Inoue, T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl. Acad. Sci. USA 1991, 88, 184–188. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, M.A.; Doherty, E.A.; Knitt, D.S.; Doudna, J.A.; Herschlag, D. The P5abc peripheral element facilitates preorganization of the Tetrahymena group I ribozyme for catalysis. Biochemistry 2000, 39, 2639–2651. [Google Scholar] [CrossRef]
- Doudna, J.A.; Cech, T.R. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA 1995, 1, 36–45. [Google Scholar]
- Ikawa, Y.; Shiraishi, H.; Inoue, T. Trans-activation of the Tetrahymena ribozyme by its P2-2.1 domains. J. Biochem. 1998, 123, 528–533. [Google Scholar] [CrossRef]
- Rahman, M.M.; Matsumura, S.; Ikawa, Y. Effects of molecular crowding on a bimolecular group I ribozyme and its derivative that self-assembles to form ribozyme oligomers. Biochem. Biophys. Res. Commun. 2018, 507, 136–141. [Google Scholar] [CrossRef]
- Pan, T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry 1995, 34, 902–909. [Google Scholar] [CrossRef]
- Loria, A.; Pan, T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA 1996, 2, 551–563. [Google Scholar]
- Kim, H.; Poelling, R.R.; Leeper, T.C.; Meyer, M.A.; Schmidt, F.J. In vitro transactivation of Bacillus subtilis RNase P RNA. FEBS Lett. 2001, 506, 235–238. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Matsumura, S.; Furuta, H.; Ikawa, Y. Tecto-GIRz: Engineered group I ribozyme the catalytic ability of which can be controlled by self-dimerization. ChemBioChem 2016, 17, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Ikawa, Y.; Matsumura, S. Rational engineering of a modular group I ribozyme to control its activity by self-dimerization. Methods Mol. Biol. 2017, 1632, 325–340. [Google Scholar] [PubMed]
- Kiyooka, R.; Akagi, J.; Hidaka, K.; Sugiyama, H.; Endo, M.; Matsumura, S.; Ikawa, Y. Catalytic RNA nano-objects formed by self-assembly of group I ribozyme dimers serving as unit structures. J. Biosci. Bioeng. 2020, 130, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Gooding, A.R.; Cech, T.R. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell 2004, 16, 351–362. [Google Scholar]
- Klein, D.J.; Schmeing, T.M.; Moore, P.B.; Steitz, T.A. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001, 20, 4214–4221. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Hirata, Y.; Tominaga, Y.; Furuta, H.; Matsumura, S.; Ikawa, Y. Heterodimerization of group I ribozymes enabling exon recombination through pairs of cooperative trans-splicing reactions. ChemBioChem 2017, 18, 1659–1667. [Google Scholar] [CrossRef]
- Ikawa, Y.; Moriyama, S.; Furuta, H. Facile syntheses of BODIPY derivatives for fluorescent labeling of the 3’ and 5’ ends of RNAs. Anal. Biochem. 2008, 378, 166–170. [Google Scholar] [CrossRef]
- Ohno, H.; Kobayashi, T.; Kabata, R.; Endo, K.; Iwasa, T.; Yoshimura, S.H.; Takeyasu, K.; Inoue, T.; Saito, H. Synthetic RNA-protein complex shaped like an equilateral triangle. Nat. Nanotechnol. 2011, 6, 116–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohuchi, S.J.; Sagawa, F.; Sakamoto, T.; Inoue, T. A trifunctional, triangular RNA-protein complex. FEBS Lett. 2015, 589, 2424–2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Furuta, H.; Ikawa, Y. Installation of orthogonality to the interface that assembles two modular domains in the Tetrahymena group I ribozyme. J. Biosci. Bioeng. 2014, 117, 407–412. [Google Scholar] [CrossRef]
- Rahman, M.M.; Matsumura, S.; Ikawa, Y. Artificial RNA motifs expand the programmable assembly between RNA modules of a bimolecular ribozyme leading to application to RNA nanostructure design. Biology 2017, 6, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oi, H.; Fujita, D.; Suzuki, Y.; Sugiyama, H.; Endo, M.; Matsumura, S.; Ikawa, Y. Programmable formation of catalytic RNA triangles and squares by assembling modular RNA enzymes. J. Biochem. 2017, 161, 451–462. [Google Scholar] [CrossRef]
- Fujita, Y.; Furushima, R.; Ohno, H.; Sagawa, F.; Inoue, T. Cell-surface receptor control that depends on the size of a synthetic equilateral-triangular RNA-protein complex. Sci. Rep. 2014, 4, 6422. [Google Scholar] [CrossRef] [Green Version]
- Osada, E.; Suzuki, Y.; Hidaka, K.; Ohno, H.; Sugiyama, H.; Endo, M.; Saito, H. Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications. ACS Nano. 2014, 8, 8130–8140. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Kobayashi, T.; Hara, T.; Fujita, Y.; Hayashi, K.; Furushima, R.; Inoue, T. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 2010, 6, 71–78. [Google Scholar] [CrossRef]
- Wroblewska, L.; Kitada, T.; Endo, K.; Siciliano, V.; Stillo, B.; Saito, H.; Weiss, R. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat. Biotechnol. 2015, 33, 839–841. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, S.; Ono, H.; Kawasaki, S.; Kuang, Y.; Fujita, Y.; Saito, H. Synthetic RNA-based logic computation in mammalian cells. Nat. Commun. 2018, 9, 4847. [Google Scholar] [CrossRef]
- Ohno, H.; Akamine, S.; Saito, H. Synthetic mRNA-based systems in mammalian cells. Adv. Biosyst. 2020, 4, e1900247. [Google Scholar] [CrossRef] [PubMed]
- Ennifar, E.; Nikulin, A.; Tishchenko, S.; Serganov, A.; Nevskaya, N.; Garber, M.; Ehresmann, B.; Ehresmann, C.; Nikonov, S.; Dumas, P. The crystal structure of UUCG tetraloop. J. Mol. Biol. 2000, 304, 35–42. [Google Scholar] [CrossRef]
- Hall, K.B. Mighty tiny. RNA 2015, 21, 630–631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Gurtu, V.; Kain, S.R. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem. Biophys. Res. Commun. 1996, 227, 707–711. [Google Scholar] [CrossRef]
- Campbell, T.B.; Cech, T.R. Mutations in the Tetrahymena ribozyme internal guide sequence: Effects on docking of the P1 helix into the catalytic core and correlation with catalytic activity. Biochemistry 1996, 35, 11493–11502. [Google Scholar] [CrossRef]
- Ikawa, Y.; Matsumura, S. Engineered group I ribozymes as RNA-based modular tools to control gene expression. In Applied RNA Bioscience; Masuda, S., Izawa., S., Eds.; Springer: Berlin, German, 2018; pp. 203–220. [Google Scholar]
- Müller, U.F. Design and experimental evolution of trans-splicing group I intron ribozymes. Molecules 2017, 22, 75. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.H.; Han, S.R.; Lee, S.W. Therapeutic applications of group I intron-based trans-splicing ribozymes. Wiley Interdiscip. Rev. RNA 2018, 9, e1466. [Google Scholar] [CrossRef]
- Furukawa, A.; Maejima, T.; Matsumura, S.; Ikawa, Y. Characterization of an RNA receptor motif that recognizes a GCGA tetraloop. Biosci. Biotechnol. Biochem. 2016, 80, 1386–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef]
- Furukawa, A.; Tanaka, T.; Furuta, H.; Matsumura, S.; Ikawa, Y. Use of a fluorescent aptamer RNA as an exonic sequence to analyze self-splicing ability of a group I intron from structured RNAs. Biology 2016, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Kappel, K.; Zhang, K.; Su, Z.; Watkins, A.M.; Kladwang, W.; Li, S.; Pintilie, G.; Topkar, V.V.; Rangan, R.; Zheludev, I.N.; et al. Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures. Nat. Methods 2020, 17, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, S.; Ikawa, Y.; Inoue, T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003, 31, 5544–5551. [Google Scholar] [CrossRef] [Green Version]
- Goody, T.A.; Melcher, S.E.; Norman, D.G.; Lilley, D.M. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA 2004, 10, 254–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Huang, L.; Lilley, D.M.; Harbury, P.B.; Herschlag, D. The solution structural ensembles of RNA kink-turn motifs and their protein complexes. Nat. Chem. Biol. 2016, 12, 146–152. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akagi, J.; Yamada, T.; Hidaka, K.; Fujita, Y.; Saito, H.; Sugiyama, H.; Endo, M.; Matsumura, S.; Ikawa, Y. An RNA Triangle with Six Ribozyme Units Can Promote a Trans-Splicing Reaction through Trimerization of Unit Ribozyme Dimers. Appl. Sci. 2021, 11, 2583. https://doi.org/10.3390/app11062583
Akagi J, Yamada T, Hidaka K, Fujita Y, Saito H, Sugiyama H, Endo M, Matsumura S, Ikawa Y. An RNA Triangle with Six Ribozyme Units Can Promote a Trans-Splicing Reaction through Trimerization of Unit Ribozyme Dimers. Applied Sciences. 2021; 11(6):2583. https://doi.org/10.3390/app11062583
Chicago/Turabian StyleAkagi, Junya, Takahiro Yamada, Kumi Hidaka, Yoshihiko Fujita, Hirohide Saito, Hiroshi Sugiyama, Masayuki Endo, Shigeyoshi Matsumura, and Yoshiya Ikawa. 2021. "An RNA Triangle with Six Ribozyme Units Can Promote a Trans-Splicing Reaction through Trimerization of Unit Ribozyme Dimers" Applied Sciences 11, no. 6: 2583. https://doi.org/10.3390/app11062583
APA StyleAkagi, J., Yamada, T., Hidaka, K., Fujita, Y., Saito, H., Sugiyama, H., Endo, M., Matsumura, S., & Ikawa, Y. (2021). An RNA Triangle with Six Ribozyme Units Can Promote a Trans-Splicing Reaction through Trimerization of Unit Ribozyme Dimers. Applied Sciences, 11(6), 2583. https://doi.org/10.3390/app11062583





