Technique Evolutions for Microorganism Detection in Complex Samples: A Review
Abstract
:1. Introduction
2. Current Detection Methods
2.1. Methods Based on Growth Monitoring
2.1.1. Measurement of Gas Production
2.1.2. Electrochemical Methods
2.1.3. Bioluminescence
2.1.4. Microcalorimetry
2.1.5. Turbidimetry
2.2. Individual Cells Detection Methods
2.2.1. Solid Phase Cytometry
2.2.2. Flow Cytometry
2.3. Cellular Components Detection and Analytical Methods
2.3.1. Immunological Methods
2.3.2. Infrared Spectroscopy
2.3.3. Mass Spectrometry
2.3.4. Nucleic Acid Amplification Techniques
3. Developments in Innovative Detection Methods
3.1. Ligands for Classical Detection Techniques Improvement
3.1.1. Broad-Spectrum Ligand
3.1.2. The Most Promising Ligands
Aptamer and DNAzyme
Antimicrobial Peptides
3.2. Improvements and Developments in Analytical Methods Requiring Sampling
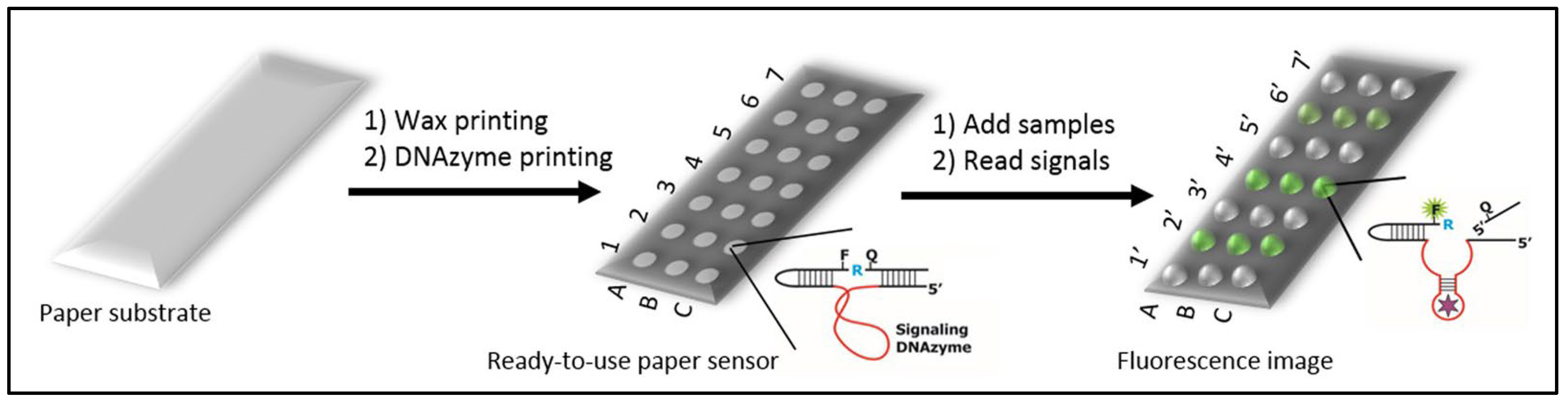
3.2.1. Paper Sensors
3.2.2. Microfluidics
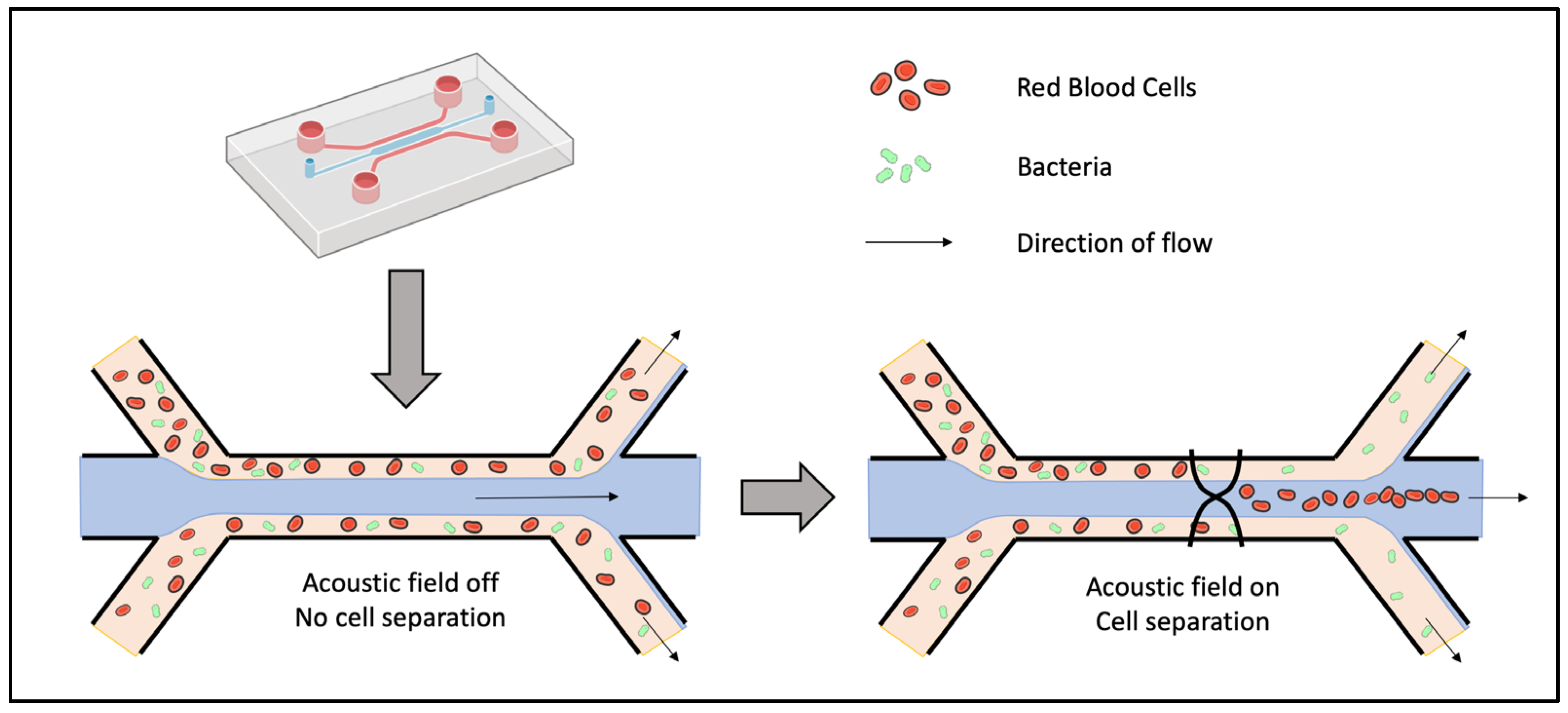
Sorting by Acoustophoresis
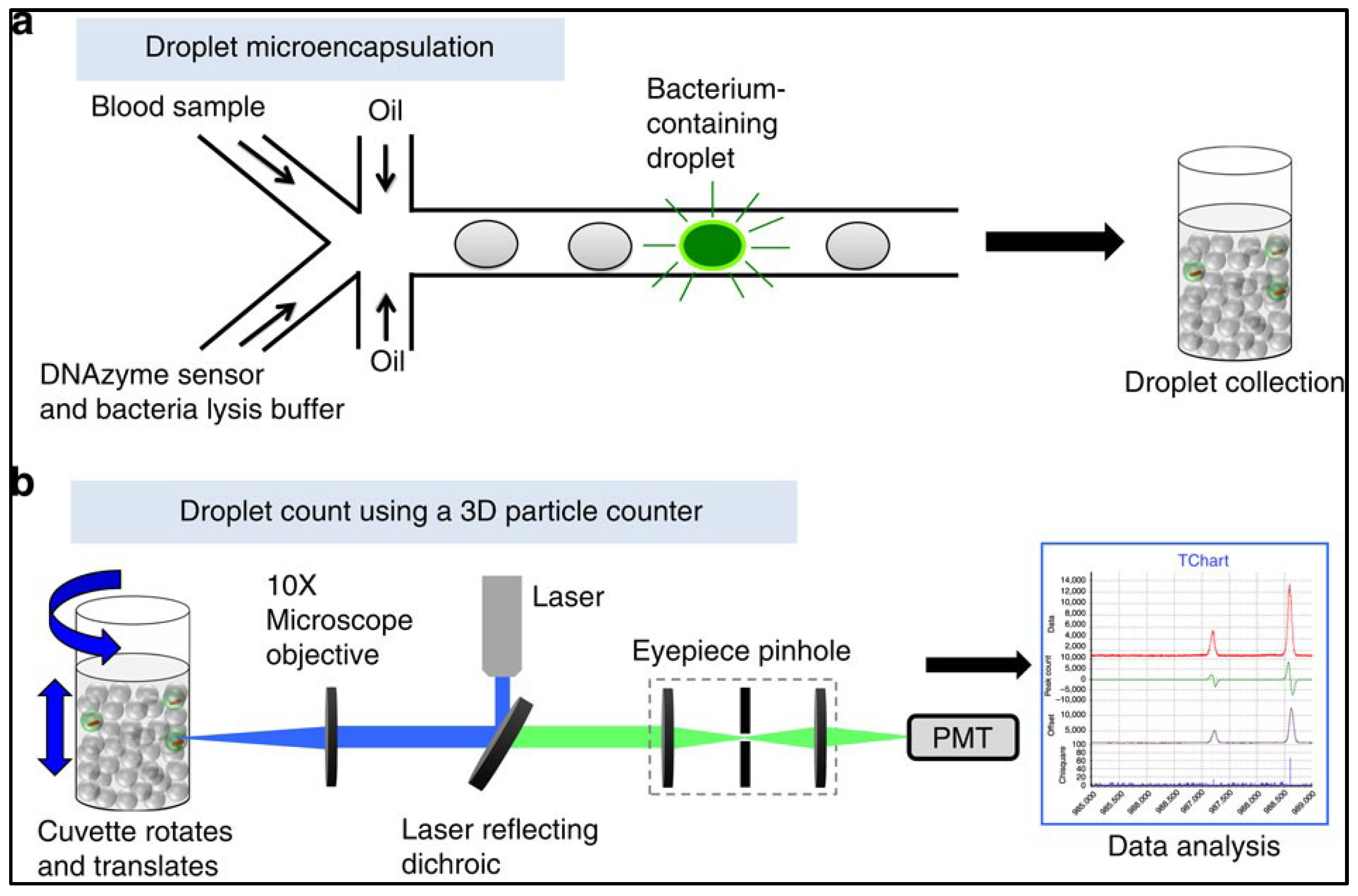
Microdroplets and 3D Particle Counter
3.3. Development of Physical and Computer Analysis Methods
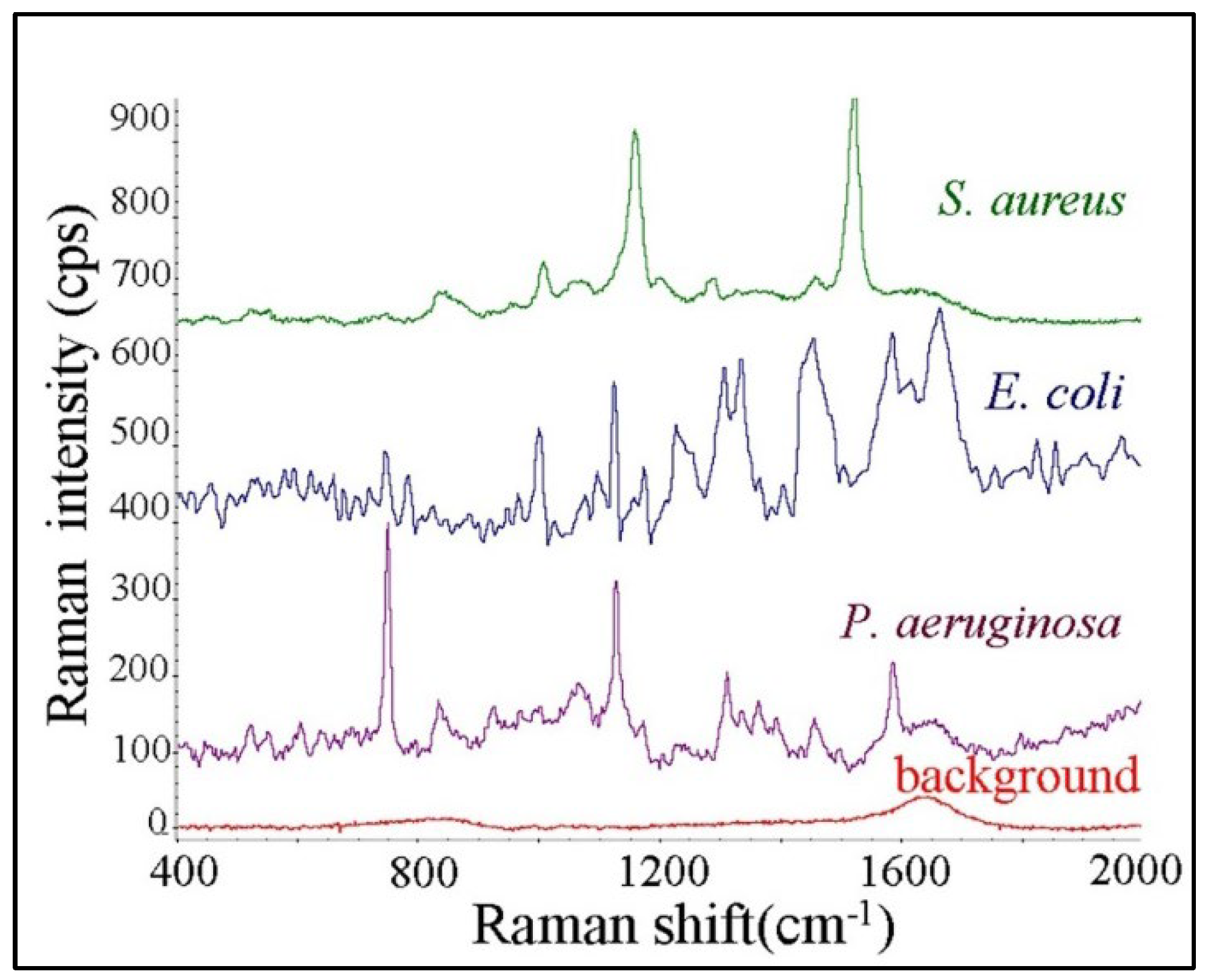
3.3.1. Raman Spectroscopy
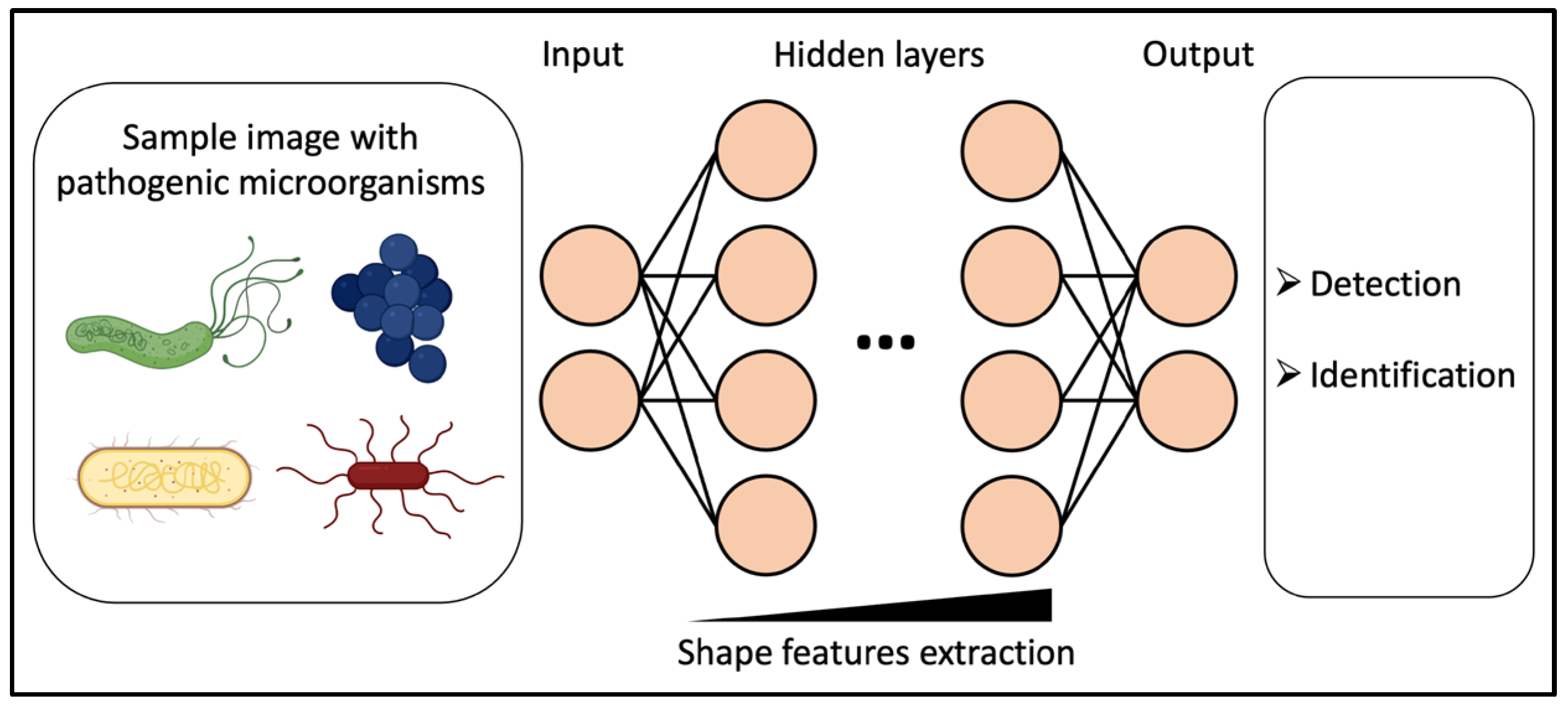
3.3.2. Deep Learning for Microscopy-Based Sampling Methods
3.4. Improvements and Developments in Real-Time and Online Analysis Techniques
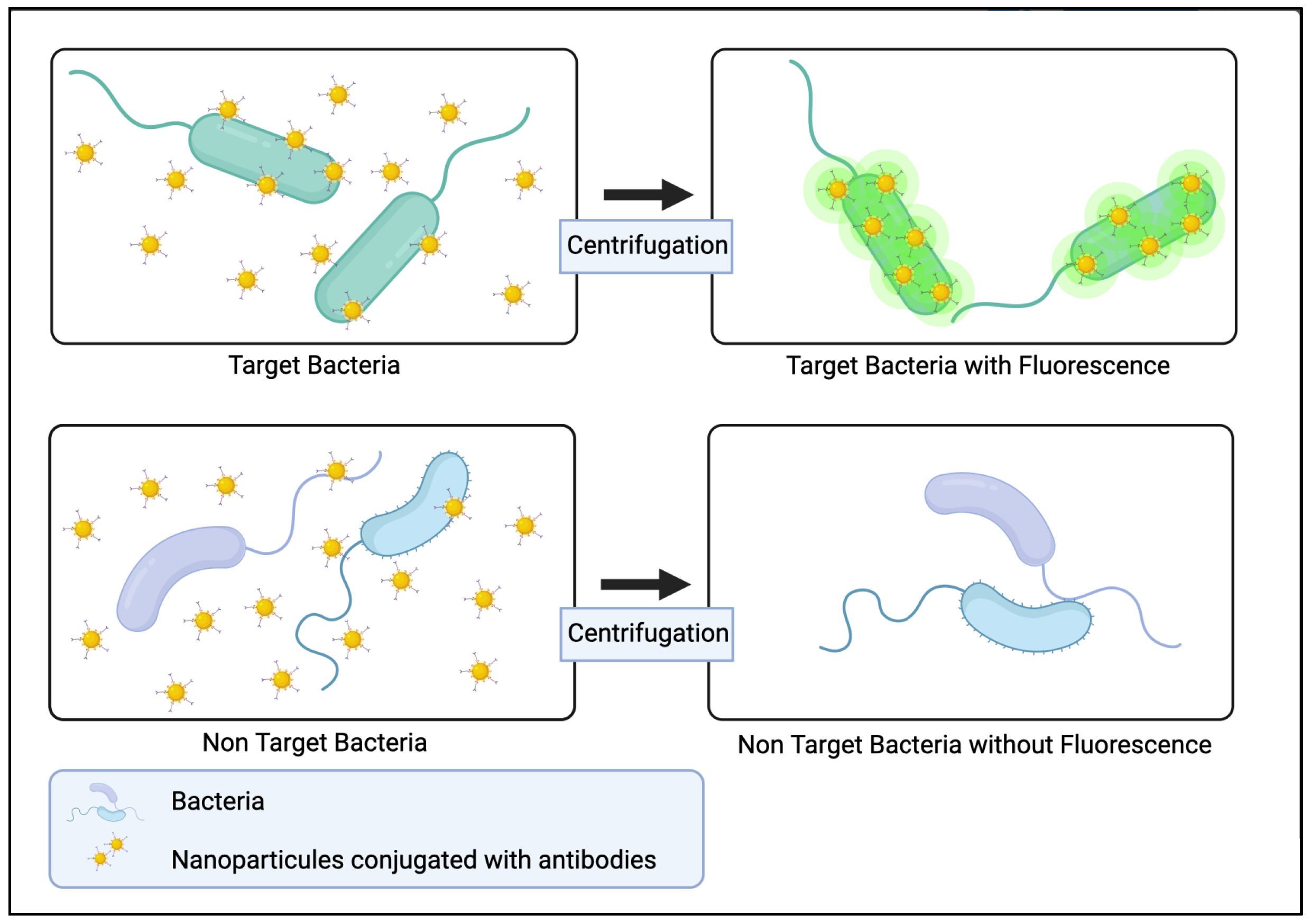
3.4.1. Bio-Conjugated Nanoparticles
3.4.2. Surface Plasmon Resonance
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hameed, S.; Xie, L.; Ying, Y. Conventional and Emerging Detection Techniques for Pathogenic Bacteria in Food Science: A Review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Green, M.S.; LeDuc, J.; Cohen, D.; Franz, D.R. Confronting the Threat of Bioterrorism: Realities, Challenges, and Defensive Strategies. Lancet Infect. Dis. 2019, 19, e2–e13. [Google Scholar] [CrossRef]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A Review of Methods for the Detection of Pathogenic Microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.H.; Sibbald, W.J. Definitions for Sepsis and Organ Failure and Guidelines for the Use of Innovative Therapies in Sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [Green Version]
- Chun, K.; Syndergaard, C.; Damas, C.; Trubey, R.; Mukindaraj, A.; Qian, S.; Jin, X.; Breslow, S.; Niemz, A. Sepsis Pathogen Identification. J. Lab. Autom. 2015, 20, 539–561. [Google Scholar] [CrossRef]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and Clinical Management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef] [Green Version]
- Hocquet, D.; Sauget, M.; Roussel, S.; Malugani, C.; Pouthier, F.; Morel, P.; Gbaguidi-Haore, H.; Bertrand, X.; Grenouillet, F. Validation of an Automated Blood Culture System for Sterility Testing of Cell Therapy Products. Cytotherapy 2014, 16, 692–698. [Google Scholar] [CrossRef]
- Váradi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the Detection and Identification of Pathogenic Bacteria: Past, Present, and Future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef]
- Monteiro, A.C.; Fortaleza, C.M.; Ferreira, A.M.; Cavalcante Rde, S.; Mondelli, A.L.; Bagagli, E.; da Cunha Mde, L. Comparison of Methods for the Identification of Microorganisms Isolated from Blood Cultures. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 45. [Google Scholar] [CrossRef] [Green Version]
- Buszewski, B.; Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V. Identification of Microorganisms by Modern Analytical Techniques. J. AOAC Int. 2017, 100, 1607–1623. [Google Scholar] [CrossRef]
- Kumar, H.; Bhardwaj, K.; Kaur, T.; Nepovimova, E.; Kuča, K.; Kumar, V.; Bhatia, S.K.; Dhanjal, D.S.; Chopra, C.; Singh, R.; et al. Detection of Bacterial Pathogens and Antibiotic Residues in Chicken Meat: A Review. Foods 2020, 9, 1504. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, T.C.; Wilson, M.; Turner, J.; DiGuiseppi, J.; Willert, M.; Mirrett, S.; Reller, L. BacT/Alert: An Automated Colorimetric Microbial Detection System. J. Clin. Microbiol. 1990, 28, 1608–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramamurthy, T.; Ghosh, A.; Pazhani, G.P.; Shinoda, S. Current Perspectives on Viable but Non-Culturable (VBNC) Pathogenic Bacteria. Front. Public Health 2014, 2, 103. [Google Scholar] [CrossRef] [PubMed]
- Wideman, N.E.; Oliver, J.D.; Crandall, P.G.; Jarvis, N.A. Detection and Potential Virulence of Viable but Non-Culturable (VBNC) Listeria Monocytogenes: A Review. Microorganisms 2021, 9, 194. [Google Scholar] [CrossRef]
- Furst, A.L.; Francis, M.B. Impedance-Based Detection of Bacteria. Chem. Rev. 2018, 119, 700–726. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen Detection with Electrochemical Biosensors: Advantages, Challenges and Future Perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef]
- Spaeth, S.; Tran, Q.; Liu, Z. Evaluation of an ATP-Bioluminescence Rapid Microbial Screening Method for In-Process Biologics. PDA J. Pharm. Sci. Technol. 2018, 72, 574–583. [Google Scholar] [CrossRef]
- Fricke, C.; Harms, H.; Maskow, T. Rapid Calorimetric Detection of Bacterial Contamination: Influence of the Cultivation Technique. Front. Microbiol. 2019, 10, 2530. [Google Scholar] [CrossRef] [Green Version]
- Wacogne, B.; Legrand, D.; Azzopardi, C.-L.; Pieralli, C.; Frelet-Barrand, A. Optical Spectroscopy Methods to Monitor Cells and Bacteria Concentrations and to Detect Contamination During Cell Culture: Application to the Fabrication of ATMPs. In Biomedical Engineering Systems and Technologies; Ye, X., Soares, F., De Maria, E., Gómez Vilda, P., Cabitza, F., Fred, A., Gamboa, H., Eds.; Communications in Computer and Information Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 1400, pp. 53–75. ISBN 978-3-030-72378-1. [Google Scholar]
- Hatiboruah, D.; Devi, D.Y.; Namsa, N.D.; Nath, P. Turbidimetric Analysis of Growth Kinetics of Bacteria in the Laboratory Environment Using Smartphone. J. Biophotonics 2020, 13, e201960159. [Google Scholar] [CrossRef]
- Chen, Z.; Yang, T.; Yang, H.; Li, T.; Nie, L.; Mou, X.; Deng, Y.; He, N.; Li, Z.; Wang, L.; et al. A Portable Multi-Channel Turbidity System for Rapid Detection of Pathogens by Loop-Mediated Isothermal Amplification. J. Biomed. Nanotechnol. 2018, 14, 198–205. [Google Scholar] [CrossRef]
- Paczesny, J.; Richter, Ł.; Hołyst, R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses 2020, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Péter, B.; Farkas, E.; Kurunczi, S.; Szittner, Z.; Bősze, S.; Ramsden, J.J.; Szekacs, I.; Horvath, R. Review of Label-Free Monitoring of Bacteria: From Challenging Practical Applications to Basic Research Perspectives. Biosensors 2022, 12, 188. [Google Scholar] [CrossRef] [PubMed]
- Sibilo, R.; Pérez, J.M.; Tebbenjohanns, F.; Hurth, C.; Pruneri, V. Surface Cytometer for Fluorescent Detection and Growth Monitoring of Bacteria over a Large Field-of-View. Biomed. Opt. Express 2019, 10, 2101–2116. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.; Wilkinson, M.G. Application of Flow Cytometry to the Detection of Pathogenic Bacteria. Curr. Issues Mol. Biol. 2017, 23, 21–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zand, E.; Froehling, A.; Schoenher, C.; Zunabovic-Pichler, M.; Schlueter, O.; Jaeger, H. Potential of Flow Cytometric Approaches for Rapid Microbial Detection and Characterization in the Food Industry—A Review. Foods 2021, 10, 3112. [Google Scholar] [CrossRef] [PubMed]
- Lemarchand, K.; Parthuisot, N.; Catala, P.; Lebaron, P. Comparative Assessment of Epifluorescence Microscopy, Flow Cytometry and Solid-Phase Cytometry Used in the Enumeration of Specific Bacteria in Water. Aquat. Microb. Ecol. 2001, 25, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Lequin, R.M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [Green Version]
- Lecellier, A.; Gaydou, V.; Mounier, J.; Hermet, A.; Castrec, L.; Barbier, G.; Ablain, W.; Manfait, M.; Toubas, D.; Sockalingum, G. Implementation of an FTIR Spectral Library of 486 Filamentous Fungi Strains for Rapid Identification of Molds. Food Microbiol. 2015, 45, 126–134. [Google Scholar] [CrossRef]
- Kochan, K.; Bedolla, D.E.; Perez-Guaita, D.; Adegoke, J.A.; Chakkumpulakkal Puthan Veettil, T.; Martin, M.; Roy, S.; Pebotuwa, S.; Heraud, P.; Wood, B.R. Infrared Spectroscopy of Blood. Appl. Spectrosc. 2021, 75, 611–646. [Google Scholar] [CrossRef]
- Hou, T.-Y.; Chiang-Ni, C.; Teng, S.-H. Current Status of MALDI-TOF Mass Spectrometry in Clinical Microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR Past, Present and Future. BioTechniques 2020, 69, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Benevides Lima, L.; Mesquita, F.P.; Brasil de Oliveira, L.L.; Andréa da Silva Oliveira, F.; Elisabete Amaral de Moraes, M.; Souza, P.F.N.; Montenegro, R.C. True or False: What Are the Factors That Influence COVID-19 Diagnosis by RT-QPCR? Expert Rev. Mol. Diagn. 2022, 22, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Gdoura, M.; Abouda, I.; Mrad, M.; Ben Dhifallah, I.; Belaiba, Z.; Fares, W.; Chouikha, A.; Khedhiri, M.; Layouni, K.; Touzi, H.; et al. SARS-CoV2 RT-PCR Assays: In Vitro Comparison of 4 WHO Approved Protocols on Clinical Specimens and Its Implications for Real Laboratory Practice through Variant Emergence. Virol. J. 2022, 19, 54. [Google Scholar] [CrossRef]
- Li, J.; Macdonald, J.; Stetten, F. von Review: A Comprehensive Summary of a Decade Development of the Recombinase Polymerase Amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glökler, J.; Lim, T.S.; Ida, J.; Frohme, M. Isothermal Amplifications—A Comprehensive Review on Current Methods. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 543–586. [Google Scholar] [CrossRef]
- Leonardo, S.; Toldrà, A.; Campàs, M. Biosensors Based on Isothermal DNA Amplification for Bacterial Detection in Food Safety and Environmental Monitoring. Sensors 2021, 21, 602. [Google Scholar] [CrossRef]
- Etchebarne, B.E.; Li, Z.; Stedtfeld, R.D.; Nicholas, M.C.; Williams, M.R.; Johnson, T.A.; Stedtfeld, T.M.; Kostic, T.; Khalife, W.T.; Tiedje, J.M.; et al. Evaluation of Nucleic Acid Isothermal Amplification Methods for Human Clinical Microbial Infection Detection. Front. Microbiol. 2017, 8, 2211. [Google Scholar] [CrossRef] [Green Version]
- Zanoli, L.M.; Spoto, G. Isothermal Amplification Methods for the Detection of Nucleic Acids in Microfluidic Devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef] [Green Version]
- Templier, V.; Roupioz, Y. On the Challenges of Detecting Whole Staphylococcus Aureus Cells with Biosensors. J. Appl. Microbiol. 2017, 123, 1056–1067. [Google Scholar] [CrossRef] [Green Version]
- Templier, V.; Livache, T.; Boisset, S.; Maurin, M.; Slimani, S.; Mathey, R.; Roupioz, Y. Biochips for Direct Detection and Identification of Bacteria in Blood Culture-Like Conditions. Sci. Rep. 2017, 7, 9457. [Google Scholar] [CrossRef]
- Xu, J.; Chau, Y.; Lee, Y. Phage-Based Electrochemical Sensors: A Review. Micromachines 2019, 10, 855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pala, L.; Sirec, T.; Spitz, U. Modified Enzyme Substrates for the Detection of Bacteria: A Review. Molecules 2020, 25, 3690. [Google Scholar] [CrossRef] [PubMed]
- Bollella, P.; Katz, E. Enzyme-Based Biosensors: Tackling Electron Transfer Issues. Sensors 2020, 20, 3517. [Google Scholar] [CrossRef] [PubMed]
- Yunus, G. Chapter 42—Biosensors: An Enzyme-Based Biophysical Technique for the Detection of Foodborne Pathogens. In Enzymes in Food Biotechnology; Kuddus, M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 723–738. ISBN 978-0-12-813280-7. [Google Scholar]
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Melaine, F.; Coilhac, C.; Roupioz, Y.; Buhot, A. A Nanoparticle-Based Thermo-Dynamic Aptasensor for Small Molecule Detection. Nanoscale 2016, 8, 16947–16954. [Google Scholar] [CrossRef]
- Jafari, M.; Rezaei, M.; Kalantari, H.; Tabarzad, M.; Daraei, B. DNAzyme-Aptamer or Aptamer-DNAzyme Paradigm: Biochemical Approach for Aflatoxin Analysis. Biotechnol. Appl. Biochem. 2018, 65, 274–280. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Kaur, H. Recent Developments in Cell-SELEX Technology for Aptamer Selection. Biochim. Biophys. Acta BBA-Gen. Subj. 2018, 1862, 2323–2329. [Google Scholar] [CrossRef]
- McConnell, E.M.; Morrison, D.; Rincon, M.A.R.; Salena, B.J.; Li, Y. Selection and Applications of Synthetic Functional DNAs for Bacterial Detection. TrAC Trends Anal. Chem. 2020, 124, 115785. [Google Scholar] [CrossRef]
- Zhang, K.; Li, H.; Wang, W.; Cao, J.; Gan, N.; Han, H. Application of Multiplexed Aptasensors in Food Contaminants Detection. ACS Sens. 2020, 5, 3721–3738. [Google Scholar] [CrossRef]
- Jamal, R.B.; Shipovskov, S.; Ferapontova, E.E. Electrochemical Immuno-and Aptamer-Based Assays for Bacteria: Pros and Cons over Traditional Detection Schemes. Sensors 2020, 20, 5561. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Aguirre, S.D.; Mok, W.W.; Li, Y. Developing Fluorogenic RNA-Cleaving DNAzymes for Biosensing Applications. In Ribozymes; Springer: Totowa, NJ, USA, 2012; pp. 395–418. [Google Scholar]
- Micura, R.; Höbartner, C. Fundamental Studies of Functional Nucleic Acids: Aptamers, Riboswitches, Ribozymes and DNAzymes. Chem. Soc. Rev. 2020, 49, 7331–7353. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ding, W.; Wang, C.; Wu, H.; Tian, X.; Lyu, M.; Wang, S. DNAzyme Biosensors for the Detection of Pathogenic Bacteria. Sens. Actuators B Chem. 2021, 331, 129422. [Google Scholar] [CrossRef]
- Ali, M.M.; Aguirre, S.D.; Lazim, H.; Li, Y. Fluorogenic DNAzyme Probes as Bacterial Indicators. Angew. Chem. Int. Ed. 2011, 50, 3751–3754. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-K.; Ali, M.M.; Zhang, K.; Huang, S.S.; Peterson, E.; Digman, M.A.; Gratton, E.; Zhao, W. Rapid Detection of Single Bacteria in Unprocessed Blood Using Integrated Comprehensive Droplet Digital Detection. Nat. Commun. 2014, 5, 5427. [Google Scholar] [CrossRef] [Green Version]
- Debiais, M.; Lelievre, A.; Smietana, M.; Müller, S. Splitting Aptamers and Nucleic Acid Enzymes for the Development of Advanced Biosensors. Nucleic Acids Res. 2020, 48, 3400–3422. [Google Scholar] [CrossRef]
- Shin, H.-S.; Gedi, V.; Kim, J.-K.; Lee, D. Detection of Gram-Negative Bacterial Outer Membrane Vesicles Using DNA Aptamers. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Reffuveille, F.; de la Fuente-Núñez, C.; Mansour, S.; Hancock, R.E.W. A Broad-Spectrum Antibiofilm Peptide Enhances Antibiotic Action against Bacterial Biofilms. Antimicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef] [Green Version]
- De la Fuente-Núñez, C.; Reffuveille, F.; Mansour, S.C.; Reckseidler-Zenteno, S.L.; Hernández, D.; Brackman, G.; Coenye, T.; Hancock, R.E.W. D-Enantiomeric Peptides That Eradicate Wild-Type and Multidrug-Resistant Biofilms and Protect against Lethal Pseudomonas Aeruginosa Infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Broad-Spectrum Anti-Biofilm Peptide That Targets a Cellular Stress Response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [Green Version]
- Templier, V.; Roux, A.; Roupioz, Y.; Livache, T. Ligands for Label-Free Detection of Whole Bacteria on Biosensors: A Review. TrAC Trends Anal. Chem. 2016, 79, 71–79. [Google Scholar] [CrossRef]
- Silva, R.R.; Avelino, K.Y.; Ribeiro, K.L.; Franco, O.L.; Oliveira, M.D.; Andrade, C.A. Optical and Dielectric Sensors Based on Antimicrobial Peptides for Microorganism Diagnosis. Front. Microbiol. 2014, 5, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lachowicz, J.I.; Szczepski, K.; Scano, A.; Casu, C.; Fais, S.; Orrù, G.; Pisano, B.; Piras, M.; Jaremko, M. The Best Peptidomimetic Strategies to Undercover Antibacterial Peptides. Int. J. Mol. Sci. 2020, 21, 7349. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardoux, É.; Boturyn, D.; Roupioz, Y. Antimicrobial Peptides as Probes in Biosensors Detecting Whole Bacteria: A Review. Molecules 2020, 25, 1998. [Google Scholar] [CrossRef] [PubMed]
- Lillehoj, P.B.; Kaplan, C.W.; He, J.; Shi, W.; Ho, C.-M. Rapid, Electrical Impedance Detection of Bacterial Pathogens Using Immobilized Antimicrobial Peptides. J. Lab. Autom. 2014, 19, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Malvano, F.; Pilloton, R.; Albanese, D. A Novel Impedimetric Biosensor Based on the Antimicrobial Activity of the Peptide Nisin for the Detection of Salmonella Spp. Food Chem. 2020, 325, 126868. [Google Scholar] [CrossRef]
- Kim, H.J.; Kwon, C.; Lee, B.S.; Noh, H. One-Step Sensing of Foodborne Pathogenic Bacteria Using a 3D Paper-Based Device. Analyst 2019, 144, 2248–2255. [Google Scholar] [CrossRef]
- Díaz-Amaya, S.; Zhao, M.; Lin, L.-K.; Ostos, C.; Allebach, J.P.; Chiu, G.T.-C.; Deering, A.J.; Stanciu, L.A. Inkjet Printed Nanopatterned Aptamer-Based Sensors for Improved Optical Detection of Foodborne Pathogens. Small Weinh. Bergstr. Ger. 2019, 15, e1805342. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, J.; Wan, K.; Jiang, D.; Jin, C. Miniaturized Paper-Supported 3D Cell-Based Electrochemical Sensor for Bacterial Lipopolysaccharide Detection. ACS Sens. 2020, 5, 1325–1335. [Google Scholar] [CrossRef]
- Bisha, B.; Adkins, J.; Jokerst, J.; Chandler, J.; Pérez-Méndez, A.; Coleman, S.; Sbodio, A.; Suslow, T.; Danyluk, M.; Henry, C.; et al. Colorimetric Paper-Based Detection of Escherichia Coli, Salmonella Spp., and Listeria Monocytogenes from Large Volumes of Agricultural Water. J. Vis. Exp. JoVE 2014, 88, 51414. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Lantigua, D.; Meka, A.; Taing, S.; Pandher, M.; Camci-Unal, G. Paper-Based Sensors: Emerging Themes and Applications. Sensors 2018, 18, 2838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.M.; Wolfe, M.; Tram, K.; Gu, J.; Filipe, C.D.M.; Li, Y.; Brennan, J.D. A DNAzyme-Based Colorimetric Paper Sensor for Helicobacter Pylori. Angew. Chem. Int. Ed. Engl. 2019, 58, 9907–9911. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Brown, C.L.; Jahanshahi-Anbuhi, S.; Kannan, B.; Li, Y.; Filipe, C.D.M.; Brennan, J.D. A Printed Multicomponent Paper Sensor for Bacterial Detection. Sci. Rep. 2017, 7, 12335. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; You, T.; Jang, H.; Ryu, H.; Lee, E.-S.; Oh, M.-H.; Huh, Y.S.; Kim, S.M.; Jeon, T.-J. Aptamer-Conjugated Polydiacetylene Colorimetric Paper Chip for the Detection of Bacillus Thuringiensis Spores. Sensors 2020, 20, 3124. [Google Scholar] [CrossRef]
- Li, S.; Ma, F.; Bachman, H.; Cameron, C.E.; Zeng, X.; Huang, T.J. Acoustofluidic Bacteria Separation. J. Micromech. Microeng. 2017, 27, 015031. [Google Scholar] [CrossRef]
- Ohlsson, P.; Petersson, K.; Augustsson, P.; Laurell, T. Acoustic Impedance Matched Buffers Enable Separation of Bacteria from Blood Cells at High Cell Concentrations. Sci. Rep. 2018, 8, 9156. [Google Scholar] [CrossRef]
- Hilton, S.H.; Hall, C.; Nguyen, H.T.; Everitt, M.L.; DeShong, P.; White, I.M. Phenotypically Distinguishing ESBL-Producing Pathogens Using Paper-Based Surface Enhanced Raman Sensors. Anal. Chim. Acta 2020, 1127, 207–216. [Google Scholar] [CrossRef]
- Narang, R.; Mohammadi, S.; Ashani, M.M.; Sadabadi, H.; Hejazi, H.; Zarifi, M.H.; Sanati-Nezhad, A. Sensitive, Real-Time and Non-Intrusive Detection of Concentration and Growth of Pathogenic Bacteria Using Microfluidic-Microwave Ring Resonator Biosensor. Sci. Rep. 2018, 8, 15807. [Google Scholar] [CrossRef]
- Srinivasan, V.; Pamula, V.K.; Fair, R.B. An Integrated Digital Microfluidic Lab-on-a-Chip for Clinical Diagnostics on Human Physiological Fluids. Lab. Chip 2004, 4, 310. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.-Y.; Lee, W.-C.; Kung, C.-T.; Li, L.-C.; Lee, C.-T.; Fu, L.-M. Recent Advances in Microfluidic Devices for Contamination Detection and Quality Inspection of Milk. Micromachines 2021, 12, 558. [Google Scholar] [CrossRef] [PubMed]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic Lab-on-a-Chip Platforms: Requirements, Characteristics and Applications. Chem. Soc. Rev. 2010, 39, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haeberle, S.; Zengerle, R. Microfluidic Platforms for Lab-on-a-Chip Applications. Lab. Chip 2007, 7, 1094. [Google Scholar] [CrossRef]
- Burklund, A.; Zhang, J.X.J. Microfluidics-Based Organism Isolation from Whole Blood: An Emerging Tool for Bloodstream Infection Diagnosis. Ann. Biomed. Eng. 2019, 47, 1657–1674. [Google Scholar] [CrossRef]
- Luo, F.; Li, Z.; Dai, G.; Lu, Y.; He, P.; Wang, Q. Simultaneous Detection of Different Bacteria by Microchip Electrophoresis Combined with Universal Primer-Duplex Polymerase Chain Reaction. J. Chromatogr. A 2020, 1615, 460734. [Google Scholar] [CrossRef]
- Ohlsson, P.; Evander, M.; Petersson, K.; Mellhammar, L.; Lehmusvuori, A.; Karhunen, U.; Soikkeli, M.; Seppä, T.; Tuunainen, E.; Spangar, A.; et al. Integrated Acoustic Separation, Enrichment, and Microchip Polymerase Chain Reaction Detection of Bacteria from Blood for Rapid Sepsis Diagnostics. Anal. Chem. 2016, 88, 9403–9411. [Google Scholar] [CrossRef] [Green Version]
- Dow, P.; Kotz, K.; Gruszka, S.; Holder, J.; Fiering, J. Acoustic Separation in Plastic Microfluidics for Rapid Detection of Bacteria in Blood Using Engineered Bacteriophage. Lab. Chip 2018, 18, 923–932. [Google Scholar] [CrossRef]
- Hedde, P.N.; Bouzin, M.; Abram, T.J.; Chen, X.; Toosky, M.N.; Vu, T.; Li, Y.; Zhao, W.; Gratton, E. Rapid Isolation of Rare Targets from Large Fluid Volumes. Sci. Rep. 2020, 10, 12458. [Google Scholar] [CrossRef]
- Kang, D.-K.; Gong, X.; Cho, S.; Kim, J.; Edel, J.B.; Chang, S.-I.; Choo, J.; deMello, A.J. 3D Droplet Microfluidic Systems for High-Throughput Biological Experimentation. Anal. Chem. 2015, 87, 10770–10778. [Google Scholar] [CrossRef]
- Zhang, K.; Kang, D.-K.; Ali, M.M.; Liu, L.; Labanieh, L.; Lu, M.; Riazifar, H.; Nguyen, T.N.; Zell, J.A.; Digman, M.A.; et al. Digital Quantification of MiRNA Directly in Plasma Using Integrated Comprehensive Droplet Digital Detection. Lab. Chip 2015, 15, 4217–4226. [Google Scholar] [CrossRef] [PubMed]
- Burklund, A.; Petryk, J.D.; Hoopes, P.J.; Zhang, J.X.J. Microfluidic Enrichment of Bacteria Coupled to Contact-Free Lysis on a Magnetic Polymer Surface for Downstream Molecular Detection. Biomicrofluidics 2020, 14, 034115. [Google Scholar] [CrossRef] [PubMed]
- AlMasoud, N.; Muhamadali, H.; Chisanga, M.; AlRabiah, H.; Lima, C.A.; Goodacre, R. Discrimination of Bacteria Using Whole Organism Fingerprinting: The Utility of Modern Physicochemical Techniques for Bacterial Typing. Analyst 2021, 146, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Rebrošová, K.; Šiler, M.; Samek, O.; Růžička, F.; Bernatová, S.; Holá, V.; Ježek, J.; Zemánek, P.; Sokolová, J.; Petráš, P. Rapid Identification of Staphylococci by Raman Spectroscopy. Sci. Rep. 2017, 7, 14846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorenz, B.; Wichmann, C.; Stöckel, S.; Rösch, P.; Popp, J. Cultivation-Free Raman Spectroscopic Investigations of Bacteria. Trends Microbiol. 2017, 25, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Strola, S.A.; Baritaux, J.-C.; Schultz, E.; Simon, A.C.; Allier, C.; Espagnon, I.; Jary, D.; Dinten, J.-M. Single Bacteria Identification by Raman Spectroscopy. J. Biomed. Opt. 2014, 19, 111610. [Google Scholar] [CrossRef]
- Cheng, I.-F.; Chang, H.-C.; Chen, T.-Y.; Hu, C.; Yang, F.-L. Rapid (<5 Min) Identification of Pathogen in Human Blood by Electrokinetic Concentration and Surface-Enhanced Raman Spectroscopy. Sci. Rep. 2013, 3, 2365. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and Label-Free Based Surface-Enhanced Raman Scattering for Pathogen Bacteria Detection: A Review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef]
- Yang, E.; Li, D.; Yin, P.; Xie, Q.; Li, Y.; Lin, Q.; Duan, Y. A Novel Surface-Enhanced Raman Scattering (SERS) Strategy for Ultrasensitive Detection of Bacteria Based on Three-Dimensional (3D) DNA Walker. Biosens. Bioelectron. 2021, 172, 112758. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, H.; Ye, T.; Juhas, M. Deep Learning for Imaging and Detection of Microorganisms. Trends Microbiol. 2021, 29, 569–572. [Google Scholar] [CrossRef]
- Xu, J.; Yi, X.; Jin, G.; Peng, D.; Fan, G.; Xu, X.; Chen, X.; Yin, H.; Cooper, J.M.; Huang, W.E. High-Speed Diagnosis of Bacterial Pathogens at the Single Cell Level by Raman Microspectroscopy with Machine Learning Filters and Denoising Autoencoders. ACS Chem. Biol. 2022, 17, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-End Lung Cancer Screening with Three-Dimensional Deep Learning on Low-Dose Chest Computed Tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Matuszewski, D.J.; Sintorn, I.-M. Reducing the U-Net Size for Practical Scenarios: Virus Recognition in Electron Microscopy Images. Comput. Methods Programs Biomed. 2019, 178, 31–39. [Google Scholar] [CrossRef]
- Zieliński, B.; Plichta, A.; Misztal, K.; Spurek, P.; Brzychczy-Włoch, M.; Ochońska, D. Deep Learning Approach to Bacterial Colony Classification. PLoS ONE 2017, 12, e0184554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.; Ermon, S.; Dionne, J. Rapid Identification of Pathogenic Bacteria Using Raman Spectroscopy and Deep Learning. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.W.; Zaidi, N.A.; Rao, A.A.; Blank, R.; Vellekoop, M.J.; Lang, W. A Fungus Spores Dataset and a Convolutional Neural Network Based Approach for Fungus Detection. IEEE Trans. Nanobiosci. 2018, 17, 281–290. [Google Scholar] [CrossRef]
- Li, S.; Yang, Q.; Jiang, H.; Cortés-Vecino, J.A.; Zhang, Y. Parasitologist-Level Classification of Apicomplexan Parasites and Host Cell with Deep Cycle Transfer Learning (DCTL). Bioinformatics 2020, 36, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Park, B.; Eady, M.; Ouyang, Q.; Chen, K. Single-Cell Classification of Foodborne Pathogens Using Hyperspectral Microscope Imaging Coupled with Deep Learning Frameworks. Sens. Actuators B Chem. 2020, 309, 127789. [Google Scholar] [CrossRef]
- Wang, H.; Koydemir, H.C.; Qiu, Y.; Bai, B.; Zhang, Y.; Jin, Y.; Tok, S.; Yilmaz, E.C.; Gumustekin, E.; Rivenson, Y.; et al. Early Detection and Classification of Live Bacteria Using Time-Lapse Coherent Imaging and Deep Learning. Light Sci. Appl. 2020, 9, 1–17. [Google Scholar] [CrossRef]
- Mocan, T.; Matea, C.T.; Pop, T.; Mosteanu, O.; Buzoianu, A.D.; Puia, C.; Iancu, C.; Mocan, L. Development of Nanoparticle-Based Optical Sensors for Pathogenic Bacterial Detection. J. Nanobiotechnol. 2017, 15, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Hilliard, L.R.; Mechery, S.J.; Wang, Y.; Bagwe, R.P.; Jin, S.; Tan, W. A Rapid Bioassay for Single Bacterial Cell Quantitation Using Bioconjugated Nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 15027–15032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, A.M.; Fan, Z.; Sinha, S.S.; Shi, Y.; Le, L.; Bai, F.; Ray, P.C. Bio-Conjugated Gold Nanoparticle Based SERS Probe for Ultrasensitive Identification of Mosquito-Borne Viruses Using Raman Fingerprinting. J. Phys. Chem. C Nanomater. Interfaces 2015, 119, 23669–23775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for Whole-Cell Bacterial Detection. Clin. Microbiol. Rev. 2014, 27, 631–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocková, M.; Slabỳ, J.; Špringer, T.; Homola, J. Advances in Surface Plasmon Resonance Imaging and Microscopy and Their Biological Applications. Annu. Rev. Anal. Chem. 2019, 12, 151–176. [Google Scholar] [CrossRef] [PubMed]
- Piliarik, M.; Vaisocherová, H.; Homola, J. Surface Plasmon Resonance Biosensing. Methods Mol. Biol. Clifton N.J. 2009, 503, 65–88. [Google Scholar] [CrossRef]
- Dudak, F.C.; Boyaci, I.H. Rapid and Label-Free Bacteria Detection by Surface Plasmon Resonance (SPR) Biosensors. Biotechnol. J. 2009, 4, 1003–1011. [Google Scholar] [CrossRef]
- Desmet, C.; Vindas, K.; Alvarado Meza, R.; Garrigue, P.; Voci, S.; Sojic, N.; Maziz, A.; Courson, R.; Malaquin, L.; Leichle, T.; et al. Multiplexed Remote SPR Detection of Biological Interactions through Optical Fiber Bundles. Sensors 2020, 20, 511. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Park, B.; Chen, J.; He, X. Rapid and Label-Free Immunosensing of Shiga Toxin Subtypes with Surface Plasmon Resonance Imaging. Toxins 2020, 12, 280. [Google Scholar] [CrossRef]
- Filion-Côté, S.; Melaine, F.; Kirk, A.G.; Tabrizian, M. Monitoring of Bacterial Film Formation and Its Breakdown with an Angular-Based Surface Plasmon Resonance Biosensor. Analyst 2017, 142, 2386–2394. [Google Scholar] [CrossRef]
- Nair, S.; Gomez-Cruz, J.; Manjarrez-Hernandez, Á.; Ascanio, G.; Sabat, R.G.; Escobedo, C. Rapid Label-Free Detection of Intact Pathogenic Bacteria in Situ via Surface Plasmon Resonance Imaging Enabled by Crossed Surface Relief Gratings. Analyst 2020, 145, 2133–2142. [Google Scholar] [CrossRef]
- Park, B.; Wang, B.; Chen, J. Label-Free Immunoassay for Multiplex Detections of Foodborne Bacteria in Chicken Carcass Rinse with Surface Plasmon Resonance Imaging. Foodborne Pathog. Dis. 2021, 18, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Pardoux, É.; Roux, A.; Mathey, R.; Boturyn, D.; Roupioz, Y. Antimicrobial Peptide Arrays for Wide Spectrum Sensing of Pathogenic Bacteria. Talanta 2019, 203, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Melaine, F.; Saad, M.; Faucher, S.; Tabrizian, M. Selective and High Dynamic Range Assay Format for Multiplex Detection of Pathogenic Pseudomonas Aeruginosa, Salmonella Typhimurium, and Legionella Pneumophila RNAs Using Surface Plasmon Resonance Imaging. Anal. Chem. 2017, 89, 7802–7807. [Google Scholar] [CrossRef]
- Bouguelia, S.; Roupioz, Y.; Slimani, S.; Mondani, L.; Casabona, M.G.; Durmort, C.; Vernet, T.; Calemczuk, R.; Livache, T. On-Chip Microbial Culture for the Specific Detection of Very Low Levels of Bacteria. Lab. Chip 2013, 13, 4024–4032. [Google Scholar] [CrossRef] [Green Version]
- Shimanoe, K.; Endo, S.; Matsuyama, T.; Wada, K.; Okamoto, K. Localized Surface Plasmon Resonance in Deep Ultraviolet Region below 200 Nm Using a Nanohemisphere on Mirror Structure. Sci. Rep. 2021, 11, 5169. [Google Scholar] [CrossRef]
- Takemura, K. Surface Plasmon Resonance (SPR)- and Localized SPR (LSPR)-Based Virus Sensing Systems: Optical Vibration of Nano- and Micro-Metallic Materials for the Development of Next-Generation Virus Detection Technology. Biosensors 2021, 11, 250. [Google Scholar] [CrossRef]
- Azzouz, A.; Hejji, L.; Kim, K.-H.; Kukkar, D.; Souhail, B.; Bhardwaj, N.; Brown, R.J.C.; Zhang, W. Advances in Surface Plasmon Resonance–Based Biosensor Technologies for Cancer Biomarker Detection. Biosens. Bioelectron. 2022, 197, 113767. [Google Scholar] [CrossRef] [PubMed]




| Flow Rate | Dilution Factor | Time to Process 1 mL | Red Blood Cells Removal | Bacteria Recovery | Optimized for |
|---|---|---|---|---|---|
| 400 µL/min | 100 | 4 h | >99.9% | 99.7% | Bacteria recovery |
| 100 µL/min | 5 | 50 min | 99.99% | 75% | Blood cell removal |
| 400 µL/min | 5 | 12.5 min | >99% | 90% | Throughput |
| Specifications | IC 3D System | Blood Culture |
|---|---|---|
| Specimen types | Diluted blood | Blood |
| Sample volume | Microliters to milliliters | Milliliters |
| Culture enrichment | No | Yes |
| Time to results | <90 min, yes or no <4 h, quantitative | 10 h–20 h |
| Limit of detection (CFU mL−1) | 1–10 | ~100 |
| Selective | Yes | No |
| Quantitative | Yes | No |
| Techniques | LOD (UFC/mL) | Analysis Time | Needed for Sampling | Real-Time Analysis | Specificity | Advantage | Disadvantage | Cost |
|---|---|---|---|---|---|---|---|---|
| Blood culture | 10–100 | 20 h | Yes | No | Not specific |
|
| + |
| Raman spectroscopy | 1 | 10 min | Yes | No | Database |
|
| +++ |
| Paper Sensor | 100 | 1–8 h | Yes | No | Design Ligand |
|
| + |
| Microfluidics and DNAzyme | 1 | 90 min | Yes | No | Design DNAzyme |
|
| ++ |
| Bio-conjugated nanoparticles | 1–10 | 20 min | No | Yes | Design Antibody |
|
| ++ |
| SPRi | 10 | 15 h | No | Yes | Design AMP |
|
| ++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prada, P.; Brunel, B.; Reffuveille, F.; Gangloff, S.C. Technique Evolutions for Microorganism Detection in Complex Samples: A Review. Appl. Sci. 2022, 12, 5892. https://doi.org/10.3390/app12125892
Prada P, Brunel B, Reffuveille F, Gangloff SC. Technique Evolutions for Microorganism Detection in Complex Samples: A Review. Applied Sciences. 2022; 12(12):5892. https://doi.org/10.3390/app12125892
Chicago/Turabian StylePrada, Pierre, Benjamin Brunel, Fany Reffuveille, and Sophie C. Gangloff. 2022. "Technique Evolutions for Microorganism Detection in Complex Samples: A Review" Applied Sciences 12, no. 12: 5892. https://doi.org/10.3390/app12125892
APA StylePrada, P., Brunel, B., Reffuveille, F., & Gangloff, S. C. (2022). Technique Evolutions for Microorganism Detection in Complex Samples: A Review. Applied Sciences, 12(12), 5892. https://doi.org/10.3390/app12125892






