Micro-Computed Tomography Soft Tissue Biological Specimens Image Data Visualization
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials, Devices, and Software
2.2. Methods
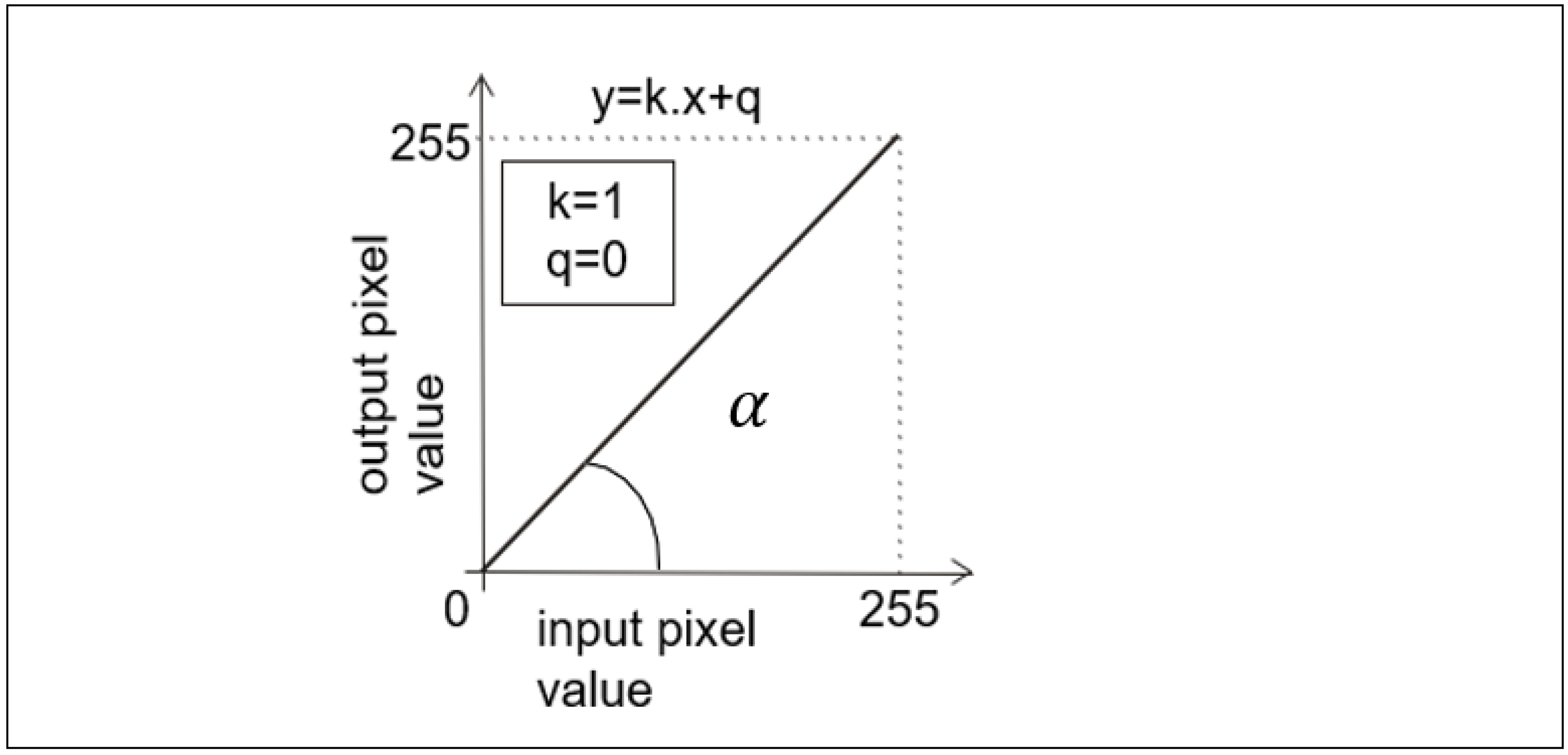
2.2.1. Transfer Functions, Resp. Look-up-Table Concept
2.2.2. Transfer Functions Related to the Hounsfield Units (HU)
2.2.3. Transfer Functions Design Possibilities
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Ethanol


Appendix A.2. Aurovist





Appendix A.3. Omnipaque




Appendix A.4. Potassium iodide (KI) in 20% Ethanol Solution





Appendix A.5. Mercox (Resin)


Appendix B
| Parameter | Value | Physical Unit |
|---|---|---|
| object-source distance | 310 | mm |
| detector-source distance | 360 | mm |
| anode voltage | 35 | kV |
| emission current | 1 | mA |
| pixel projection dimension | 96 | μm |
| exposure time | 500 | ms |
| degree of rotation (increm.) | 1.8 | |
| binning 1 | 500 × 500 | pixels |
| Parameter | Value 50E5D | Value 100E5D | Value F5D | Value 50E6D | Value 80E6D | Value 100E6D | Physical Unit |
|---|---|---|---|---|---|---|---|
| object-source distance | 40.97 | 40.97 | 40.97 | 40.97 | 40.97 | 40.97 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 50 | 50 | 50 | 50 | 50 | 50 | kV |
| emission current | 200 | 200 | 200 | 200 | 200 | 200 | μA |
| pixel projection dimension | 11.6 | 11.6 | 11.6 | 11.6 | 11.6 | 11.6 | μm |
| exposure time | 50 | 50 | 50 | 50 | 50 | 50 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value F6D | Value 80E7D | Value 100E7D | Value 80E8D | Value 80E9D | Value A80E10D | Physical Unit |
|---|---|---|---|---|---|---|---|
| object-source distance | 40.97 | 48.39 | 48.39 | 57.22 | 71.47 | 92.67 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 50 | 50 | 50 | 65 | 70 | 100 | kV |
| emission current | 200 | 200 | 200 | 153 | 142 | 100 | μA |
| pixel projection dimension | 11.6 | 13.7 | 13.7 | 16.2 | 20.24 | 26.24 | μm |
| exposure time | 50 | 50 | 50 | 36 | 36 | 35 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | ° |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value A80E10D | Value 80E10-11D | Value 80E10-11D | Value 50E11D | Value 80E12D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 71.47 | 60.75 | 67.9 | 92.67 | 80.4 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 75 | 75 | 75 | 75 | 75 | kV |
| emission current | 133 | 133 | 133 | 133 | 133 | μA |
| pixel projection dimension | 20.24 | 17.2 | 19.23 | 26.24 | 22.77 | μm |
| exposure time | 36 | 36 | 36 | 36 | 500 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | ° |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value A80E12D | Value A80E12D | Value 50E13D | Value 50E14D | Value 100E14D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 92.67 | 82.19 | 92.67 | 96.48 | 123.29 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 75 | 75 | 75 | 75 | 80 | kV |
| emission current | 133 | 133 | 133 | 133 | 125 | μA |
| pixel projection dimension | 26.24 | 23.27 | 26.24 | 27.32 | 34.91 | μm |
| exposure time | 500 | 500 | 500 | 500 | 32 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 50E15D | Value 50E15D | Value 50E16D | Value 100E18D | Value 100E18D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 146.51 | 142.94 | 178.67 | 169.74 | 176.89 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 80 | 80 | 80 | 80 | 80 | kV |
| emission current | 125 | 125 | 125 | 125 | 125 | μA |
| pixel projection dimension | 41.48 | 40.47 | 50.59 | 48.06 | 50.08 | μm |
| exposure time | 32 | 32 | 32 | 33 | 33 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | ° |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 100E19D | Value 100E19D | Value 100E19D | Value 100E19D | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 173.31 | 132.22 | 92.67 | 141.15 | mm |
| detector-source distance | 287 | 286 | 286 | 286 | mm |
| anode voltage | 40 | 80 | 100 | 100 | kV |
| emission current | 250 | 125 | 100 | 100 | μA |
| pixel projection dimension | 49.07 | 37.44 | 26.24 | 39.97 | μm |
| exposure time | 35 | 35 | 35 | 35 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 8D | Value 9D | Value 10D | Value 11D | Value 12D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 155.45 | 146.51 | 151.87 | 137.58 | 153.66 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 80 | 80 | 80 | 80 | 80 | kV |
| emission current | 125 | 125 | 125 | 125 | 125 | μA |
| pixel projection dimension | 44.01 | 41.48 | 43 | 38.95 | 43.51 | μm |
| exposure time | 36 | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 13D | Value 14D | Value 15D | Value 15D | Value 16D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 146.51 | 148.30 | 155.45 | 158.93 | 144.73 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 80 | 80 | 80 | 80 | 80 | kV |
| emission current | 125 | 125 | 125 | 125 | 125 | μA |
| pixel projection dimension | 41.48 | 41.49 | 44.01 | 45 | 40.98 | μm |
| exposure time | 36 | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 17D | Value 18D | Value 18D | Value 19D | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 159.02 | 157.23 | 167.95 | 150.09 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | mm |
| anode voltage | 95 | 80 | 80 | 80 | kV |
| emission current | 105 | 125 | 125 | 125 | μA |
| pixel projection dimension | 45.03 | 44.52 | 47.55 | 42.50 | μm |
| exposure time | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 50E8D | Value 50E17D | Value 100E8D | Value 100E16D | Value 100E37D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 63.57 | 56.51 | 54.74 | 52.98 | 49.45 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 30 | 40 | 30 | 35 | 40 | kV |
| emission current | 212 | 250 | 212 | 231 | 250 | μA |
| pixel projection dimension | 18.00 | 16.00 | 15.50 | 15.00 | 14.00 | μm |
| exposure time | 58 | 35 | 58 | 38 | 35 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 50E7D | Value 50E16D | Value 100E7D | Value 100E16D | Value 100E37D | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 28.26 | 28.26 | 30.02 | 28.26 | 28.26 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 35 | 35 | 35 | 35 | 40 | kV |
| emission current | 231 | 231 | 231 | 231 | 250 | μA |
| pixel projection dimension | 8.00 | 8.00 | 8.50 | 8.00 | 8.00 | μm |
| exposure time | 35 | 38 | 35 | 38 | 35 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 17D | Value 18D | Value 18D | Value 19D | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 159.02 | 157.23 | 167.95 | 150.09 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | mm |
| anode voltage | 95 | 80 | 80 | 80 | kV |
| emission current | 105 | 125 | 125 | 125 | μA |
| pixel projection dimension | 45.03 | 44.52 | 47.55 | 42.50 | μm |
| exposure time | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 17D | Value 18D | Value 18D | Value 19D | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 159.02 | 157.23 | 167.95 | 150.09 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | mm |
| anode voltage | 95 | 80 | 80 | 80 | kV |
| emission current | 105 | 125 | 125 | 125 | μA |
| pixel projection dimension | 45.03 | 44.52 | 47.55 | 42.50 | μm |
| exposure time | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | ° |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 17D | Value 18D | Value 18D | Value 19D | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 159.02 | 157.23 | 167.95 | 150.09 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | mm |
| anode voltage | 95 | 80 | 80 | 80 | kV |
| emission current | 105 | 125 | 125 | 125 | μA |
| pixel projection dimension | 45.03 | 44.52 | 47.55 | 42.50 | μm |
| exposure time | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value 17D | Value 18D | Value 18D | Value 19D | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 159.02 | 157.23 | 167.95 | 150.09 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | mm |
| anode voltage | 95 | 80 | 80 | 80 | kV |
| emission current | 105 | 125 | 125 | 125 | μA |
| pixel projection dimension | 45.03 | 44.52 | 47.55 | 42.50 | μm |
| exposure time | 36 | 36 | 36 | 36 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | ° |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value A-Mouse | Value M-Rabbit | Value M-Rat | Physical Unit |
|---|---|---|---|---|
| object-source distance | 92.67 | 148.30 | 62.54 | mm |
| detector-source distance | 286 | 286 | 286 | mm |
| anode voltage | 42 | 46 | 34 | kV |
| emission current | 238 | 217 | 227 | μA |
| pixel projection dimension | 26.24 | 41.99 | 17.71 | μm |
| exposure time | 99 | 35 | 41 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | ° |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
| Parameter | Value O-Liver | Value O-Lungs | Value O-Spleen | Value O-Heart | Physical Unit |
|---|---|---|---|---|---|
| object-source distance | 58.96 | 33.95 | 28.59 | 33.95 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | mm |
| anode voltage | 40 | 54 | 54 | 40 | kV |
| emission current | 238 | 185 | 185 | 238 | μA |
| pixel projection dimension | 16.70 | 19.22 | 16.19 | 9.61 | μm |
| exposure time | 35 | 50 | 50 | 35 | ms |
| degree of rotation (increm.) | 0.2 | 0.4 | 0.4 | 0.2 | |
| binning 1 | 1 × 1 | 2 × 2 | 2 × 2 | 1 × 1 | pixels |
| Parameter | Value KI-Liver | Value KI-Kidney | Value KI-Brain | Value KI-Heart | Value KI-Leg | Physical Unit |
|---|---|---|---|---|---|---|
| object-source distance | 107.21 | 32.16 | 117.93 | 39.31 | 178.67 | mm |
| detector-source distance | 286 | 286 | 286 | 286 | 286 | mm |
| anode voltage | 80 | 80 | 80 | 80 | 80 | kV |
| emission current | 125 | 125 | 125 | 125 | 125 | μA |
| pixel projection dimension | 30.36 | 9.11 | 33.39 | 11.13 | 50.59 | μm |
| exposure time | 35 | 35 | 35 | 35 | 35 | ms |
| degree of rotation (increm.) | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| binning 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | 1 × 1 | pixel |
References
- Liguori, C.; Frauenfelder, G.; Massaroni, C.; Saccomandi, P.; Giurazza, F.; Pitocco, F.; Marano, R.; Schena, E. Emerging clinical applications of computed tomography. Med. Devices 2015, 8, 265–278. [Google Scholar] [CrossRef]
- Stock, S.R. Microcomputed Tomography: Methodology and Applications, 2nd ed.; CRC Press/Taylor and Francis: Boca Raton, FL, USA, 2020. [Google Scholar]
- Vickerton, P.; Jarvis, J.; Jeffery, N. Concentration-dependent specimen shrinkage in iodine-enhanced microCT. J. Anat. 2013, 223, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.D.E.; Zanette, I.; Noel, P.B.; Cardoso, M.B.; Kimm, M.A.; Pfeiffer, F. Three-dimensional non-destructive soft-tissue visualization with X-ray staining micro-tomography. Sci. Rep. 2015, 5, 14088. [Google Scholar] [CrossRef] [PubMed]
- Metscher, B.D. MicroCT for comparative morphology: Simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 2009, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Patzelt, M.; Mrzilkova, J.; Dudak, J.; Krejci, F.; Zemlicka, J.; Karch, J.; Musil, V.; Rosina, J.; Sykora, V.; Horehledova, B.; et al. Ethanol fixation method for heart and lung imaging in micro-CT. Jpn. J. Radiol. 2019, 37, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Dudak, J.; Zemlicka, J.; Karch, J.; Patzelt, M.; Mrzilkova, J.; Zach, P.; Hermanova, Z.; Kvacek, J.; Krejci, F. High-contrast X-ray micro-radiography and micro-CT of ex-vivo soft tissue murine organs utilizing ethanol fixation and large area photon-counting detector. Sci. Rep. 2016, 6, 30385. [Google Scholar] [CrossRef] [PubMed]
- McNitt-Gray, M.F.; Taira, R.K.; Johnson, S.L.; Razavi, M. An automatic method for enhancing the display of different tissue densities in digital chest radiographs. J. Digit. Imaging 1993, 6, 95–104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seeram, E.; Seeram, D. Image Postprocessing in Digital Radiology—A Primer for Technologists. J. Med. Imaging Radiat. Sci. 2008, 39, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Alshipli, M.; Kabir, N.A.; Hashim, R.; Marashdeh, M.W.; Tajuddin, A.A. Measurement of attenuation coefficients and CT numbers of epoxy resin and epoxy-based Rhizophora spp. particleboards in computed tomography energy range. Radiat. Phys. Chem. 2018, 149, 41–48. [Google Scholar] [CrossRef]
- Phywe XR 4.0 Expert Unit, X-ray Unit, 35 kV: Germany: PHYWE Systeme GmbH. Available online: https://repository.curriculab.net/files/bedanl.pdf/09057.99/0905799e.pdf (accessed on 18 July 2021).
- XR 4.0 Software Measure CT Manual. PHYWE. Available online: https://repository.curriculab.net/files/bedanl.pdf/14421.61/1442161e.pdf (accessed on 18 July 2021).
- CTvox Quick Start Guide. Bruker. Available online: https://www.bruker.com/fileadmin/user_upload/3-Service/Support/Preclinical_Imaging/X-Ray-uCT_Software/CTvox_QuickStartGuide.pdf (accessed on 18 July 2021).
- Key Features of Dragonfly. Available online: https://www.theobjects.com/dragonfly/features.html (accessed on 18 July 2021).
- NRecon User Manual. Available online: https://umanitoba.ca/faculties/health_sciences/medicine/units/cacs/sam/media/NReconUserManual.pdf (accessed on 18 July 2021).
- MIPS (Medical/Microscopy Image Processing Software). Available online: http://webzam.fbmi.cvut.cz/hozman/ (accessed on 20 February 2022).
- DICOM PS3.3 2021e—Information Object Definitions. C.11 LUT and Presentation States. Available online: https://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.11.2.htm (accessed on 28 November 2021).
- Weiss, S.; Westermann, R. Differentiable Direct Volume Rendering. IEEE Trans. Vis. Comput. Graph. 2022, 28, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Skyscan CT-Volume Manual. Available online: https://umanitoba.ca/faculties/health_sciences/medicine/units/cacs/sam/media/CTVol_UserManual.pdf (accessed on 18 July 2021).
- Wolfram Research, Inc. Centroid of triangle–Wolfram Alpha. Wolfram Research, Inc., Champaign, IL. Available online: https://www.wolframalpha.com/input?i=centroid+of+triangle (accessed on 6 April 2022).
- Stakat. Let’s Master Statistics Together. Spearman’s Rho. Available online: https://statkat.com/stat-tests/spearmans-rho.php#5 (accessed on 25 April 2022).
- Real Statistics Using Excel. Spearman’s Rank Correlation Hypothesis Testing. Available online: https://www.real-statistics.com/correlation/spearmans-rank-correlation/spearmans-rank-correlation-detailed/ (accessed on 25 April 2022).
- Gaspar, B.; Mrzilkova, J.; Hozman, J.; Zach, P.; Lahutsina, A.; Morozova, A.; Guarnieri, G.; Riedlova, J. Data Availability Statement: There Are Available All LUTs as Images and *.tf Files and Relevant Images as Well. Available online: http://webzam.fbmi.cvut.cz/hozman/LUT.htm (accessed on 25 April 2022).












| Parameter | Value p-Value | Note |
|---|---|---|
| X′—x coordinate of centroid 1 | 0.2605 | liver |
| Ethanol solution concentration [%] | 0.0010 | liver |
| Storage time of the sample in Ethanol solution [days] | 0.0010 | liver |
| X′—x coordinate of centroid 1 | 0.0223 | kidneys |
| Ethanol solution concentration [%] | 0.0010 | kidneys |
| Storage time of the sample in Ethanol solution [days] | 0.0055 | kidneys |
| Solution and Soft Tissue | Value X1 1 [-] | Value X2 2 [-] | Value Y2 3 [-] | Value X3 4 [-] | Value X′ 5 [-] | Value c 6 [%] | Value d 7 [Days] |
|---|---|---|---|---|---|---|---|
| Ethanol, liver | 0.2461 | 0.4303 | 0.1898 | 0.4320 | 0.3695 | 50 | 8 |
| Ethanol, liver | 0.1200 | 0.1994 | 0.1551 | 0.2004 | 0.1733 | 50 | 17 |
| Ethanol, liver | 0.1476 | 0.223 | 0.1343 | 0.2241 | 0.1982 | 50 | 17 |
| Ethanol, liver | 0.1329 | 0.1827 | 0.1828 | 0.1838 | 0.1665 | 50 | 17 |
| Ethanol, liver | 0.2410 | 0.3718 | 0.2465 | 0.3735 | 0.3288 | 100 | 8 |
| Ethanol, liver | 0.1610 | 0.2374 | 0.1801 | 0.2385 | 0.2123 | 100 | 16 |
| Ethanol, liver | 0.2771 | 0.4010 | 0.4626 | 0.4028 | 0.3603 | 100 | 8 |
| Ethanol, liver | 0.1103 | 0.1467 | 0.2147 | 0.1906 | 0.1492 | 100 | 16 |
| Ethanol, liver | 0.2031 | 0.2048 | 0.3019 | 0.2995 | 0.2358 | 100 | 8 |
| Ethanol, liver | 0.1153 | 0.1687 | 0.2756 | 0.1704 | 0.1515 | 100 | 16 |
| Ethanol, liver | 0.0978 | 0.0989 | 0.1842 | 0.1378 | 0.1115 | 100 | 37 |
| Ethanol, liver | 0.0836 | 0.1213 | 0.1607 | 0.1519 | 0.1189 | 100 | 37 |
| Ethanol, liver | 0.0707 | 0.1425 | 0.1233 | 0.1437 | 0.1190 | 100 | 37 |
| Solution and Soft Tissue | Value X1 1 [-] | Value X2 2 [-] | Value Y2 3 [-] | Value X3 4 [-] | Value X′ 5 [-] | Value c 6 [%] | Value d 7 [Days] |
|---|---|---|---|---|---|---|---|
| Ethanol, kidneys | 0.1279 | 0.1478 | 0.2493 | 0.1814 | 0.1524 | 50 | 16 |
| Ethanol, kidneys | 0.1170 | 0.1515 | 0.1565 | 0.1532 | 0.1406 | 50 | 7 |
| Ethanol, kidneys | 0.1112 | 0.1753 | 0.1247 | 0.1762 | 0.1542 | 50 | 16 |
| Ethanol, kidneys | 0.1188 | 0.1411 | 0.1385 | 0.1429 | 0.1343 | 100 | 7 |
| Ethanol, kidneys | 0.1055 | 0.1739 | 0.1925 | 0.1747 | 0.1514 | 100 | 16 |
| Ethanol, kidneys | 0.1170 | 0.1463 | 0.2091 | 0.1480 | 0.1371 | 100 | 7 |
| Ethanol, kidneys | 0.1117 | 0.2443 | 0.2452 | 0.2455 | 0.2005 | 100 | 16 |
| Ethanol, kidneys | 0.0669 | 0.0932 | 0.0997 | 0.0939 | 0.0847 | 100 | 7 |
| Ethanol, kidneys | 0.1849 | 0.2733 | 0.0693 | 0.2744 | 0.2442 | 100 | 37 |
| Ethanol, kidneys | 0.1979 | 0.2379 | 0.0568 | 0.3286 | 0.2548 | 100 | 37 |
| Ethanol, kidneys | 0.2002 | 0.2792 | 0.1981 | 0.3239 | 0.2678 | 100 | 37 |
| Parameter | Value 7D | Value 8D | Value 16/17D | Value 37D | Note |
|---|---|---|---|---|---|
| N day avg centroid x coord. | 0.1242 | NA | 0.1646 | 0.2556 | Ethanol, kidneys |
| CT number 1 | −379.25 | NA | −176.92 | 277.94 | Ethanol, kidneys |
| N day avg centroid x coord. | NA | 0.3236 | 0.1752 | 0.1165 | Ethanol, liver |
| CT number 1 | NA | 617.92 | −124.22 | −417.67 | Ethanol, liver |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaspar, B.; Mrzilkova, J.; Hozman, J.; Zach, P.; Lahutsina, A.; Morozova, A.; Guarnieri, G.; Riedlova, J. Micro-Computed Tomography Soft Tissue Biological Specimens Image Data Visualization. Appl. Sci. 2022, 12, 4918. https://doi.org/10.3390/app12104918
Gaspar B, Mrzilkova J, Hozman J, Zach P, Lahutsina A, Morozova A, Guarnieri G, Riedlova J. Micro-Computed Tomography Soft Tissue Biological Specimens Image Data Visualization. Applied Sciences. 2022; 12(10):4918. https://doi.org/10.3390/app12104918
Chicago/Turabian StyleGaspar, Branislav, Jana Mrzilkova, Jiri Hozman, Petr Zach, Anastasiya Lahutsina, Alexandra Morozova, Giulia Guarnieri, and Jitka Riedlova. 2022. "Micro-Computed Tomography Soft Tissue Biological Specimens Image Data Visualization" Applied Sciences 12, no. 10: 4918. https://doi.org/10.3390/app12104918
APA StyleGaspar, B., Mrzilkova, J., Hozman, J., Zach, P., Lahutsina, A., Morozova, A., Guarnieri, G., & Riedlova, J. (2022). Micro-Computed Tomography Soft Tissue Biological Specimens Image Data Visualization. Applied Sciences, 12(10), 4918. https://doi.org/10.3390/app12104918







