Abstract
As the conventional models for time series forecasting often use single-valued data (e.g., closing daily price data or the end of the day data), a large amount of information during the day is neglected. Traditionally, the fixed reference points from intervals, such as midpoints, ranges, and lower and upper bounds, are generally considered to build the models. However, as different datasets provide different information in intervals and may exhibit nonlinear behavior, conventional models cannot be effectively implemented and may not be guaranteed to provide accurate results. To address these problems, we propose the artificial neural network with convex combination (ANN-CC) model for interval-valued data. The convex combination method provides a flexible way to explore the best reference points from both input and output variables. These reference points were then used to build the nonlinear ANN model. Both simulation and real application studies are conducted to evaluate the accuracy of the proposed forecasting ANN-CC model. Our model was also compared with traditional linear regression forecasting (information-theoretic method, parametrized approach center and range) and conventional ANN models for interval-valued data prediction (regularized ANN-LU and ANN-Center). The simulation results show that the proposed ANN-CC model is a suitable alternative to interval-valued data forecasting because it provides the lowest forecasting error in both linear and nonlinear relationships between the input and output data. Furthermore, empirical results on two datasets also confirmed that the proposed ANN-CC model outperformed the conventional models.
1. Introduction
Time-series point (single-value data) forecasting normally fails to reflect the range of fluctuation or uncertainty for economic, financial, and environmental data. Moreover, the existing model of interval forecasting is still incomplete, complex, and has relatively low accuracy. Thus, interval-value data forecasting has become an important issue to be investigated [1,2]. This study comes within interval-valued time series forecasting framework by introducing the convex combination (CC) method designed to choose the reference points that better represent the interval-valued data. The CC method automatically explores the set of reference points from input and output variables to build the neural networks (NN) models. This is an enhancement and a generalization over existing methods.
Interval-valued data forecasting serves the needs of investors and data scientists who are sometimes interested in single value-data and the variability of value intervals in the data. Interval-valued data provides rich information that can help investors and data scientists make an accurate decision [3]. With the advance in data science, enormous information and big data can be collected nowadays. However, conventional forecasting methods cannot be effectively implemented to deal with this big data to yield accurate results. Furthermore, these methods are generally proposed to forecast future observations using only the point-valued data, which gives rise to a higher computation cost when dealing with big data. Moreover, it is sometimes difficult to express the real behavior of the variable using only point-valued data. Thus, interval-valued data, such as the range of temperature, stock returns, and willingness to pay (minimum and maximum), is generally used to predict uncertainty in many situations. Interval analysis suggested by Lauro and Palumbo [4] assumed that observations and estimations in the world are usually uncertain, incomprehensive, and do not precisely represent the real behavior of the data. This is quite true, as our use of point-valued data will entitle us to lose substantial information about prices or values during the day or the week. With interval-valued data, however, we can capture more realistic movement or information of the variables and can also handle big data forecasting at the same time. Thus, the interval approach should be considered to explain real data behavior under the context of big data. Interval-valued data is a special type for symbolic data analysis (SDA), composed of lower and upper bounds of an interval. The objective of SDA is to provide a way to construct aggregated data described by multivalued variables, and thereby provide an efficient way to summarize the large data sets by some reference value of symbolic data. Thus, the estimation tools for interval-valued data analysis are very intensively required in recent studies. The use of interval-data to represent uncertainty is common in various situations, such as in coalitional games where payoffs of coalitions are uncertain and could be modeled as intervals of real numbers, see, e.g., Branzei et al. [5] and Kiekintveld et al. [6].
From a methodological point of view, interval-valued data is generally transformed into point-valued data or reference point data by some techniques; then, this reference point data is furthered used as a variable in the models. One of the famous approaches for dealing with interval-valued data is the mid-point method which was introduced by Billard and Diday [7]. They analyzed these data using ordinary least squares regression on the midpoints of the intervals, namely the lower and upper bounds of the independent and dependent variables. Neto and De Carvalho [8] improved this approach by presenting a new method based on two linear regression models (center-range method). The first regression model is fitted at the midpoints of the intervals and the second one on the ranges of the intervals. It was found to be more efficient than that of Billard and Diday [7]. More recently, Souza et al. [9], Chanaim et al. [10], and Phadkantha et al. [11] argued that the mid-point and range methods are not appropriate to be the reference of the interval data as they cannot present the real behavior of the data and both are also too restrictive. Thus, Souza et al. [9] introduced the parametrized approach (PM) to the intervals of input variables. This method can choose the reference points that better represent the input intervals before building the regression. Chanaim et al. [10] and Phadkantha et al. [11] extended the PM approach by suggesting the convex combination (CC) method to get the reference points of the input and output intervals before estimating the regression model. Specifically, instead of restricting the weight of interval-valued data to be 0.5 (mid-point), , they generalized this weight to be unknown. Hence, the reference point data become , where is the weight parameter. Then, this reference point data is used as the input data in the regression model. Buansing et al. [12] also proposed the iterative, information-theoretic (IT) method for forecasting interval-valued data. Their method differs from others, as it does not assume that every point in the interval emerged from the same underlying process; there may be multiple models behind the process. They showed that the IT method provides more accurate forecasting of the upper and lower bounds when compared to the center-range method.
There is voluminous literature on stock market estimation and forecasting based on a wide variety of both linear and nonlinear models. Linear models like autoregressive (AR) and autoregressive moving average (ARMA) [13,14] and nonlinear models like threshold and Markov switching models [15,16,17,18] have been used for time series forecasting. However, the equivocal and unforeseeable nature of the time series data has brought about the difficulty in prediction [19], so artificial neural networks (ANN) models were introduced to forecast the future return of the stock. The major advantage of ANN models is their flexible nonlinear modeling capability. With ANN, there is no need to specify a particular model form. Instead, the model is adaptively formed based on the features inherent in the data. This data-driven approach is suitable for many empirical data sets where no theoretical guidance is available to suggest an appropriate data generating process. ANN provides desirable properties that some traditional linear and nonlinear regression models lack, such as being noise tolerant. The structure of the ANN model is inspired by real-life information-processing abilities of the human brain. Key attributes of the brain’s information network include a nonlinear, parallel information processing structure and multiple connections between information nodes [20].
In the recent decade, several studies have indicated the higher performance of the ANN models in forecasting compared to that of regression models. Zhang et al. [21] and Leung et al. [22] examined various prediction models based on multivariate classification techniques and compared both neural network and classical forecasting models. Their experiment results suggested that the probabilistic neural network can outperform the level estimation models, including adaptive exponential smoothing, vector autoregression with Kalman filter updating, multivariate transfer function, and multilayered feedforward neural network, in terms of the prediction accuracy. In a more recent study, Cao et al. [23] demonstrated the accuracy of ANN in predicting Shanghai Stock Exchange (SHSE) movement by comparing the neural networks and linear models under the capital asset pricing model (CAPM) and Fama and French’s three-factor contexts, and they found that neural networks outperformed the linear models.
As we mentioned above, this paper comes within the framework of interval-valued data forecasting by ANN. To the best of our knowledge, work-related to interval-valued data forecasting by neural networks is somewhat limited. Some attempts along this line include San Roque et al. [24], Maia et al. [25], Maia and De Carvalho [26], and Yang et al. [27]. Maia et al. [25] proposed the ANN-Center method to predict the interval value by using the midpoint of the interval as the input. Later, Maia and De Carvalho [26] introduced the ANN-LU method by using ANN-Center and ANN-Range (predicting the difference between upper and lower bounds) for predicting the lower and upper bounds of intervals separately. Yang et al. [27] suggested that ANN-LU may face the unnatural interval crossing problem when the predicted lower bounds of intervals are larger than the predicted upper ones and vice versa and thereby leading to an invalid interval prediction. Hence, they introduced the regularized ANN-LU (RANN-LU) model for interval-valued time series forecasting.
Although ANN-center, ANN-LU, and RANN-LU are found to be superior to the classical linear regression models for interval-valued data prediction of Billard and Diday [7], these models rely either on midpoint (ANN-Center) or lower and upper bounds of the interval for forecasting, which may not be a good input for predicting the future value of an interval. For example, in the case of RANN-LU, if we predict the future value of the upper bound and lower bound separately, the prediction may not be reliable, since the whole information of the interval is not taken into account [27,28]. Although ANN-Center and ANN-LU consider both upper and lower bounds in the prediction process, the prediction still relies on the midpoint of the interval indicating the symmetric weight between lower and upper bounds. To overcome these problems, in this study, we introduce the convex combination (CC) approach to ANN for predicting interval-valued data, say, lower and upper bounds. Our model is a generalization of the ANN-Center, allowing the weight to be more flexible and not to be fixed at 0.5 (asymmetric weight).
The novelty of the proposed ANN-CC can be summarized in the following two aspects: First, our method can construct prediction intervals based on the CC approach and ANN models. More specifically, this study proposes the novel CC method for interval ANN modeling. In this approach, the intervals of input and output variables are parametrized through the convex combination of lower and upper bounds. The proposed ANN-CC is a promising alternative to the existing approaches. With optimal reference points that better represent the intervals, we are able to find an efficient solution that improves the prediction accuracy of the lower and upper bounds. Second, to the best of our knowledge, there is no study extending the CC approach for interval ANN modeling. Our proposed method fills in such a literature gap and can capture both linear and nonlinear patterns within interval-valued data.
The rest of this paper is organized as follows. Section 2 gives a brief review of interval-valued prediction methods. Section 3 presents the proposed ANN with the convex combination method. In Section 4, we provide a simulation study to assess the performance of our proposed method; Section 5 describes the data used in this study. The analytical results are presented in Section 5. Section 6 provides the conclusion of this study.
2. Reviews of Existing Methods
2.1. Linear Regression Based on Center Method
Let and , , are the lower and upper bounds of the intervals, respectively.
According to Billard and Diday [7], the center or mid-point of interval-valued explanatory variables denoted as is calculated from:
Likewise, interval-valued response variable denoted as is calculated from:
where and .
Thus, regression based on the center method can be constructed as
where is the vector of parameters. are errors that have a normal distribution. Using matrix notation, this problem can be estimated by the ordinary least squares (OLS) method under the full rank assumption:
Then, the estimates for the response lower and upper bounds are presented as and , respectively.
2.2. Linear Regression Based on Center-Range Method
Neto and De Carvalho [8] also introduced the center-range method to predict the upper and lower bounds of the dependent variable intervals. In this method, the lower and upper bounds of the interval-valued response variable are separately predicted by the mid-points and ranges of the interval-valued explanatory variables. Thus, this model is built on two linear regression models, namely the regression based on center method (Equation (3)) and the regression-based on range method:
where is half-ranges of explanatory variables, is half-ranges of response variables, is the error. Using matrix notation, the least-squares estimator parameter is given by
Then, we can predict the response lower and upper bounds as and , respectively.
2.3. Linear Regression Based on Convex Combination Method
Chanaim et al. [10] suggested that the center method may lead to the misspecification problem, as the midpoint of the intervals might not be a good reference of the intervals. To tackle this problem, they proposed employing the convex combination approach to determine the best reference point between the ranges of interval data
where is the weight parameter of the interval data with values [0, 1]. The advantage of this method lies in the flexibility to assign weights in calculating the appropriate value between intervals. Thus
where , . Using matrix notation, this problem is also estimated by the OLS method under the full rank assumption:
The response lower bound prediction is described in Equation (11), and the model to predict the response upper bound is given by Equation (12),
where
where is the range prediction and , .
2.4. Regularized Artificial Neural Network (RANN)
Yang et al. [27] introduced RANN for interval-valued data prediction. This method is able to approximate various forms of nonlinearity in the data and directly models the non-cross lower and upper bounds of intervals. In this model, the relation between the interval-valued output ( and ) and interval-valued inputs ( and ) is as follows:
where consists of 2k inputs. , and , represent the weight parameters and bias terms of the jth hidden layer neuron of input layer. , and , represent the weight parameters and bias terms of the jth hidden layer neuron of output layer. and are the activation functions. To meet non-crossing lower and upper bounds of intervals, a non-crossing regularize is introduced in the loss function as follows,
where is the regularization parameter for controlling the non-crossing strength.
3. The Proposed Method Artificial Neural Network with Convex Combination (ANN-CC)
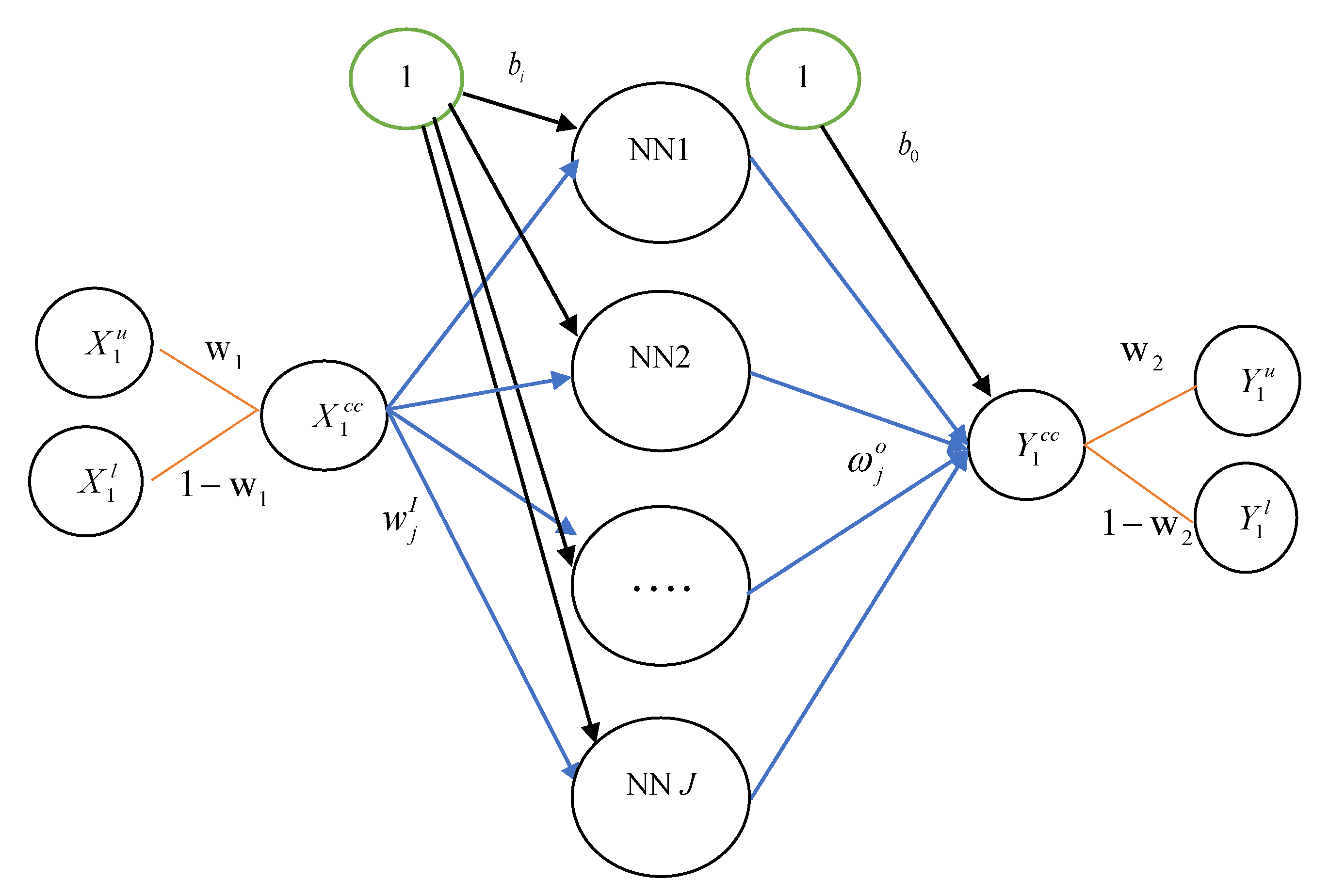
Artificial neural network (ANN) models can approximate various forms of nonlinearity in the data. There are various types of ANN, but the most popular one is the Multilayer Perceptron (MLP). ANN models have been successfully applied in a variety of fields such as accounting, economics, finance, and marketing, as well as forecasting [20,26]. In this study, we use a three-layer ANN model in which both the inputs and outputs contain the lower and upper bounds of intervals. As shown in Figure 1, suppose the interval-valued data consists of one predictor and one response. The first layer is the input layers, the middle layer is called the hidden layer, and the last layer is the output layers. All the data are expressed in the form of a lower-upper bound. Thus, we have and . For convenience, we use the notation for input variable and for output variable. Note that ANN-center is the particular case of ANN-CC if we set . Each layer contains jth neurons which are connected to one another and are also connected with all neurons in the immediate next layer. The neuron input path has a signal on its , and the strength of the path is characterized by a weight of neuron (). The neuron is modeled as summing the path weight times the input signal over all paths and adding the node bias . Note that the neural network consists of input function and output function; thus, we can express these functions as:
where is the jth hidden neuron’s input, is the weight vector between the hidden layer and the input layer. is the activation function for the hidden layer. is the bias term of input layer. Then, is transformed into output with the activation function of the output layer. Thus, the model can be written as:
where is the weight vector between the hidden layer and the output layer. is the bias term of output layer. is the activation function of the output layer. A challenge in ANN design is the selection of activation function. It is also known as transfer function, and can be basically divided into four types: tanh or hyperbolic tangent activation function (tanh), sigmoid or logistic activation function (sigmoid), linear activation function (linear), and exponential activation function (exp).
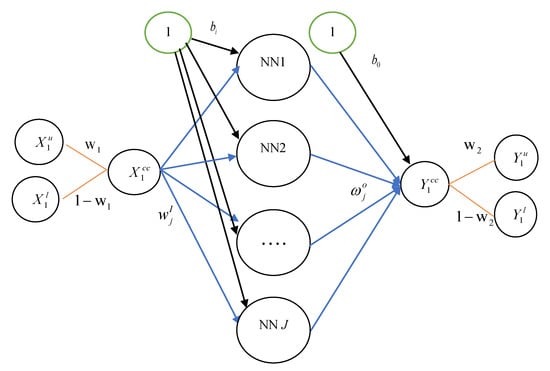
Figure 1.
The network architecture of the interval-valued data artificial neural network with convex combination (ANN-CC) model (one hidden layer is assumed.
Learning occurs through the adjustment of the path weights and node biases. The most common method used for this adjustment is backpropagation. In this method, the optimal weights and are estimated by minimizing the squared difference between the model output and the estimated. We formulate the loss function as follows,
where and are the estimated output and observed output, respectively. In addition to the weights of neuron j , our estimation also considers the weight parameter of the interval data to determine the reference of the output and input variables.
As the interval data is manipulated as a new type of numbers represented by an ordered pair of its minimum and maximum values, the numerical manipulations of interval data should follow “interval calculus” [25]. In practice, we can separately predict the lower and upper bounds of the interval using the min-max method. However, this method does not guarantee the mathematical coherence of predicted bounds. That is, the predicted lower bounds of intervals should be smaller than the predicted upper ones. Otherwise, the unnatural interval crossing problem will occur, which leads to an invalid interval prediction [27]. Furthermore, the forecasting performance can be impaired if there is not a clear dependency between the respective bounds of output and input [28,29]. Instead of restricting the weight of interval-valued data to be 0.5 (mid-point), in this study, we consider the convex combination method to get the reference point of the interval-valued data. Thus, in this estimation, the weights of neuron j are dependent on the given weight . Since there is no close form solution for this weight parameter of the interval data, we thus employ a grid search selection of that minimizes the sum of squared errors, denoted as . Then, we can rewrite our loss function in Equation (19) as
This loss function is computed using two steps of estimation. Firstly, it is important to solve a nonlinear optimization to obtain which depends on the candidate . In the second step, the loss function is then minimized with respect to the candidate . Then, we introduce another candidate to repeat the first step. After is computed for all candidates , the minimum value of is preferred. Thus, the optimal is obtained by
We note that ANN estimates over a grid search between 0 and 1. Finally, following the CC method in Section 2.3, we can predict the lower and upper bounds as follows.
where is the jth hidden neuron’s input and the range prediction is computed by
where and . By making this prediction, the predicted lower bounds of intervals should not cross over the corresponding upper bound. We also note that and have contained the information of the weight parameter of the interval data and .
In our work, the ANN-CC methodology includes the grid search for the parameter . Note that grid search was executed before estimating the weight parameters and in the ANN structure. A pseudo-code for the performed grid search and ANN-CC estimation is presented in Algorithm 1.
| Algorithm1. Pseudo code for the proposed ANN-CC with one predictor and one response |
| , where and are the set of candidate weight of input and output , respectively, within [0, 1]. |
| #Serach the optimal |
| For each in |
| Calculateand |
| #Define the loss function of ANN structure |
| , = Parameters( ) |
| # Follow the gradients until convergence |
| repeat |
| until convergence |
| end for |
| #Choose the with the lowest as the optimal |
| Calculateand |
| # Compute the loss function of ANN structure using and |
| # Follow the gradients until convergence |
| repeat |
| until convergence |
4. Simulation Study
To examine the performance of our proposed method, we conducted a simulation study. We considered two data generation processes which are different in structure: linear and nonlinear.
4.1. Linear Structure
The simple interval generation process is conducted, and one independent variable is assumed. We considered three typical data generation processes with different weight parameters. For this purpose, we considered the following three Scenarios of weight in the intervals:
Scenario 1: Center of interval data: ,
Scenario 2: Deviate from the center to the lower of the interval data: ,
Scenario 3: Deviate from the center to the upper of the interval data: ,
For each scenario, we performed 100 replications. In each simulation, we proceeded as follows.
(1) Generate the error from the normal distribution with mean zero and variance one.
(2) Generate the upper bound of the independent variable , from the uniform (1,3). Then, we computed the lower bound of the independent variable , where denotes the range between the upper and lower bounds.
(3) Compute the expected independent variable of the intervals that have been computed by
. Then, we could generate the expected dependent variables .
(4) Finally, we derived the upper and lower bounds of intervals by , where and . We note and are a random number for simulating the interval and , respectively. This guaranteed that the bounds are not crossing each other.
In this simulation study, we performed 100 replications with a sample size n = 1000 for all three scenarios. Each simulation dataset a randomly split, with 80% for training and 20% for testing. Our ANN with the convex combination method (ANN-CC) was then compared with two conventional models, namely the ANN-Center and the RANN-LU method of Yang et al. [27]. In this simulation study, four transfer functions, namely tanh or hyperbolic tangent activation function (tanh), sigmoid or logistic activation function (sigmoid), linear activation function (linear), and exponential activation function (exp) were considered. To simplify the comparison, one input layer, one output layer, one hidden layer, and one hidden neuron were assumed. We noted that the ANN-Center could be estimated by the validann package in R programming language [30]. In addition, this package also provides validation methods for the replicative, predictive, and structural validation of artificial neural network models.
To assess the performance of these models, we conducted the following measures: mean absolute error (MAE), mean squared errors (MSE), and root mean squared errors (RMSE). The formulae of MAE, MSE, and RMSE are as follows:
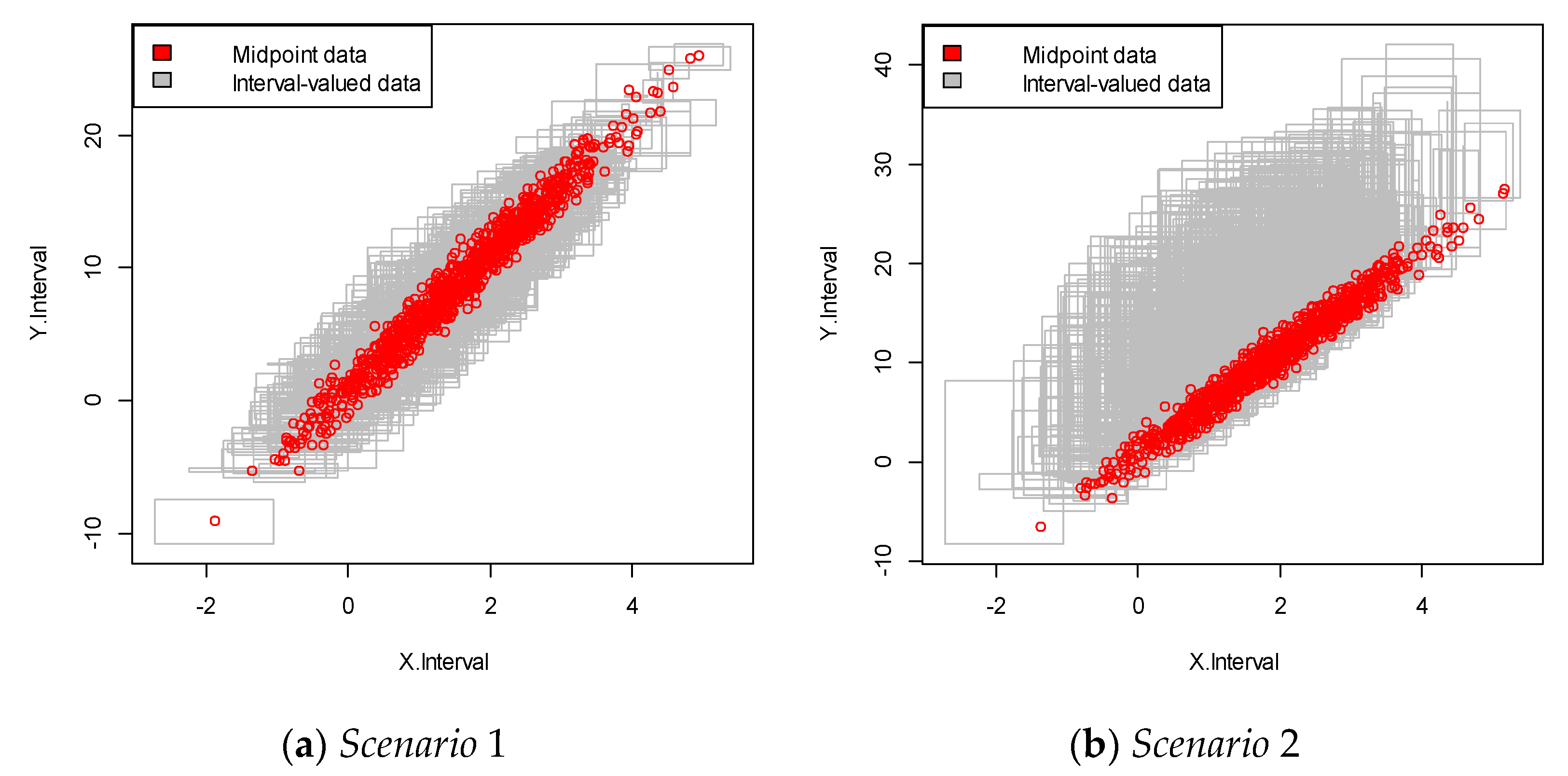
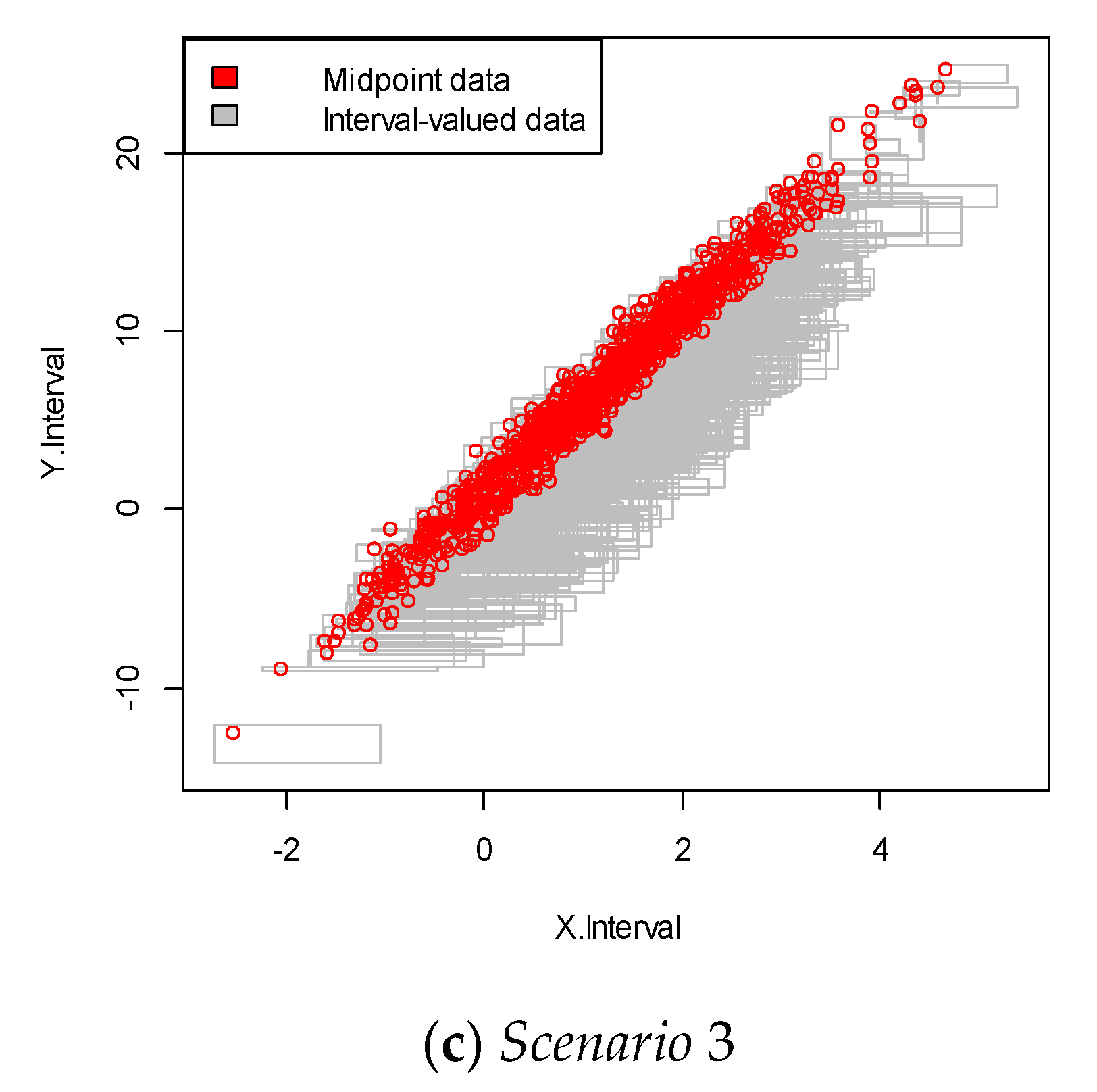
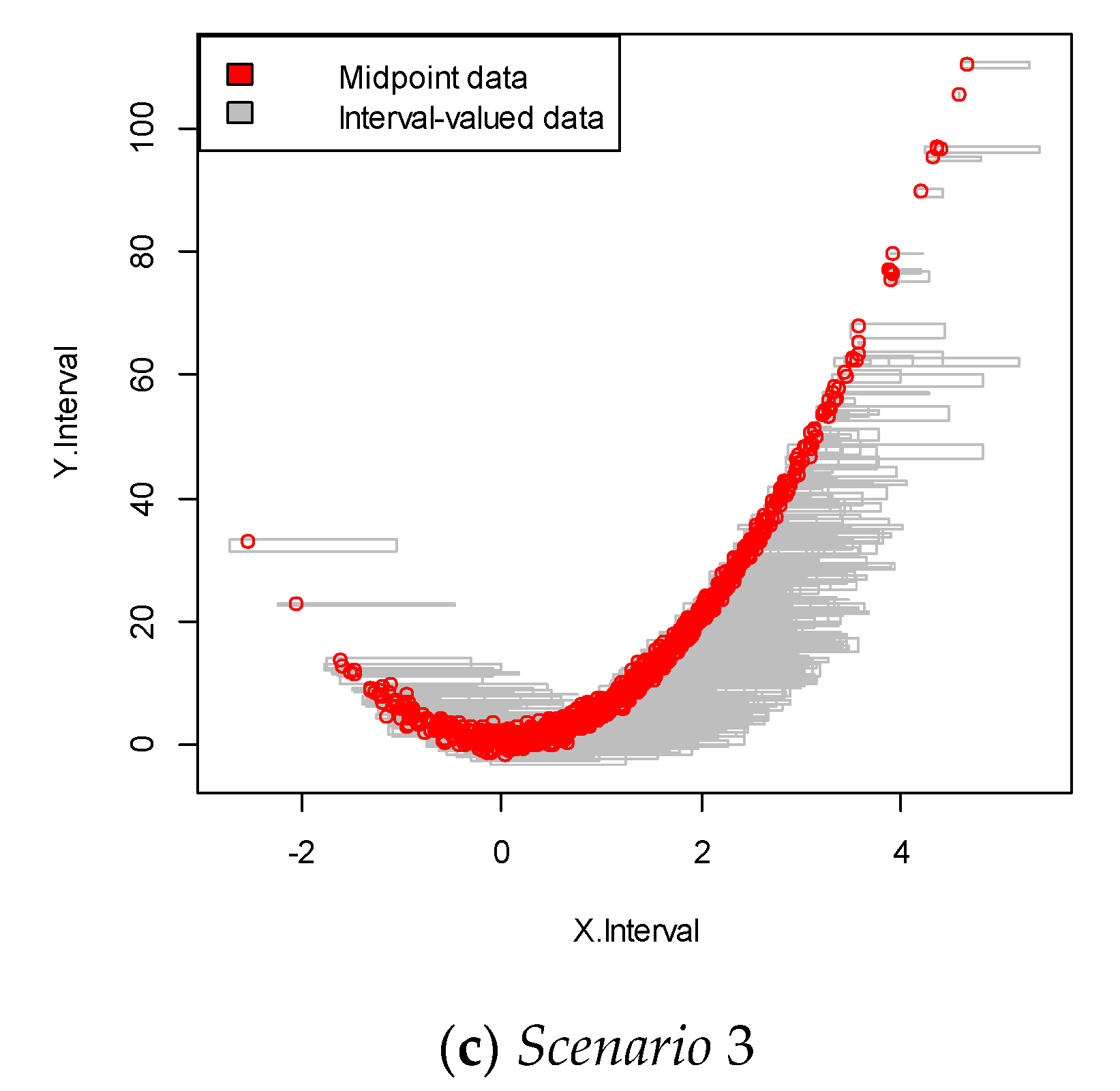
We repeated the simulation 100 times and obtained the simulation data with 100 samples. An example of each of the simulated interval-valued time series is presented in Figure 2. In this figure, each square plot represents the relationship between the interval and .
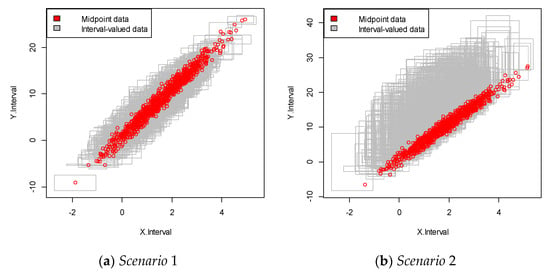
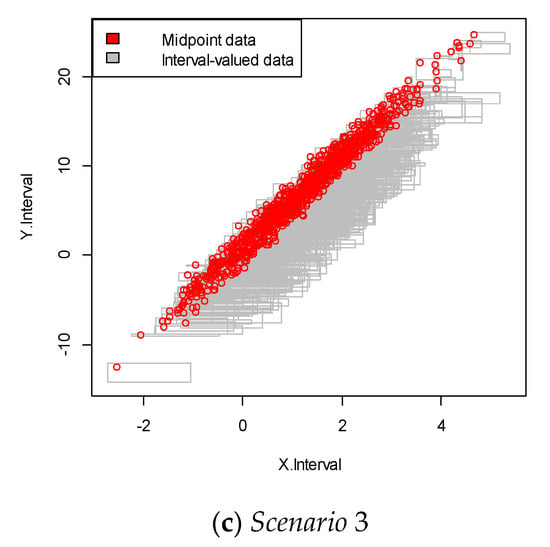
Figure 2.
Interval-valued data plot for the linear case. The red dots indicate the mid-point value within the interval.
Table 1 presents the results of 100 repetitions for the linear structure case. The MAE, MSE, and RMSE are reported. We observed that the ANN model with the CC method (ANN-CC) showed its powerful nonlinear approximation ability (tanh, sigmoid, exp) as the MAE, MSE, and RMSE values were lower than those of the ANN-Center and RANN-LU in Scenarios 2 and 3. It is also noticed that tanh function performed the best fit function for the ANN-CC model for these simulated datasets. Not surprisingly, we observed that our ANN-CC did not outperform the ANN-Center method under Scenario 1. This is due to the interval data being simulated from the midpoint. However, our ANN-CC method still performed better than RANN-LU in this scenario. In sum, from evaluating our ANN model with convex combination performance, we reached a similar conclusion for Scenarios 2 and 3. Our proposed model performed well in the simulation study, and the ANN-CC method showed high performance in all scenarios.

Table 1.
Experimental results for the linear case.
4.2. Nonlinear Structure
Similar to the linear structure, we considered three typical data generation processes with different weight parameters. Three Scenarios of weight in the intervals were as follows:
Scenario 1: Center of interval data: ,
Scenario 2: Deviate from the center to the lower of the interval: ,
Scenario 3: Deviate from the center to the upper of the interval: ,
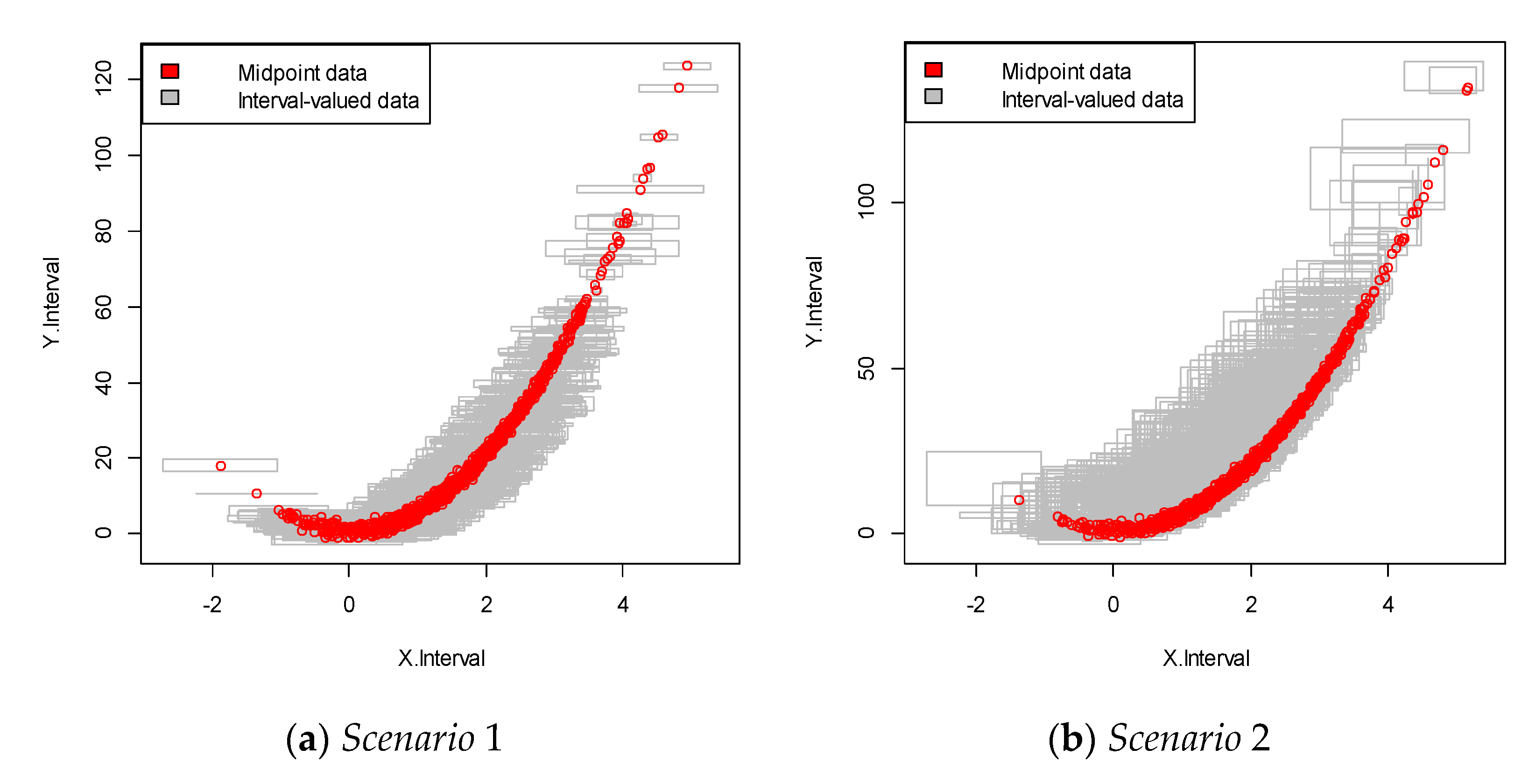
For each scenario, we performed 100 replications. This data was more complicated than that in the linear structure case, as it showed a nonlinear relationship between the dependent and independent variables, as shown in Figure 3. The results are summarized in Table 2.
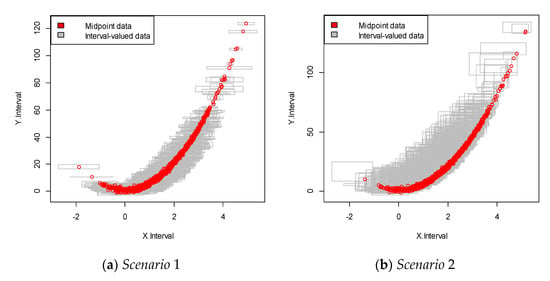
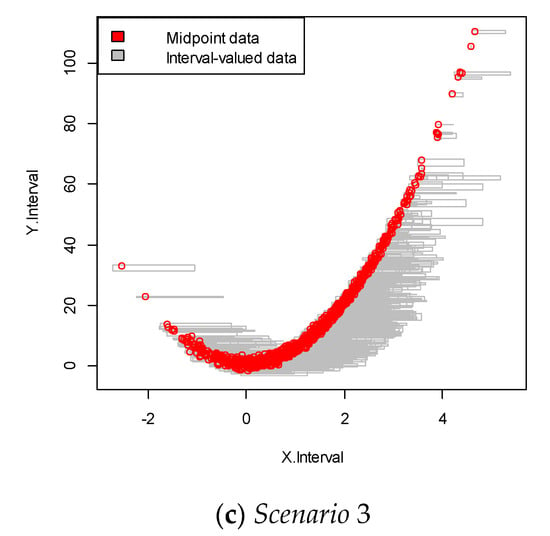
Figure 3.
Interval-valued data plot for the nonlinear case. The red dots indicate the mid-point value within the interval.

Table 2.
Experimental results for the nonlinear case.
Table 2 presents the simulation results based on the nonlinear case. Similar results were obtained, as the proposed ANN-CC showed higher performance in Scenarios 2 and 3. We observed that ANN-Center still performed poorly in Scenarios 2 and 3. The reason is simple. This model fixes the weight parameter at the center, which does not correspond to the true data generating process, thus the ANN-Center leads to higher bias of the prediction.
The experiments were carried out using an Intel Core i5-6400 CPU 2.7 GHz 4 core 16Gb RAM. The computational cost of our ANN-CC model were a bit higher than those of ANN-Center and RANN-LU. The training was the only time-consuming step for all models. It was also observed that the computation time of our proposed model was larger than ANN-Center and RANN-LU, as the additional weight of interval-valued data was estimated simultaneously during the optimization.
5. Application to Real Data
5.1. Capital Asset Pricing Model: Thai Stocks
The performance of this ANN-CC model was assessed using interval-valued returns (maximum and minimum returns) from the Stock Exchange of Thailand. In this study, we compared the forecasting accuracy of several ANN models under the context of the capital asset pricing model (CAPM). Our analysis employed lower and upper bounds of the real daily stock returns, including SET50, PTT, SCC, and CPALL, over the period from 26 October 2011, to 7 May 2019. These three companies were selected for this study because they are considered well-performing companies in terms of their share prices. Moreover, they also experienced large trade volumes in the current decade and are regarded as fast-growing and highly volatile in the Thai stock market. All data were collected from Thomson Reuters DataStream. The criteria to choose the best fit model were MAE, MSE, and RMSE.
CAPM was proposed in separate studies by Sharpe [31] and Lintner [32] to measure the risk of the individual stock against the market in terms of the beta risk . The is a measure of a stock’s risk (volatility of returns) reflected by measuring the fluctuation of its price changes relative to the overall market. In other words, it is the stock’s sensitivity to market risk. The model can be written as
where is the return of the stock , is the return of the market, is risk-free, is error term at time . If , the stock is called aggressive stock (high risk), otherwise defensive stock (low risk) of excess stock return. In this study, we preserve the interval format of the stock as
where , and are maximum, minimum, and average prices of the individual stock at time , respectively. Note that the risk-free rate and , in this empirical study, is assumed to be zero.
Again, we preserve the interval format of the market return as
where , and are maximum, minimum, and average prices of the interval of the market at time , respectively.
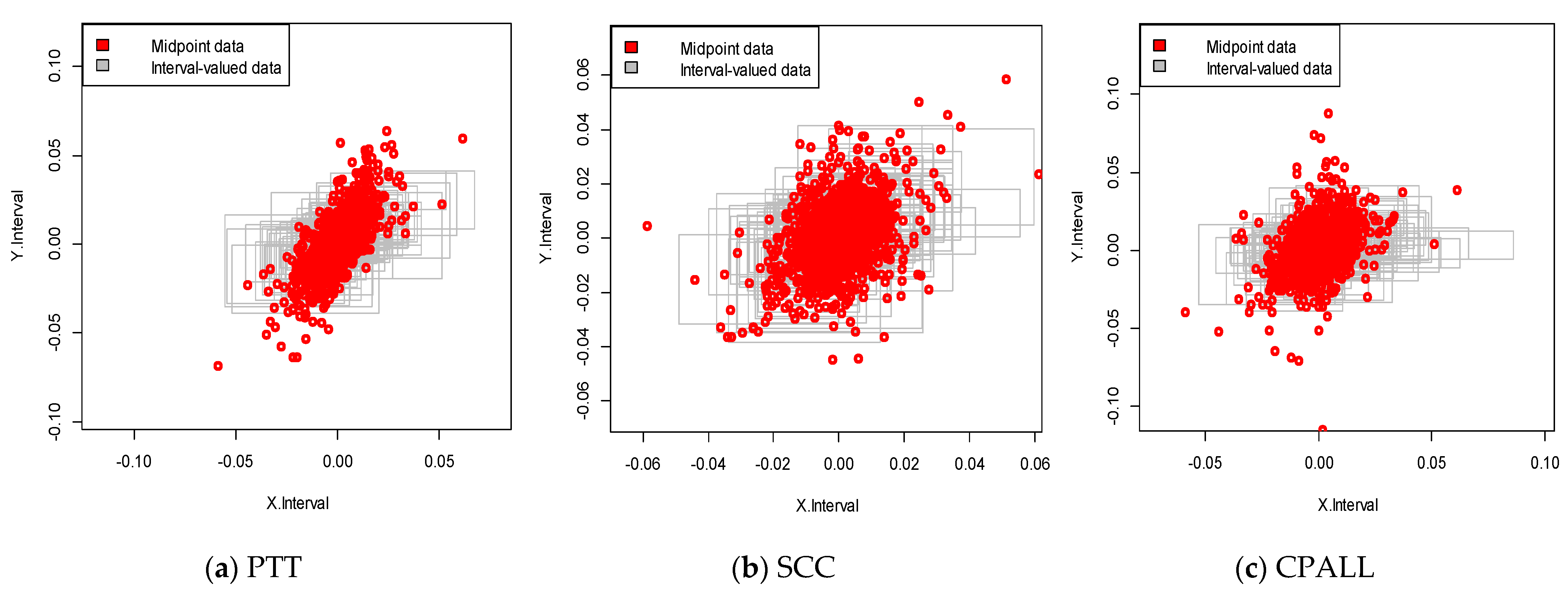
We constructed a deep neural network using stock returns from the Stock Exchange of Thailand (SET), the major stock market in Thailand. We have selected three major stocks with high market capitalization at the beginning of the sample period. We collected SET50 (proxy of the market) and three company stocks, PTT, SCC, and CPALL, over the period from 4 January 2012 to 30 December 2019. The interval-valued data were constructed from a daily range of selected price indexes, i.e., the lowest and highest trading index values for the day were calculated to define the movement on the market for that day. Then, the construction of our interval-valued prediction for these three stocks would be made based on the models developed in Section 3. We note that all the interval price series were transformed to be interval returns following Equations (34) and (35). The descriptive statistics, namely the mean, standard deviation, minimum value, and maximum value of the variables for the full sample, are summarized in Table 3. We observe that the returns exhibited negative skewness for lower bound returns and positive for upper bound returns. In addition, the values of skewness on both sides were asymmetric, indicating that the gain and the loss in the Thai stock market were quite different. For illustration, a visual relationship between the stock market and each stock is shown in Figure 4.

Table 3.
Data description.
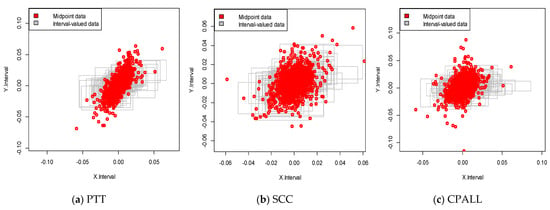
Figure 4.
Interval-valued data plots for Stock Exchange of Thailand (SET) and stocks. The red dot presents the midpoint data plot for SET and stocks.
Moreover, a unit root test was also conducted to examine whether a time series variable was nonstationary and possessed a unit root. In this study, we used the minimum Bayes factor (MBF) as a tool for making a statistical test. The MBF has a significant advantage over the p-value because the likelihood of the observed data can be expressed under each hypothesis [33]. If 1 < MBF < 1/3, 1/3 < MBF < 1/10, 1/10 < MBF < 1/30, 1/30 < MBF < 1/100, 1/100 < MBF < 1/300 and MBF < 1/300, there is a chance that the MBF favors, respectively, the weak evidence, moderate evidence, substantial evidence, strong evidence, very strong evidence, and decisive evidence for the null hypothesis. According to the results, all data series were decisive stationary, as shown by the low MBF values [33].
5.2. Comparison Results
This section shows the results of the models representing artificial neural networks for interval-valued data of Thailand’s stock market. In this empirical example, we consider the number of hidden neurons between one and three hidden neurons with four activation functions. Again, one input layer, one output layer, and one hidden layer were assumed. Thus, twelve ANN specifications were used to describe and forecast the excess return of PTT, SCC, and CPALL stock returns under the CAPM context. To evaluate the forecasting performance of the twelve ANN-class specifications presented in this section, the forecasting process was handled as follows: Each dataset was split into 80% for training and 20% for testing. Thus, both in-sample and out-of-sample forecasts were conducted in this comparison. The performance evaluation of the interval-valued data forecasting models ANN-CC under CAPM was accomplished through MAE, MSE, and RMSE.
Table 4 shows the in-sample and out-of-sample estimation results for different ANN-CC specifications. Focusing on the MAE, MSE, and RMSE, the result indicated that the interval-valued prediction results were a bit sensitive to activation functions. These activation functions were compared, and we found that the exponential activation function enabled more accurate forecasting in most cases, as the MAE, MSE, and RMSE of this activation were lower than of other activation functions. When the number of hidden nodes considered in this comparison was compared, we observed that the higher the number of hidden nodes, the lower MAE, MSE, and RMSE in some cases. Finally, we considered the weight of interval-valued data, which was calculated by the convex combination method. The results show that most of the weight parameters for interval excess stock return and interval excess market return were not equal to 0.5. Therefore, it can be said that the assumption of the center method in the neural network forecasting of Maia et al. [25] may be inappropriate for practical problems. This result confirms the reliability of the convex combination method in ANN-CC forecasting.

Table 4.
Experimental results of ANN-CC.
Furthermore, we also compared the performance of the ANN-CC models with the traditional ANN models, ANN-Center (Table 5) and RANN-LU (Table 6). Table 5 and Table 6 also provide the MAE, MSE, and RMSE of ANN-Center and RANN-LU, respectively. We note that four activations and a different number of hidden neurons were compared, and we also found that the exponential activation function provided higher performance in most cases. Furthermore, the prediction error decreased for a larger number of hidden neurons because the MAE, MSE, and RMSE of the ANN-Center and RANN-LU models were lower when the number of hidden neurons increased.

Table 5.
Experimental results of ANN-Center.

Table 6.
Experimental results of regularized ANN-LU (RANN-LU).
To make a clear explanation, the best prediction models for the three stocks are summarized in Table 7. We found that the exponential activation function was mostly selected (except for ANN-Center in the PTT case). This indicates a nonlinear pattern of these three stock companies. It is evident that the performance measures of the best specification of ANN-CC models were lower than those of the ANN-Center and RANN-LU for all stock indices, meaning that the ANN-CC model was superior to the conventional models in both in- and out-of-sample forecast performances. Additionally, in this section, we also compared the performance of our ANN-CC against the methods proposed in the literature, namely, center and center-range methods of Billard and Diday [7], PM method of Souza et al. [9], and IT method of Buansing et al. [12]. The result clearly confirmed the higher prediction performance of our proposed ANN-Center.

Table 7.
Summary of the forecasting performance and model confidence set (MCS) test of the first dataset.
A robustness check was also conducted to confirm the performance of our proposed model. As in practice, the simple loss functions, such as MAE, MSE, and RMSE, may not yield a piece of sufficient information to identify a single forecasting model as “best”. Therefore, in this study, another accuracy measure, the model confidence set (MCS), was used to evaluate forecasting performance. Hansen et al. [34] introduced the MCS test to validate the forecasting performance of forecasting models. In this study, our MCS tests were based on two loss functions, MAE, MSE, and RMSE. We note that when the p-value was higher, it meant that they were more likely to reject the null hypothesis of equal predictive ability. In other words, the greater the p-value, the better the model. For more details of the MCS test, we referred to Hansen et al. [34].
The statistical performance results for all competing models are provided in the bracket shown in Table 7. According to the MCS test results, the ANN-CC model clearly outperformed other competing models for both in- and out-of-sample forecasts. The p-values of the ANN-CC model were equal to one, but the other three models were less than the 0.10 threshold. It means that ANN-Center, RNN-LU, and linear regression forecasting models were removed in the MCS inspection process, and thus the ANN-CC was the only survivor model.
5.3. Hong Kong Air Quality Monitoring Dataset
In the second dataset, we considered the Hong Kong air quality monitoring dataset as another example. This dataset is suggested in Yang et al. [27] and can be retrieved from http://www.epd.gov.hk (accessed on 20 February 2021). They provide hourly air quality data of 16 monitoring stations in Hong Kong. In this study, we also considered the data from Central/Western Station and downloaded the hourly data ranging from 1 January 2020, to 31 December 2020. Then, we aggregated the hourly data to the minimum and maximum form according to each day’s record. There were seven air quality indicators in the database. However, we considered some of them and selected the respirable suspended particulates (RSP) as the interval-valued response variable and selected dinitrogen tetroxide (NO2) and sulfur dioxide (SO2) as the interval-valued explanatory variables.
Again, each dataset was split into 80% for training and 20% for testing. Thus, both in-sample and out-of-sample forecasts were conducted in this comparison. The performance evaluation of the interval-valued data forecasting models ANN-CC under CAPM was accomplished through MAE, MSE, and RMSE. As shown in Table 8, similar to the first example, our proposed ANN-CC model was the best in terms of MAE, MSE, RMSE, and MCS’s p-value.

Table 8.
Summary of the forecasting performance and MCS test of the second dataset.
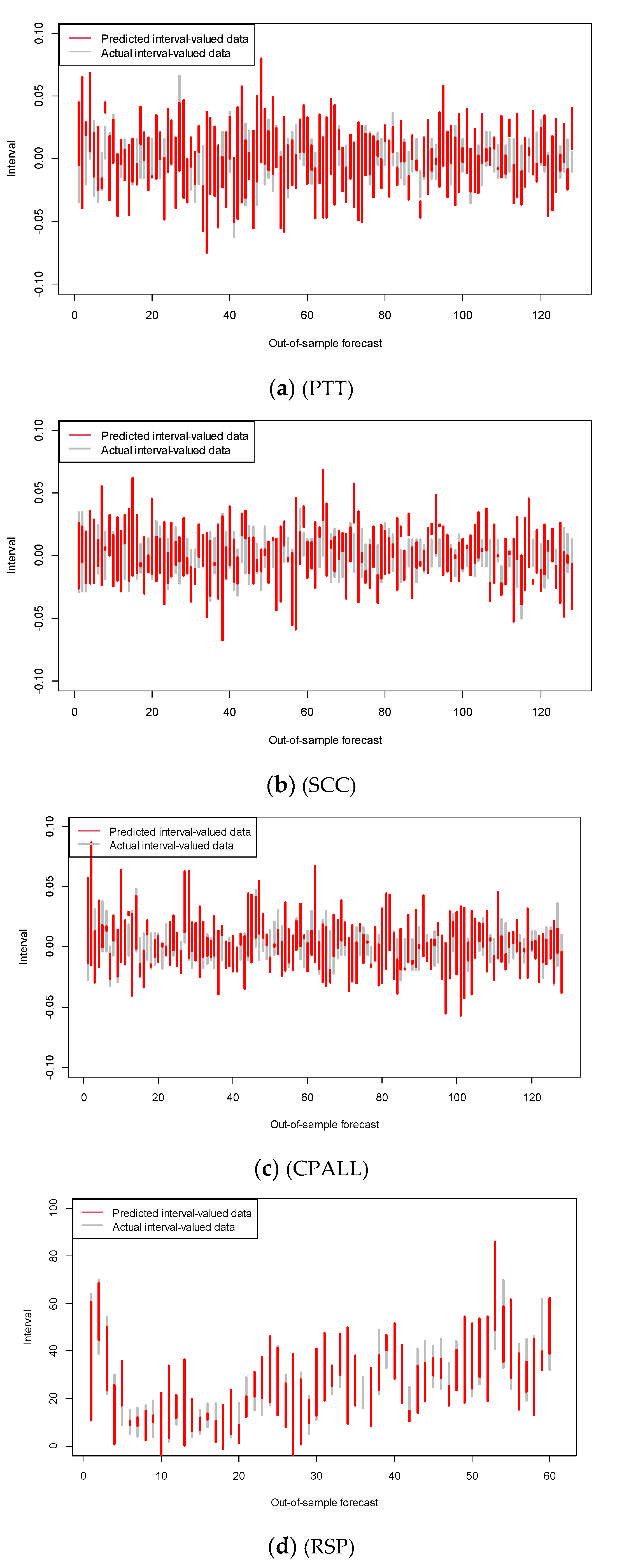
To illustrate the performance of our ANN-CC models, we show the out-of-sample forecasting result of our model on the Thai stocks and RSP in Figure 5. For clarity, only 10% of the out-of-sample forecasting result are shown. In this figure, the red vertical line segment represents a predicted interval-valued data, while the gray vertical line segment represents a predicted interval-valued data; the extremes correspond to the minimum and maximum interval values. The comparison between actual values and predicted values indicates the quality of modeling and the prediction task. The results show that the predicted values were very close to the actual values, indicating the goodness of fit of our model.
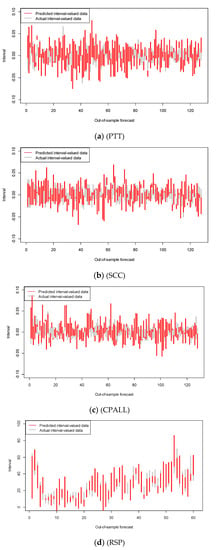
Figure 5.
The out of sample interval forecasts by ANN-CC (red) vs. the actual data (gray) for example data.
5.4. Discussion
Although the traditional models like ANN-Center and RANN-LU provide acceptable prediction results in all stocks and RSP, they still face some limitations. The ANN-Center relies on the midpoint of the data, which may not reflect real behavior of the data during the day. Likewise, the RANN-LU predicts future observation based on either lower or upper bounds, separately, clearly leaving out the information within the interval-value data. Thus, our proposed ANN-CC model is used to solve such a challenging task and consider the whole information during the interval. According to the above experimental results, we can draw the following conclusions:
- (1)
- Regarding the prediction performance, our ANN-CC was superior to other traditional models in all datasets.
- (2)
- We note that the symmetric weight within the interval data should not be . We found that the prediction result was sensitive to the weight ; thus, the weight should not be a fixed parameter.
- (3)
- Our model outperformed the ANN-LU and RANN-LU models in situations in which the interval series had linear and nonlinear behavior.
- (4)
- We also studied the sensitivity of each activation function and found that the quality of the prediction model was not very sensitive in many cases. However, careful assessment needs to be made when choosing the activation function.
- (5)
- Even though the exponential activation function seemed to be the best fit one in the ANN architecture, it was noticed that other activation functions performed well in some cases. Although the exponential activation function performed very well in the selected three stocks, it may not be reliable in other stocks or under other ANN structures.
- (6)
- However, we can draw an important conclusion that our ANN-CC is a promising model for interval-valued data forecasting. The ANN-CC method has the advantages of not assuming constraints for the weight nor fixing reference points. The ANN-CC model is adaptive and adjusts itself for the best fit. The fitted model allows the behavior analysis of response lower and upper bounds based on the variation of the reference points of input and output intervals.
6. Conclusions
This paper proposed an artificial neural network with a convex combination (ANN-CC) method for interval-valued data prediction. Simulation and experimental results on real data showed that the proposed ANN-CC model is a useful tool in interval-valued prediction tasks, especially for complicated nonlinear datasets. Moreover, the proposed ANN-CC model fills the research gap by considering interval-valued data using the convex combination method. Our proposed model was examined by comparing its performance with conventional ANN with the center method (ANN-Center) and regularized ANN-LU (RANN-LU), linear regression with the center method, center-range, PM, and IT methods. We considered three stock returns in the Thai stock market and Hong Kong air quality monitoring dataset in our empirical comparison. According to the in-sample and out-of-sample forecasts, we found that the performances of various ANN-CC specifications were not much different. However, we observed that the tanh activation function performed well in the in-sample and out-sample forecast, while the linear activation function performed relatively well in the in-sample forecast. In addition, we could also confirm the higher performance of our ANN-CC compared to that of the ANN-Center and RANN-LU. Experimental results on two real datasets also confirmed that the proposed ANN-CC model outperformed the conventional models.
In this study, a neural network with one hidden layer was assumed. However, real-world data is quite complex, and one hidden layer may not be enough to learn the data. Thus, a deep neural network (more than two hidden layers) should be more promising in the approximation performance. Another meaningful future work is to employ other deep learning methods, such as recurrent neural networks and long short-term memory. These models handle incoming data in time order and learn about the previous time to predict future value. In addition to deep learning methods, the fuzzy inference system (FIS) modeling approach for interval-valued time series forecasting [1] is also suggested for further study. Finally, our proposed model can be applied for forecasting in other areas such as environmental and medical sciences.
Author Contributions
Conceptualization, R.P. and W.Y.; methodology, W.Y.; software, W.Y.; validation, R.P., P.M. and W.Y.; formal analysis, R.P.; investigation, W.Y.; resources, P.M.; data curation, P.M.; writing—original draft preparation, W.Y.; writing—review and editing, W.Y.; visualization, R.P.; supervision, W.Y. and P.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Center of Excellence in Econometrics, Faculty of Economics, Chiang Mai University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
In this study, we used simulated data to show the performance of our model, and the simulation processes are already explained in the paper. For the real data analysis section, the data can be freely collected from Thomson Reuter DataStream. However, the data are available from the author upon request (woraphon.econ@gmail.com (accessed on 27 April 2021)).
Acknowledgments
The authors would like to thank Laxmi Worachai, Vladik Kreinovich, and Hung T. Nguyen for their helpful comments on this paper. The authors are also grateful for the financial support offered by the Center of Excellence in Econometrics, Chiang Mai University, Thailand.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Maciel, L.; Ballini, R. A fuzzy inference system modeling approach for interval-valued symbolic data forecasting. Knowl. Based Syst. 2019, 164, 139–149. [Google Scholar] [CrossRef]
- Ma, X.; Dong, Y. An estimating combination method for interval forecasting of electrical load time series. Expert Syst. Appl. 2020, 158, 113498. [Google Scholar] [CrossRef]
- Chou, J.S.; Truong, D.N.; Le, T.L. Interval forecasting of financial time series by accelerated particle swarm-optimized multi-output machine learning system. IEEE Access 2020, 8, 14798–14808. [Google Scholar] [CrossRef]
- Lauro, C.N.; Palumbo, F. Principal component analysis of interval data: A symbolic data analysis approach. Comput. Stat. 2000, 15, 73–87. [Google Scholar] [CrossRef]
- Branzei, R.; Branzei, O.; Gök, S.Z.A.; Tijs, S. Cooperative interval games: A survey. Cent. Eur. J. Oper. Res. 2010, 18, 397–411. [Google Scholar] [CrossRef]
- Kiekintveld, C.; Islam, T.; Kreinovich, V. Security games with interval uncertainty. In Proceedings of the 2013 International Conference on Autonomous Agents and Multi-Agent Systems, Paul, MN, USA, 6–10 May 2013; pp. 231–238. [Google Scholar]
- Billard, L.; Diday, E. Regression analysis for interval-valued data. In Data Analysis, Classification, and Related Methods; Springer: Berlin/Heidelberg, Germany, 2000; pp. 369–374. [Google Scholar]
- Neto, E.D.A.L.; de Carvalho, F.D.A. Centre and range method for fitting a linear regression model to symbolic interval data. Comput. Stat. Data Anal. 2008, 52, 1500–1515. [Google Scholar] [CrossRef]
- Souza, L.C.; Souza, R.M.; Amaral, G.J.; Silva Filho, T.M. A parametrized approach for linear regression of interval data. Knowl. Based Syst. 2017, 131, 149–159. [Google Scholar] [CrossRef]
- Chanaim, S.; Sriboonchitta, S.; Rungruang, C. A Convex Combination Method for Linear Regression with Interval Data. In Integrated Uncertainty in Knowledge Modelling and Decision Making. IUKM 2016; Huynh, V.N., Inuiguchi, M., Le, B., Le, B., Denoeux, T., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2016; Volume 9978, pp. 469–480. [Google Scholar] [CrossRef]
- Phadkantha, R.; Yamaka, W.; Tansuchat, R. Analysis of Risk, Rate of Return and Dependency of REITs in ASIA with Capital Asset Pricing Model. In Predictive Econometrics and Big Data. TES 2018; Kreinovich, V., Sriboonchitta, S., Chakpitak, N., Eds.; Studies in Computational Intelligence; Springer: Cham, Switzerland, 2018; Volume 753, pp. 536–548. [Google Scholar] [CrossRef]
- Buansing, T.T.; Golan, A.; Ullah, A. An information-theoretic approach for forecasting interval-valued SP500 daily returns. Int. J. Forecast. 2020, 36, 800–813. [Google Scholar] [CrossRef]
- Zhang, G.P. Time series forecasting using a hybrid ARIMA and neural network model. Neurocomputing 2003, 50, 159–175. [Google Scholar] [CrossRef]
- Mondal, P.; Shit, L.; Goswami, S. Study of effectiveness of time series modeling (ARIMA) in forecasting stock prices. Int. J. Comput. Sci. Eng. Appl. 2014, 4, 13. [Google Scholar] [CrossRef]
- McMillan, D.G. Nonlinear predictability of stock market returns: Evidence from nonparametric and threshold models. Int. Rev. Econ. Financ. 2001, 10, 353–368. [Google Scholar] [CrossRef]
- Nyberg, H. Predicting bear and bull stock markets with dynamic binary time series models. J. Bank. Financ. 2013, 37, 3351–3363. [Google Scholar] [CrossRef]
- Pastpipatkul, P.; Maneejuk, P.; Sriboonchitta, S. Markov Switching Regression with Interval Data: Application to Financial Risk via CAPM. Adv. Sci. Lett. 2017, 23, 10794–10798. [Google Scholar] [CrossRef]
- Phochanachan, P.; Pastpipatkul, P.; Yamaka, W.; Sriboonchitta, S. Threshold regression for modeling symbolic interval data. Int. J. Appl. Bus. Econ. Res. 2017, 15, 195–207. [Google Scholar]
- Hiransha, M.; Gopalakrishnan, E.A.; Menon, V.K.; Soman, K.P. NSE stock market prediction using deep-learning models. Procedia Comput. Sci. 2018, 132, 1351–1362. [Google Scholar]
- Haykin, S.; Principe, J. Making sense of a complex world [chaotic events modeling]. IEEE Signal Process. Mag. 1998, 15, 66–81. [Google Scholar] [CrossRef]
- Zhang, G.; Patuwo, B.E.; Hu, M.Y. Forecasting with artificial neural networks: The state of the art. Int. J. Forecast. 1998, 14, 35–62. [Google Scholar] [CrossRef]
- Leung, D.; Abbenante, G.; Fairlie, D.P. Protease inhibitors: Current status and future prospects. J. Med. Chem. 2000, 43, 305–341. [Google Scholar] [CrossRef]
- Cao, Q.; Leggio, K.B.; Schniederjans, M.J. A comparison between Fama and French′s model and artificial neural networks in predicting the Chinese stock market. Comput. Oper. Res. 2005, 32, 2499–2512. [Google Scholar] [CrossRef]
- San Roque, A.M.; Maté, C.; Arroyo, J.; Sarabia, Á. iMLP: Applying multi-layer perceptrons to interval-valued data. Neural Process. Lett. 2007, 25, 157–169. [Google Scholar] [CrossRef]
- Maia, A.L.S.; de Carvalho, F.D.A.; Ludermir, T.B. Forecasting models for interval-valued time series. Neurocomputing 2008, 71, 3344–3352. [Google Scholar] [CrossRef]
- Maia, A.L.S.; de Carvalho, F.D.A. Holt’s exponential smoothing and neural network models for forecasting interval-valued time series. Int. J. Forecast. 2011, 27, 740–759. [Google Scholar] [CrossRef]
- Yang, Z.; Lin, D.K.; Zhang, A. Interval-valued data prediction via regularized artificial neural network. Neurocomputing 2019, 331, 336–345. [Google Scholar] [CrossRef]
- Mir, M.; Nasirzadeh, F.; Kabir, H.D.; Khosravi, A. Neural Network-based interval forecasting of construction material prices. J. Build. Eng. 2021, 39, 102288. [Google Scholar]
- Moore, R.E. Methods and Applications of Interval Analysis; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 1979. [Google Scholar]
- Humphrey, G.B.; Maier, H.R.; Wu, W.; Mount, N.J.; Dandy, G.C.; Abrahart, R.J.; Dawson, C.W. Improved validation framework and R-package for artificial neural network models. Environ. Model. Softw. 2017, 92, 82–106. [Google Scholar] [CrossRef]
- Sharpe, W.F. Capital asset prices: A theory of market equilibrium under conditions of risk. J. Financ. 1964, 19, 425–442. [Google Scholar]
- Lintner, J. Security prices, risk, and maximal gains from diversification. J. Financ. 1965, 20, 587–615. [Google Scholar]
- Maneejuk, P.; Yamaka, W. Significance test for linear regression: How to test without P-values? J. Appl. Stat. 2021, 48, 827–845. [Google Scholar] [CrossRef]
- Hansen, P.R.; Lunde, A.; Nason, J.M. The model confidence set. Econometrica 2011, 79, 453–497. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).