Daucus carota L. Seed Inoculation with a Consortium of Bacteria Improves Plant Growth, Soil Fertility Status and Microbial Community
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Media
2.2. IAA Production and HPLC-FL Analysis
2.3. ACC Deaminase Activity
2.4. Phosphate Solubilization Capability
2.5. Inoculum Preparation and Seed Treatment
2.6. Field Experiments and Plant Sampling
2.7. Soil Analyses
2.8. Statistical Analysis
3. Results
3.1. IAA Production
3.2. Trp and Indole Derivates Characterization
3.3. ACC Deaminase Activity
3.4. Phosphate Solubilization Capability
3.5. Field Experiment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khoshru, B.; Mitra, D.; Khoshmanzar, E.; Myo, E.M.; Uniyal, N.; Mahakur, B.; Mohapatra, P.K.; Panneerselvam, P.; Boutaj, H.; Alizadeh, M.; et al. Current scenario and future prospects of plant growth-promoting rhizobacteria: An economic valuable resource for the agriculture revival under stressful conditions. J. Plant Nutr. 2020, 43, 3062–3092. [Google Scholar] [CrossRef]
- Yamada, Y.; Hoshino, K.; Ishikawa, T. The phylogeny of acetic acid bacteria based on the partial sequences of 16S ribosomal RNA: The elevation of the subgenus Gluconoacetobacter to the generic level. Biosci. Biotechnol. Biochem. 1997, 61, 1244–1251. [Google Scholar] [CrossRef]
- Baldani, J.I.; Baldani, V.L.D.; Seldin, L.; Dobereiner, J. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen-fixing bacterium. Int. J. Syst. Bacteriol. 1986, 36, 86–93. [Google Scholar] [CrossRef]
- Coenye, T.; Mahenthiralingam, E.; Henry, D.; LiPuma, J.J.; Laevens, S.; Gillis, M.; Speert, D.P.; Vandamme, P. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 2001, 51, 1481–1490. [Google Scholar] [CrossRef]
- Abdelrazek, S.; Choudhari, S.; Thimmapuram, J.; Simon, P.; Colley, M.; Mengiste, T.; Hoagland, L. Changes in the core endophytic mycobiome of carrot taproots in response to crop management and genotype. Sci. Rep. 2020, 10, 13685. [Google Scholar] [CrossRef]
- Peruzzi, A.; Raffaelli, M.; Ginanni, M.; Borelli, M. Physical weed control in organic carrot production. In Proceedings of the 6th EWRS Workshop on Physical and Cultural Weed Control, Lillehammer, Norway, 8–10 March 2004; pp. 24–38. [Google Scholar]
- Pellegrini, M.; Spera, D.M.; Ercole, C.; Del Gallo, M. Allium cepa L. inoculation with a consortium of plant growth-promoting bacteria: Effects on plants, soil, and the autochthonous microbial community. Microorganisms 2021, 9, 639. [Google Scholar] [CrossRef]
- Pace, L.; Pacioni, G.; Spano, L. In vitro propagation of Artemisia petrosa ssp. eriantha: Potential for the preservation of an endangered species. Plant Biosyst. 2004, 138, 291–294. [Google Scholar] [CrossRef]
- Pagnani, G.; Pellegrini, M.; Galieni, A.; D’Egidio, S.; Matteucci, F.; Ricci, A.; Stagnari, F.; Sergi, M.; Lo Sterzo, C.; Pisante, M.; et al. Plant growth-promoting rhizobacteria (PGPR) in Cannabis sativa ‘Finola’ cultivation: An alternative fertilization strategy to improve plant growth and quality characteristics. Ind. Crops Prod. 2018, 123, 75–83. [Google Scholar] [CrossRef]
- Botta, A.L.; Santacecilia, A.; Ercole, C.; Cacchio, P.; Del Gallo, M. In vitro and in vivo inoculation of four endophytic bacteria on Lycopersicon esculentum. New Biotechnol. 2013, 30, 666–674. [Google Scholar] [CrossRef]
- Pagnani, G.; Galieni, A.; Stagnari, F.; Pellegrini, M.; Del Gallo, M.; Pisante, M. Open field inoculation with PGPR as a strategy to manage fertilization of ancient Triticum genotypes. Biol. Fertil. Soils 2020, 56, 111–124. [Google Scholar] [CrossRef]
- Maheshwari, D.K.; Dheeman, S.; Agarwal, M. Phytohormone-producing PGPR for sustainable agriculture. In Bacterial Metabolites in Sustainable Agroecosystem. Sustainable Development and Biodiversity; Maheshwari, D., Ed.; Springer: Cham, Switzerland, 2015; pp. 159–182. [Google Scholar]
- Strzelczyk, E.; Pokojska-Burdziej, A. Production of auxins and gibberellin-like substances by mycorrhizal fungi, bacteria and actinomycetes isolated from soil and the mycorrhizosphere of pine (Pinus silvestris L.). Plant Soil 1984, 81, 185–194. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Gupta, G.; Parihar, S.S.; Ahirwar, N.K.; Snehi, S.K.; Singh, V. Plant growth promoting rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015, 07. [Google Scholar] [CrossRef]
- Suleman, M.; Yasmin, S.; Rasul, M.; Yahya, M.; Atta, B.M.; Mirza, M.S. Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS ONE 2018, 13, e0204408. [Google Scholar] [CrossRef] [PubMed]
- Kalayu, G. Phosphate solubilizing microorganisms: Promising approach as biofertilizers. Int. J. Agron. 2019, 2019, 4917256. [Google Scholar] [CrossRef]
- Forni, C.; Duca, D.; Glick, B.R. Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 2017, 410, 335–356. [Google Scholar] [CrossRef]
- Pellegrini, M.; Ercole, C.; Di Zio, C.; Matteucci, F.; Pace, L.; Del Gallo, M. In vitro and in planta antagonistic effects of plant growth-promoting rhizobacteria consortium against soilborne plant pathogens of Solanum tuberosum and Solanum lycopersicum. FEMS Microbiol. Lett. 2020, 367, 099. [Google Scholar] [CrossRef]
- Van de Poel, B.; Van Der Straeten, D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: More than just the precursor of ethylene! Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Danish, S.; Zafar-ul-Hye, M. Co-application of ACC-deaminase producing PGPR and timber-waste biochar improves pigments formation, growth and yield of wheat under drought stress. Sci. Rep. 2019, 9, 5999. [Google Scholar] [CrossRef] [PubMed]
- Maxton, A.; Singh, P.; Masih, S.A. ACC deaminase-producing bacteria mediated drought and salt tolerance in Capsicum annuum. J. Plant Nutr. 2018, 41, 574–583. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Rossi, M.J.; Soares, C.R.F.S.; McConkey, B.J.; Glick, B.R. New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE 2014, 9, e99168. [Google Scholar] [CrossRef]
- Neelakanta, G.; Sultana, H. The use of metagenomic approaches to analyze changes in microbial communities. Microbiol. Insights 2013, 6, 10819. [Google Scholar] [CrossRef]
- Stefanowicz, A. The biolog plates technique as a tool in ecological studies of microbial communities. Pol. J. Environ. Stud. 2006, 15, 669–676. [Google Scholar]
- Martinez-Drets, G.; Del Gallo, M.; Burpee, C.; Burris, R.H. Catabolism of carbohydrates and organic acids and expression of nitrogenase by azospirilla. J. Bacteriol. 1984, 159, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, V.A.; Döbereiner, J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil 1988, 108, 23–31. [Google Scholar] [CrossRef]
- Döbereiner, J. The genera Azospirillum and Herbaspirillum. In The Prokaryotes: A Handbook on the Biology of Bacteria Ecophysiology Isolation Identification Applications; Balows, A., Triiper, H.G., Dworkin, M., Harder, W., Schleifer, K.H., Eds.; Springer: New York, NY, USA, 1992; pp. 2236–2253. [Google Scholar]
- Palleroni, N.J. Introduction to the Family Pseudomonadaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 1981; pp. 655–665. [Google Scholar]
- Forni, C.; Riov, J.; Grilli Caiola, M.; Tel-Or, E. Indole-3-acetic acid (IAA) production by Arthrobacter species isolated from Azolla. J. Gen. Microbiol. 1992, 138, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Gupta, V. Methods for the determination of limit of detection and limit of quantitation of the analytical methods. Chron. Young Sci. 2011, 2, 21. [Google Scholar] [CrossRef]
- Brígido, C.; Duan, J.; Glick, B.R. Methods to study 1-aminocyclopropane-1-carboxylate (ACC) deaminase in plant growth-promoting bacteria. In Handbook for Azospirillum; Springer: Cham, Switzerland, 2015; pp. 287–305. [Google Scholar]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Olsen, S.R.; Sommers, L.E. Phosphorus. In Methods of Soil Analysis; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Meier, U. Growth Stages of Mono- and Dicotyledonous Plants. BBCH Monograph; Open Agrar Repositorium: Quedlinburg, Germany, 2018; ISBN 978-3-95547-071-5. [Google Scholar]
- Tavarini, S.; Degl’Innocenti, E.; Remorini, D.; Massai, R.; Guidi, L. Antioxidant capacity, ascorbic acid, total phenols and carotenoids changes during harvest and after storage of Hayward kiwifruit. Food Chem. 2008, 107, 282–288. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Ministero delle Politiche Agricole Alimentari e Forestali. Approvazione dei “Metodi Ufficiali di Analisi Chimica del Suolo”; Istituto Poligrafico dello Stato: Roma, Italy, 1999; pp. 1–222. (In Italian)
- Weber, K.P.; Legge, R.L. One-dimensional metric for tracking bacterial community divergence using sole carbon source utilization patterns. J. Microbiol. Methods 2009, 79, 55–61. [Google Scholar] [CrossRef]
- Garland, J.L. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 2006, 24, 289–300. [Google Scholar] [CrossRef]
- Available online: https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 1 March 2021).
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, B.; Simpson, G.; Solymos, P.; et al. The Vegan Package. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 28 July 2020).
- Wahyudi, A.T.; Priyanto, J.A.; Afrista, R.; Kurniati, D.; Astuti, R.I.; Akhdiya, A. Plant growth promoting activity of actinomycetes isolated from soybean rhizosphere. Online J. Biol. Sci. 2019, 19, 1–8. [Google Scholar] [CrossRef]
- Radwan, T.-S.-D.; Mohamed, Z.; Reis, V. Production of indole-3-acetic acid by different strains of Azospirillum and Herbaspirillum spp. Symbiosis 2002, 32, 39–54. [Google Scholar]
- Simonetti, A.E.; Roberts, I.N.; Marcela, S.; Gutierrez-boem, F.H.; Gomez, F.M.; Ruiz, J.A. A novel Burkholderia ambifaria strain able to degrade the mycotoxin fusaric acid and to inhibit Fusarium spp. growth. Microbiol. Res. 2018, 206, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Salgado, T.; Fuentes-Ramirez, L.E.; Tapia-Hernandez, A.; Mascarua-Esparza, M.A.; Martinez-Romero, E.; Caballero-Mellado, J. Coffea arabica L., a new host plant for Acetobacter diazotrophicus, and isolation of other nitrogen-fixing acetobacteria. Appl. Environ. Microbiol. 1997, 63, 3676–3683. [Google Scholar] [CrossRef]
- Bar, T.; Okon, Y. Conversion of tryptophan, indole-3-pyruvic acid, indole-3-lactic acid and indole to indole-3-acetic acid by Azospirillum brasilense Sp7. In Azospirillum VI and Related Microorganisms; Springer: Berlin/Heidelberg, Germany, 1995; pp. 347–359. [Google Scholar]
- Molina, R.; Rivera, D.; Mora, V.; López, G.; Rosas, S.; Spaepen, S.; Vanderleyden, J.; Cassán, F. Regulation of IAA biosynthesis in Azospirillum brasilense under environmental stress conditions. Curr. Microbiol. 2018, 75, 1408–1418. [Google Scholar] [CrossRef]
- Vande Broek, A.; Lambrecht, M.; Eggermont, K.; Vanderleyden, J. Auxins upregulated expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J. Bacteriol. 1999, 181, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Pedraza, R.O. Recent advances in nitrogen-fixing acetic acid bacteria. Int. J. Food Microbiol. 2008, 125, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, E.P.; Soares, C.D.P.; Galvão, P.G.; Imada, E.L.; Simões-Araújo, J.L.; Rouws, L.F.; de Oliveira, A.L.; Vidal, M.S.; Baldani, J.I. Identification of genes involved in indole-3-acetic acid biosynthesis by Gluconacetobacter diazotrophicus PAL5 strain using transposon mutagenesis. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, F.O.; Monteiro, R.A.; Wassem, R.; Cruz, L.M.; Ayub, R.A.; Colauto, N.B.; Fernandez, M.A.; Fungaro, M.H.P.; Grisard, E.C.; Hungria, M.; et al. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 2011, 7, e1002064. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.; Laevens, S.; Bevivino, A.; Dalmastri, C.; Tabacchioni, S.; Vandamme, P.; Chiarini, L. Burkholderia cepacia complex: Distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 2001, 3, 137–143. [Google Scholar] [CrossRef]
- Pace, L.; Pellegrini, M.; Palmieri, S.; Rocchi, R.; Lippa, L.; Del Gallo, M. Plant growth-promoting rhizobacteria for in vitro and ex vitro performance enhancement of Apennines’ Genepì (Artemisia umbelliformis subsp. eriantha), an endangered phytotherapeutic plant. Vitr. Cell. Dev. Biol. Plant 2020, 56, 134–142. [Google Scholar] [CrossRef]
- Onofre-Lemus, J.; Hernández-Lucas, I.; Girard, L.; Caballero-Mellado, J. ACC (1-aminocyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Appl. Environ. Microbiol. 2009, 75, 6581–6590. [Google Scholar] [CrossRef]
- Nukui, N.; Minamisawa, K.; Ayabe, S.-I.; Aoki, T. Expression of the 1-aminocyclopropane-1-carboxylic acid deaminase gene requires symbiotic nitrogen-fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl. Environ. Microbiol. 2006, 72, 4964–4969. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Saleh-Lakha, S.; Glick, B.R. The effect of native and ACC deaminase-containing Azospirillum brasilense Cd1843 on the rooting of carnation cuttings. Can. J. Microbiol. 2005, 51, 511–514. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R.; Hernandez, J.P. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant Soil 2014, 378, 1–33. [Google Scholar] [CrossRef]
- Orhan, E.; Esitken, A.; Ercisli, S.; Turan, M.; Sahin, F. Effects of plant growth promoting rhizobacteria (PGPR) on yield, growth and nutrient contents in organically growing raspberry. Sci. Hortic. (Amst.) 2006, 111, 38–43. [Google Scholar] [CrossRef]
- Ullah, A.; Bano, A. Role of PGPR in the reclamation and revegetation of saline land. Pak. J. Bot. 2019, 51, 27–35. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Luo, Y.; Freitas, H. Phytoextraction of heavy metal polluted soils using Sedum plumbizincicola inoculated with metal mobilizing Phyllobacterium myrsinacearum RC6b. Chemosphere 2013, 93, 1386–1392. [Google Scholar] [CrossRef]
- Sprocati, A.R.; Alisi, C.; Pinto, V.; Montereali, M.R.; Marconi, P.; Tasso, F.; Turnau, K.; De Giudici, G.; Goralska, K.; Bevilacqua, M.; et al. Assessment of the applicability of a “toolbox” designed for microbially assisted phytoremediation: The case study at Ingurtosu mining site (Italy). Environ. Sci. Pollut. Res. 2014, 21, 6939–6951. [Google Scholar] [CrossRef]
- Zhang, H.-H.; Tang, M.; Chen, H.; Zheng, C.-L. Effects of inoculation with ectomycorrhizal fungi on microbial biomass and bacterial functional diversity in the rhizosphere of Pinus tabulaeformis seedlings. Eur. J. Soil Biol. 2010, 46, 55–61. [Google Scholar] [CrossRef]
- Siddikee, M.; Zereen, M.; Li, C.-F.; Dai, C.-C. Endophytic fungus Phomopsis liquidambari and different doses of N-fertilizer alter microbial community structure and function in rhizosphere of rice. Sci. Rep. 2016, 6, 32270. [Google Scholar] [CrossRef]
- Hou, J.; Liu, W.; Wang, B.; Wang, Q.; Luo, Y.; Franks, A.E. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.N.; Goswami, M.P.; Bhattacharyya, L.H. Perspective of beneficial microbes in agriculture under changing climatic scenario: A review. J. Phytol. 2016, 8, 26. [Google Scholar] [CrossRef]
- Zhang, L.-N.; Wang, D.-C.; Hu, Q.; Dai, X.-Q.; Xie, Y.-S.; Li, Q.; Liu, H.-M.; Guo, J.-H. Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 2019, 10, 1668. [Google Scholar] [CrossRef] [PubMed]
- Farina, R.; Beneduzi, A.; Ambrosini, A.; de Campos, S.B.; Lisboa, B.B.; Wendisch, V.; Vargas, L.K.; Passaglia, L.M.P. Diversity of plant growth-promoting rhizobacteria communities associated with the stages of canola growth. Appl. Soil Ecol. 2012, 55, 44–52. [Google Scholar] [CrossRef]
- Trabelsi, D.; Mhamdi, R. Microbial inoculants and their impact on soil microbial communities: A review. BioMed Res. Int. 2013, 2013, 863240. [Google Scholar] [CrossRef]
- Piromyou, P.; Buranabanyat, B.; Tantasawat, P.; Tittabutr, P.; Boonkerd, N.; Teaumroong, N. Effect of plant growth promoting rhizobacteria (PGPR) inoculation on microbial community structure in rhizosphere of forage corn cultivated in Thailand. Eur. J. Soil Biol. 2011, 47, 44–54. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef]
- Ren, H.; Huang, B.; Fernández-García, V.; Miesel, J.; Yan, L.; Lv, C. Biochar and rhizobacteria amendments improve several soil properties and bacterial diversity. Microorganisms 2020, 8, 502. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Z.; Kowalchuk, G.A.; Xu, Z.; Fu, X.; Kuramae, E.E. Succession of the resident soil microbial community in response to periodic inoculations. Appl. Environ. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wagg, C.; Hautier, Y.; Pellkofer, S.; Banerjee, S.; Schmid, B.; van der Heijden, M.G. Diversity and asynchrony in soil microbial communities stabilizes ecosystem functioning. eLife 2021, 10. [Google Scholar] [CrossRef]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Reich, P.B.; Trivedi, C.; Eldridge, D.J.; Abades, S.; Alfaro, F.D.; Bastida, F.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Multiple elements of soil biodiversity drive ecosystem functions across biomes. Nat. Ecol. Evol. 2020, 4, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Czerpak, R.; Dobrzyń, P.; Krotke, A.; Kicińska, E. The effect of auxins and salicylic acid on chlorophyll and carotenoid contents in Wolffia arrhiza (L.) Wimm. (Lemnaceae) growing on media of various trophicities. Pol. J. Environ. Stud. 2002, 11, 231–235. [Google Scholar]
- Raklami, A.; Bechtaoui, N.; Tahiri, A.; Anli, M. Use of rhizobacteria and mycorrhizae consortium in the open field as a strategy for improving crop nutrition, productivity and soil fertility. Front. Microbiol. 2019, 10, 1106. [Google Scholar] [CrossRef] [PubMed]
- Venieraki, A.; Dimou, M.; Pergalis, P.; Kefalogianni, I.; Chatzipavlidis, I.; Katinakis, P. The genetic diversity of culturable nitrogen-fixing bacteria in the rhizosphere of wheat. Microb. Ecol. 2011, 61, 277–285. [Google Scholar] [CrossRef]
- Boddey, R.M.; Urquiaga, S.; Alves, B.J.R.; Reis, V. Endophytic nitrogen fixation in sugarcane: Present knowledge and future applications. Plant Soil 2003, 252, 139–149. [Google Scholar] [CrossRef]
- Badawi, F.S.F.; Biomy, A.M.M.; Desoky, A.H. Peanut plant growth and yield as influenced by co-inoculation with Bradyrhizobium and some rhizo-microorganisms under sandy loam soil conditions. Ann. Agric. Sci. 2011, 56, 17–25. [Google Scholar] [CrossRef]
- Urana, R.; Singh, N.; Sharma, P. Effects of PGPR on growth and photosynthetic pigment of Trigonella foenum-graceum and Brassica juncea in PAH-contaminated soil. SN Appl. Sci. 2019, 1, 761. [Google Scholar] [CrossRef]
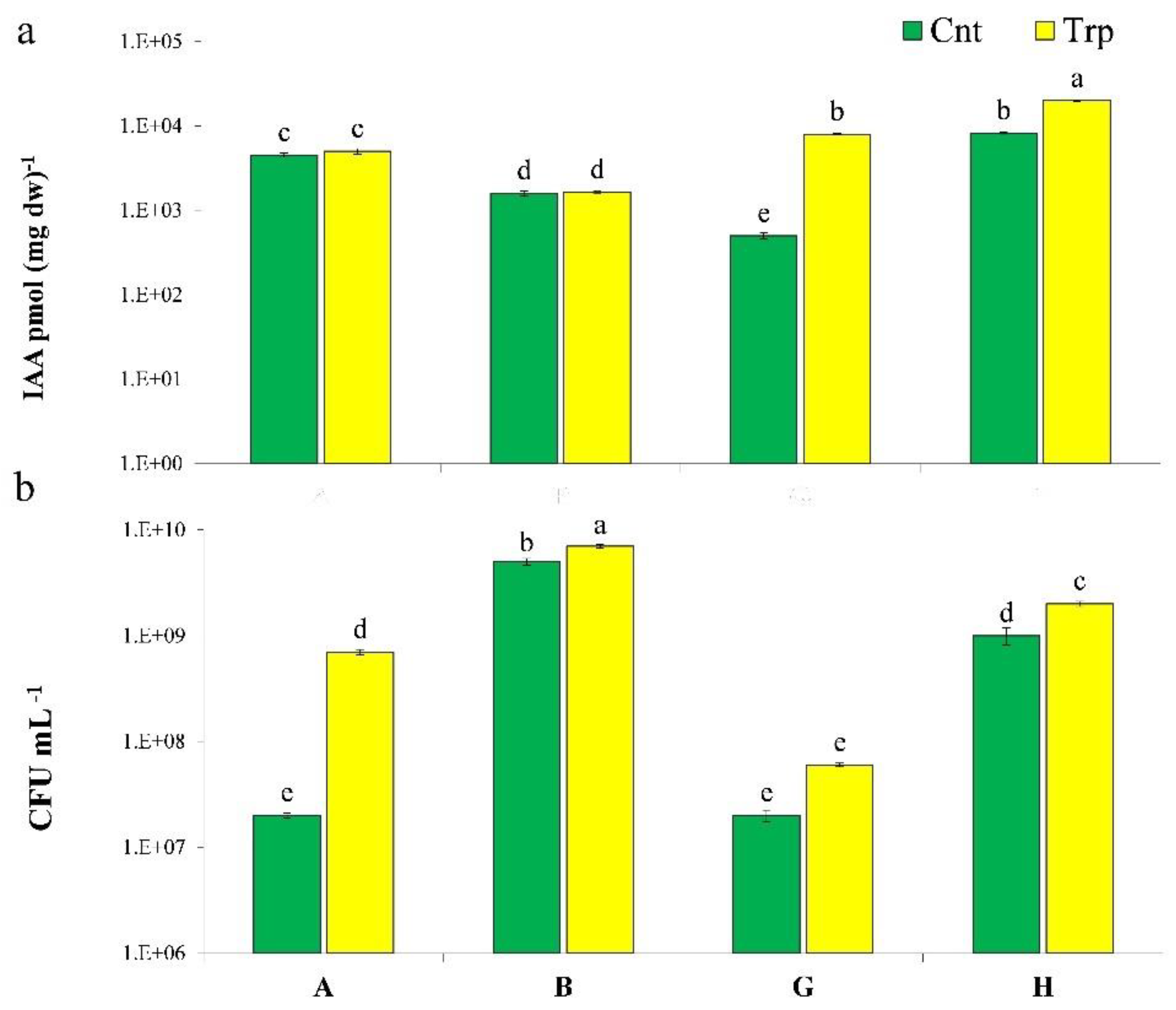
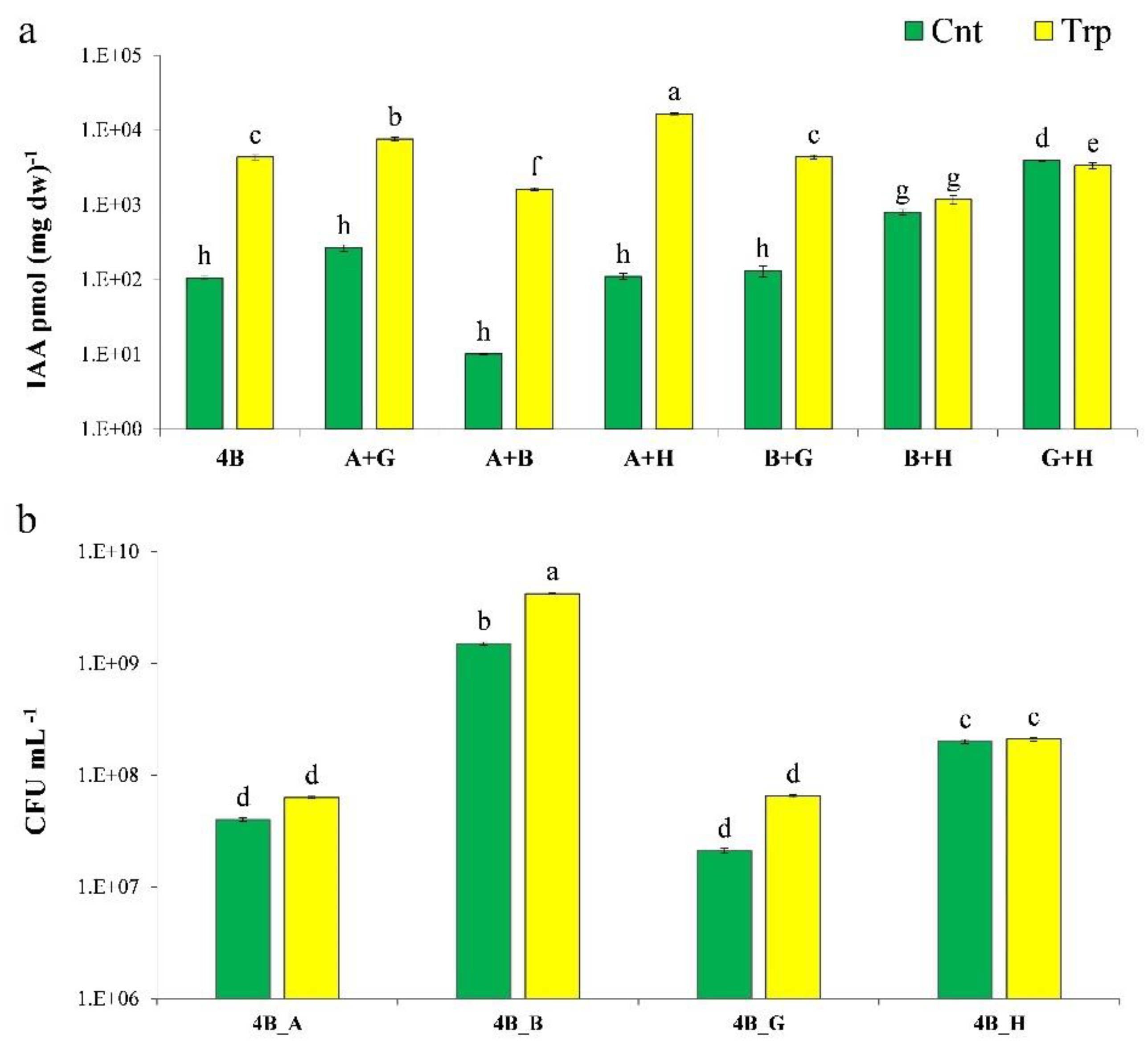

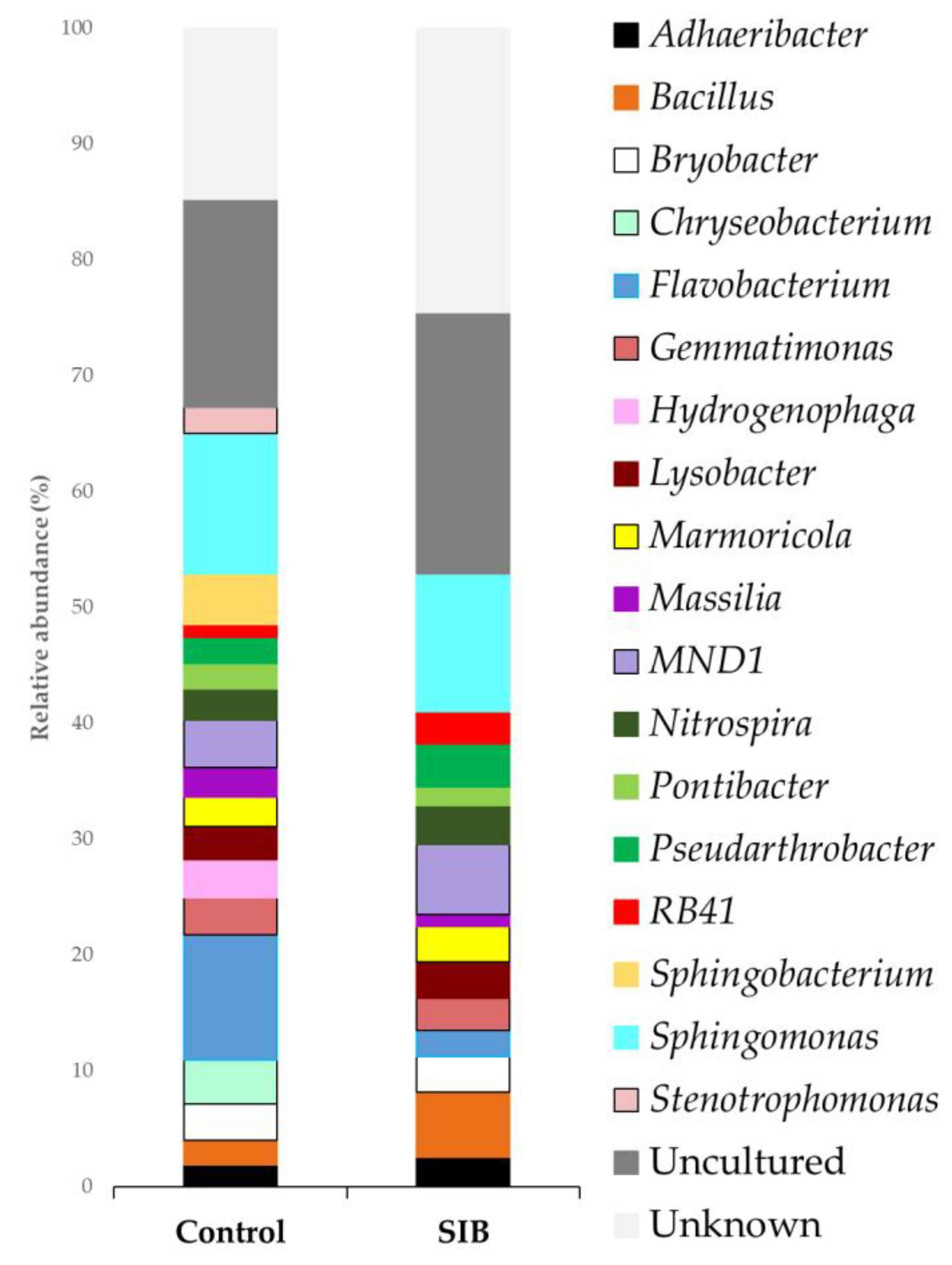
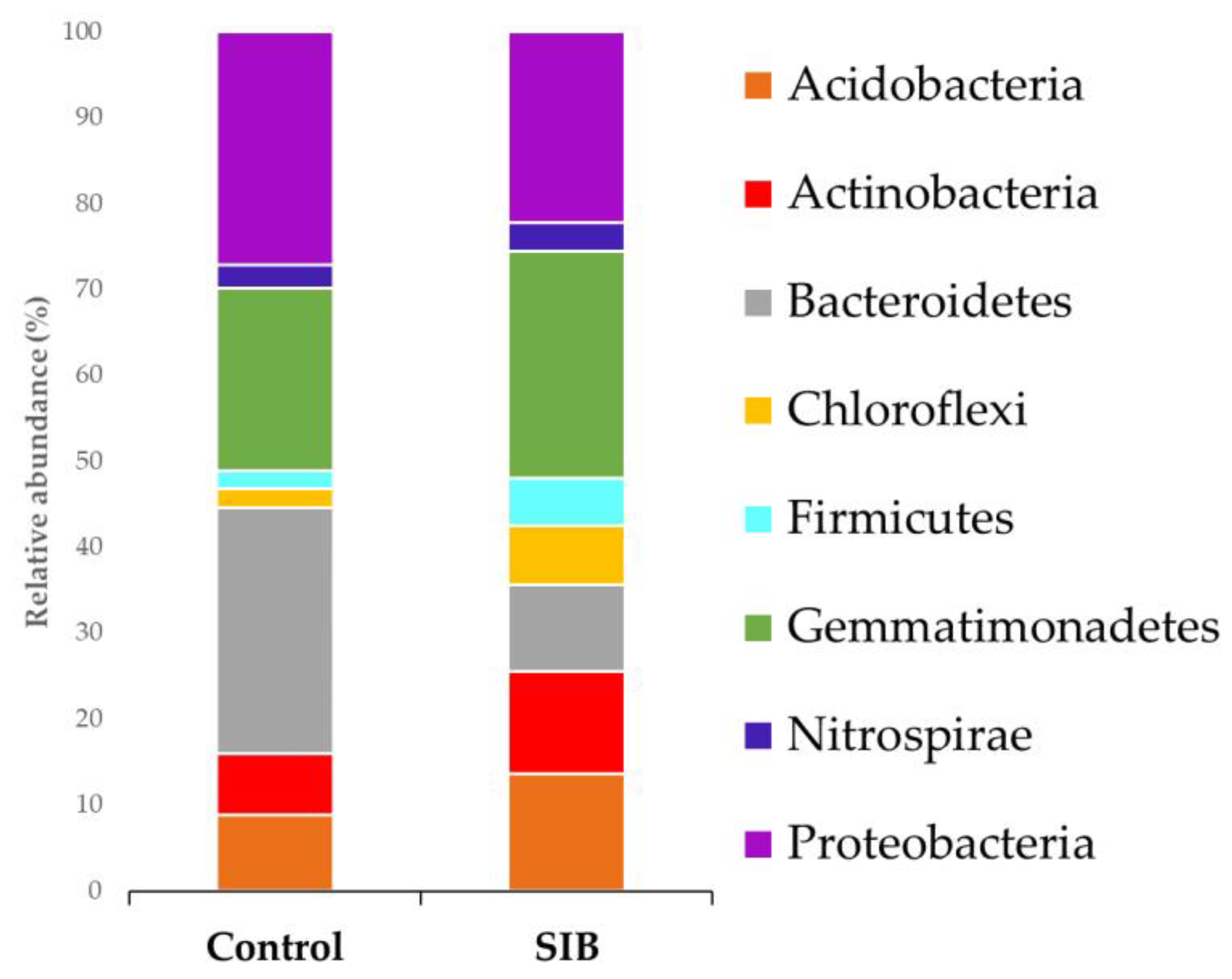
| Bacteria | Trp | IAC | IAM |
|---|---|---|---|
| nmol (mg dry wt cells)−1 | nmol (mg dry wt cells)−1 | nmol (mg dry wt cells)−1 | |
| A_C | 2.23 f | 0.93 de | - |
| A_T | 1.05 hi | 0.50 fg | <LOQ/LOD |
| B_C | 0.44 j–m | 0.40 gh | - |
| B_T | 0.80 ij | 0.29 h | - |
| G_C | 1.20 g–h | <LOQ | - |
| G_T | 4.30 d | 0.82 e | |
| H_C | 3.68 e | 8.10 a | - |
| H_T | Nq | Nq | Nq |
| 4B_C | 0.52 j–l | 0.37 gh | 0.09 c |
| 4B_T | 4.86 c | 1.02 cd | - |
| A+G_C | 0.05 mn | 0.32 h | - |
| A+G_T | 12.97 b | 2.04 b | - |
| A+B_C | - | - | - |
| A+B_T | 18.02 a | - | 2.66 a |
| A+H_C | - | 0.10 i | - |
| A+H_T | 0.59 jk | 0.62 f | 0.91 b |
| B+G_C | 0.22 k–n | <LOQ | |
| B+G_T | 2.16 f | 0.33 h | 0.10 c |
| B+H_C | 1.50 g | 1.10 c | - |
| B+H_T | 0.13 l–n | 0.33 h | - |
| G+H_C | - | - | - |
| G+H_T | - | - | 0.06 c |
| F-test | * | ** | ** |
| LSD | 0.40 | 0.15 | 0.16 |
| Parameter | Treatment | F Test | LSD | ||
|---|---|---|---|---|---|
| PS | Control | SIB | |||
| pH | 7.67 a | 7.65 a | 7.52 b | * | 0.09 |
| Total N (g Kg−1) | 2.05 b | 1.95 c | 2.15 a | ** | 0.09 |
| TOC (g Kg−1) | 16.67 b | 14.33 c | 18.67 a | ** | 1.15 |
| OM (g Kg−1) | 28.67 b | 24.65 c | 32.11 a | ** | 1.99 |
| EC (μs cm−1) | 0.44 a | 0.38 b | 0.33 c | ** | 0.03 |
| Na (mg Kg−1) | 31.50 a | 30.50 a | 17.25 b | ** | 3.07 |
| Ca (mg Kg−1) | 3055.50 a | 2997.75 b | 3082.25 a | ** | 31.61 |
| Mg (mg Kg−1) | 150.15 a | 127.25 b | 145.25 a | ** | 7.91 |
| K (mg Kg−1) | 421.00 a | 284.00 c | 311.50 b | ** | 18.46 |
| Available P (mg P2O5 Kg−1) | 300.25 b | 155.00 c | 350.75 a | ** | 9.42 |
| Parameter | Treatment | F Test | LSD | ||
|---|---|---|---|---|---|
| Control | SIB | ||||
| CLPP | AWCD | 1.35 | 1.47 | * | 6.03 |
| H | 3.28 | 3.31 | * | 1.91 | |
| R | 29 | 30 | n.s. | - | |
| NGS | H | 6.07 | 6.11 | - | - |
| Simpson 1-D | 0.9968 | 0.9971 | - | - | |
| Chao-1 | 571.6 | 587 | - | - | |
| Parameter | D.S. | Treatment | F Test | LSD | |
|---|---|---|---|---|---|
| Control | SIB | ||||
| Root Length (cm) | 41 | 15.1 | 15.9 | n.s. | - |
| 45 | 18.9 | 20.6 | n.s. | - | |
| 49 | 16.6 b | 17.7 a | * | 1.05 | |
| A.P. Length (cm) | 41 | 27.0 | 30.7 | n.s. | - |
| 45 | 54.1 | 59.3 | n.s. | - | |
| 49 | 56.7 b | 60.7 a | * | 2.94 | |
| Total (cm) | 41 | 42.1 b | 46.6 a | * | 3.88 |
| 45 | 73.0 b | 79.9 a | * | 4.16 | |
| 49 | 73.4 b | 78.4 a | * | 3.98 | |
| DW Root (%) | 41 | 9.0 b | 9.8 a | * | 0.71 |
| 45 | 10.9 b | 11.7 a | * | 0.60 | |
| 49 | 12.9 b | 13.6 a | ** | 0.23 | |
| DW A.P. (%) | 41 | 4.6 b | 5.1 a | * | 0.35 |
| 45 | 6.1 b | 6.6 a | * | 0.45 | |
| 49 | 8.2 b | 8.8 a | * | 0.54 | |
| Car (mg 100 g−1FW) | 41 | 2.4 | 2.6 | n.s. | - |
| 45 | 9.5 b | 9.8 a | * | 0.15 | |
| 49 | 12.2 b | 12.7 a | * | 0.41 | |
| Chl a + b (mg 100 g−1FW) | 41 | 373.6 | 376.1 | n.s. | - |
| 45 | 393.6 b | 395.3 a | * | 2.13 | |
| 49 | 401.4 b | 403.6 a | * | 1.40 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellegrini, M.; Pagnani, G.; Rossi, M.; D’Egidio, S.; Gallo, M.D.; Forni, C. Daucus carota L. Seed Inoculation with a Consortium of Bacteria Improves Plant Growth, Soil Fertility Status and Microbial Community. Appl. Sci. 2021, 11, 3274. https://doi.org/10.3390/app11073274
Pellegrini M, Pagnani G, Rossi M, D’Egidio S, Gallo MD, Forni C. Daucus carota L. Seed Inoculation with a Consortium of Bacteria Improves Plant Growth, Soil Fertility Status and Microbial Community. Applied Sciences. 2021; 11(7):3274. https://doi.org/10.3390/app11073274
Chicago/Turabian StylePellegrini, Marika, Giancarlo Pagnani, Massimiliano Rossi, Sara D’Egidio, Maddalena Del Gallo, and Cinzia Forni. 2021. "Daucus carota L. Seed Inoculation with a Consortium of Bacteria Improves Plant Growth, Soil Fertility Status and Microbial Community" Applied Sciences 11, no. 7: 3274. https://doi.org/10.3390/app11073274
APA StylePellegrini, M., Pagnani, G., Rossi, M., D’Egidio, S., Gallo, M. D., & Forni, C. (2021). Daucus carota L. Seed Inoculation with a Consortium of Bacteria Improves Plant Growth, Soil Fertility Status and Microbial Community. Applied Sciences, 11(7), 3274. https://doi.org/10.3390/app11073274










