Abstract
Three-dimensional bone shape reconstruction is a fundamental step for any subject-specific musculo-skeletal model. Typically, medical images are processed to reconstruct bone surfaces via slice-by-slice contour identification. Freeware software packages are available, but commercial ones must be used for the necessary certification in clinics. The commercial software packages also imply expensive hardware and demanding training, but offer valuable tools. The aim of the present work is to report the performance of five commercial software packages (Mimics®, AmiraTM, D2PTM, SimplewareTM, and Segment 3D PrintTM), particularly the time to import and to create the model, the number of triangles of the mesh, and the STL file size. DICOM files of three different computed tomography scans from five different human anatomical areas were utilized for bone shape reconstruction by using each of these packages. The same operator and the same hosting hardware were used for these analyses. The computational time was found to be different between the packages analyzed, probably because of the pre-processing implied in this operation. The longer “time-to-import” observed in one software is likely due to the volume rendering during uploading. A similar number of triangles per megabyte (approximately 20 thousand) was observed for the five commercial packages. The present work showed the good performance of these software packages, with the main features being better than those analyzed previously in freeware packages.
1. Introduction
Three-dimensional (3D) reconstruction of bone models is fundamental for musculo-skeletal biomechanics, particularly for subject-specific modeling [1,2,3,4]. Exact 3D bone morphology is becoming essential in orthopedics for the custom design and surgical planning of joint replacements [5,6,7,8]. In this context, also the recent large progress in 3D printing is contributing to the huge number of exploitations in orthopedics and traumatology, since this additive technology enables the cheap manufacturing of custom-made prostheses and implants, along with relevant cutting jigs, designed over the exact dimension, shape, and alignment of bone and joint defects starting from patient-specific anatomy [9,10,11,12,13,14,15].
The full process from medical images to final implants also allows physical replica of patient anatomy, valuable for pre-operative planning, surgical team training, and physician-to-patient communication, in addition, of course, to musculo-skeletal and finite element modeling [16,17,18]. Typically, medical images from computed tomography (CT) are processed to be segmented, i.e., to reconstruct bone surface mesh via slice-by-slice bone contour identification [19,20,21,22]. Image segmentation is a long and critical process, which implies manual, automatic, or semi-automatic tracking of the silhouette contours of the bony structures, and, therefore, requires anatomical knowledge, computer skills, and awareness of the scopes [15,23,24]. Many dedicated software packages are offered on the market, from freeware tools with basic functions [25,26,27], running more likely on fairly performing computers, to expensive software packages with more effective segmentation algorithms and features [26,28,29], likely running on powerful computers. The optimal image segmentation software should support the user in carefully defining the bone models, and eventually providing a file to be exported in standard stereo-lithography (STL) format, with a suitable number of triangles, a uniform mesh, and a minimum overall size. The reconstruction of 3D bone models requires extensive work; a good compromise should be found for each application, between the automation of the segmentation process and the quality of the final results [25,30,31,32].
The current commercial software packages for image segmentation claim high performance and valuable technical tools, but require robust hardware, demanding training, and careful maintenance, and thus these result in expensive licenses. Hence, cheap and easy-to-use software tools [33] are still pursued and utilized. The performance of a number of freeware software programs was previously analyzed and compared while processing fifteen different human bones from five different anatomical areas; a number of valuable features and fair quality of the reconstructed bone models were found [25]. However, large differences in the number of triangles of the output meshes and in the file size were found, with the triangles per megabyte (MByte) ratio ranging from around 4 to 20 thousand [25]. Distance map analysis amongst outputs from these different free software packages revealed that root-mean-square deviations ranged from 0.13 to 2.21 mm when averaged over the five anatomical areas [25].
However, the major concern of these freeware software packages is the lack of certification as medical diagnostic devices, i.e., the official recognition to be used as appropriate preoperative software for implant design and surgical planning in the standard clinical practice [34]. Hence, the aim of the present work is to report on the performance of five commercial image segmentation packages. For a possible reasonable comparison, also the same exact CT scans of bony parts that we previously examined with freeware software packages [25] were used. From the presented original combination of these two analyses, advantages and disadvantages of commercial and freeware software packages for bone segmentation from CT scans can be established.
2. Methods
2.1. CT Scan Collection
Medical images in Digital Imaging and Communications in Medicine (DICOM) format from three different subjects were taken from a previous work [25]. In detail, for each of them, a number of anatomical complexes of the upper and the lower limb, i.e., the shoulder, elbow, and wrist for the former, and the knee and ankle for the latter, were analyzed. These fifteen scans were from CT (‘Brilliance 16-slice scanner’, Philips Medical Systems; Best, The Netherlands), with matrix size 512 × 512, voxel size 0.29 × 0.29 × 0.8 mm, layer thickness 0.5 mm, and field of view and data collection protocol set according to the specific anatomical complex to be analyzed. These technical parameters and the size of the anatomical complexes under analysis resulted in the following numbers of images, for each of the three subjects: 365, 256, and 321 for the shoulder; 353, 299, and 282 for the elbow; 239, 212, and 319 for the wrist; 204, 299, and 241 for the knee; 290, 481, and 315 for the ankle.
2.2. Segmentation Software Packages
Each dataset was analyzed with five commercial software packages for medial image segmentation (Table 1), i.e., all those available in the local area where the present analysis was performed: (1) MimicsTM Innovation Suite (v. 24.0, Materialise Inc., Leuven, Belgium), (2) AmiraTM (v. 2019.4, Thermo Fisher Scientific Inc., Waltham, MA, USA), (3) D2PTM (DICOM-to-PRINT, v. 1.0.2.2055, 3D Systems Inc., Rock Hill, SC, USA), (4) SimplewareTM (v. 2021.06, Synopsys Inc., Mountain View, CA, USA), and (5) Segment 3D PrintTM (v. 3.3 R 9056, Medviso AB, Lund, Sweden). Relevant technical requirements for each software program are reported in Table 1.

Table 1.
Technical requirements for each software package analyzed (* for best performance, multiple hard drive configuration—3 or more HDDs or SSDs—in RAID 5 mode is recommended, as reported in relevant user manual).
2.3. Medical Image Segmentation Process
All analyses were executed on the same computer (platform Intel® Xeon W-2123 CPU @ 3.60GHz, 64 bit, 32 GB RAM; graphics card: NVIDIA GeForce RTX 2070 SUPER) and operating system (Windows 10, Microsoft Co., Redmond, WA, USA).
The overall operational segmentation principle was very similar over each software package (Figure 1): importing the DICOM files in the three anatomical projections, followed by image segmentation using the most suitable tools offered by the software, including thresholding and minor possible manual corrections to remove isolated voxel areas. The masks defined by image segmentation embedded all bones of the overall joints. Although editing after image segmentation was allowed in all five software packages, no additional shaping or meshing tools were used. By means of built-in functions to convert the segmented masks into surface meshes, the 3D models of the bones were generated and exported in STL format files, all in binary code and in little-endian mode.
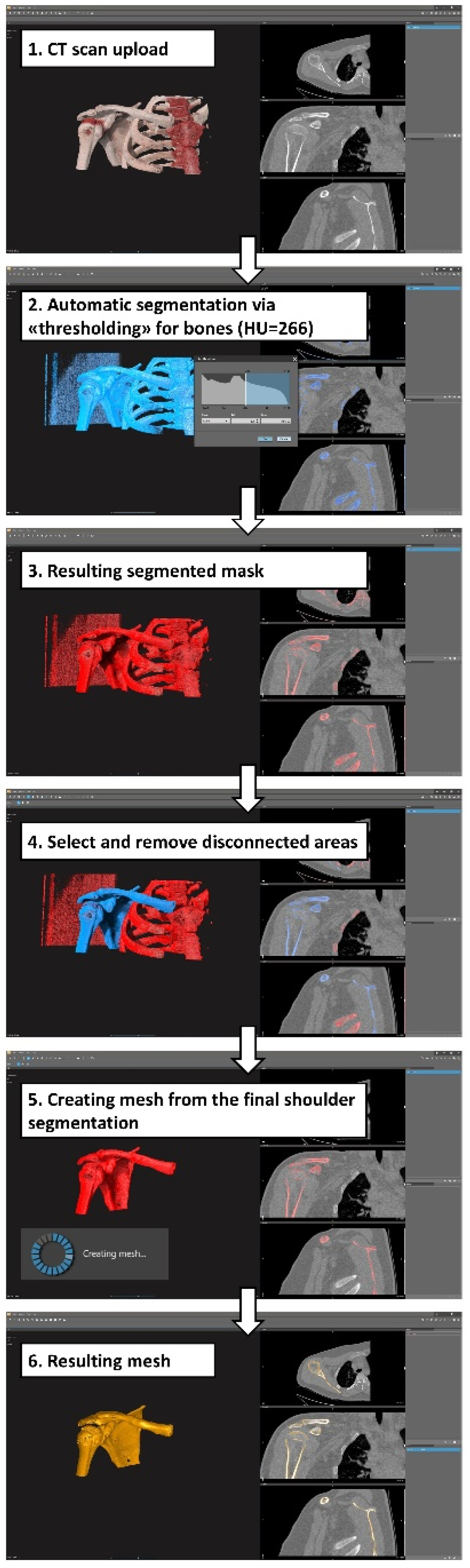
Figure 1.
A diagram for an automatic/semi-automatic workflow for the segmentation process: present exemplary screenshots obtained during this process using D2P software; a very similar workflow was followed for the other four software packages.
A radiographer, with 4 years of experience in the radiological department of an orthopedic center and 4 years of experience in 3D bone model segmentation, performed all reconstructions, using the same computer to remove possible bias; this was the same radiographer who performed a similar analysis in a previous study [25]. The necessary technical support for using the selected five software packages was provided by relevant computer scientists. All the present 3D bone reconstructions with the five packages were concentrated over a period of time of one month.
2.4. Data Collection and Processing
During the image segmentation phase, the following parameters were collected for each of the software programs utilized: DICOM time to import, time to create the model, number of model triangles, model file size, and number of model triangles per megabyte. These are reported in terms of mean ± standard deviation over the five anatomical complexes and the three subjects, along with range values (min–max).
Furthermore, the Pearson product–moment correlation coefficient (R) was also used to derive correlations between the mean number of triangles and the mean file size for the models obtained using the commercial software packages analyzed in the present study and also for those previously obtained with freeware software packages [25]. Corresponding p-values are reported for assessing significance, this being accepted at p < 0.05.
3. Results
The software features considered in the present analysis are those reported in Table 2.

Table 2.
Main features of the five software packages analyzed. The results are means ± standard deviation over the fifteen models analyzed (five anatomical areas, each from three subjects), along with range values (min–max). For the sake of comparison, the table format is similar to that reported in the previous work [25] (with the exception of information related to basic two-dimensional and 3D features, which is not reported here).
Additional features were analyzed, but because these were found to be available exactly in each package, these are not reported in Table 2 but rather listed here below: unlimited number of image slices; multiplanar visualization and representation; correspondence amongst the coronal, axial, and sagittal orientations; crop and zoom; contrast and brightness adjustments; separation of the regions of interest; linear, angular, and volumetric measures; simultaneous planar images and 3D rendering; export of images and mesh data; export in STL file format.
The computational import time of the DICOM files was found to be within the range of 1–6 s considering all analyzed software packages. The longer “time-to-import” found for the Segment 3D Print software (3.7 ± 1.1 s) is likely due to the simultaneous creation of 3D volume rendering during the uploading process. Marked differences were observed in the time to create the model, this ranging from 2.1 ± 0.4 s in Amira to 23.9 ± 13.3 s in Segment 3D Print, on average.
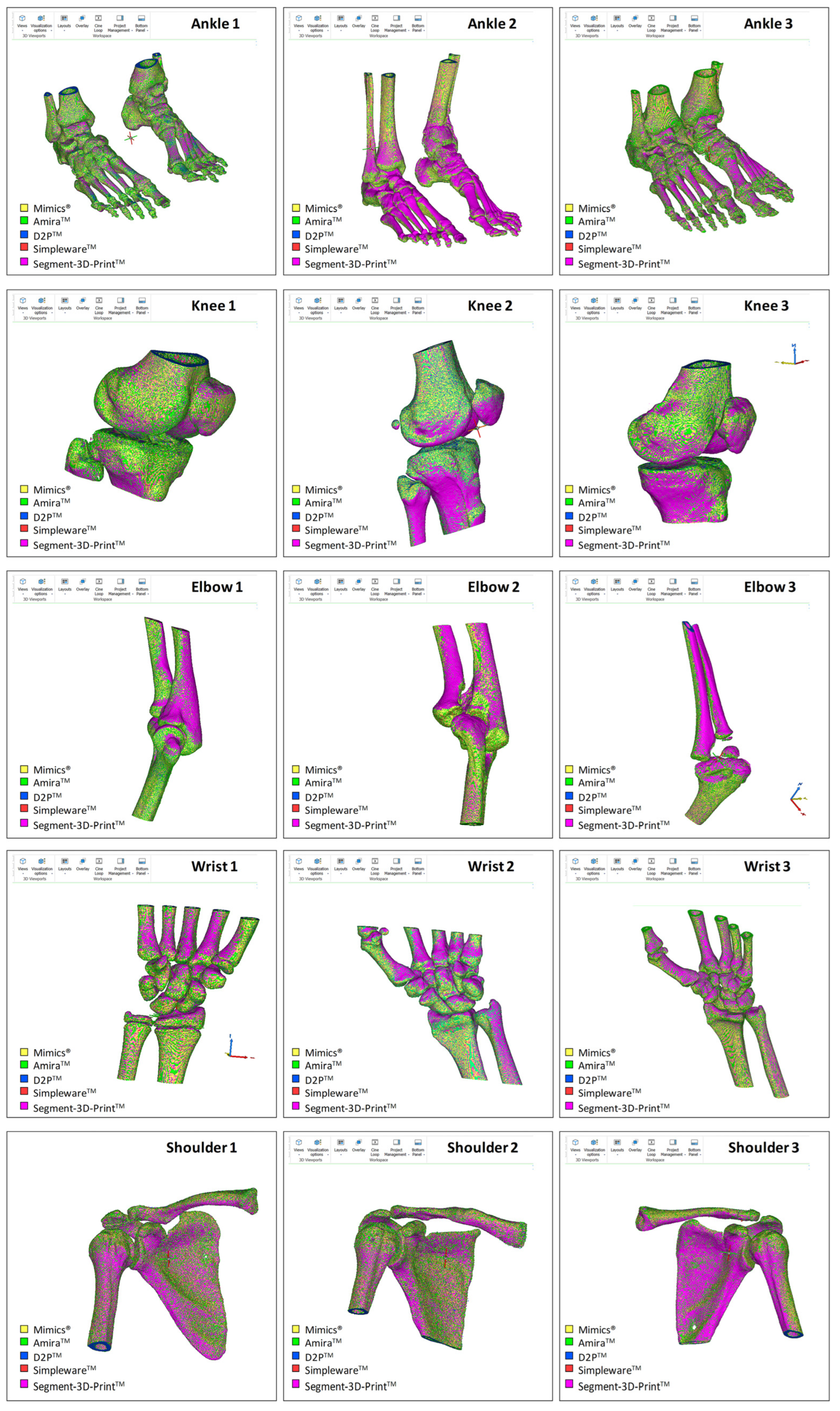
Operability was satisfactory for each of the five software packages; these were efficient enough to obtain final results for all 15 anatomical models in a few hours (Figure 2). By keeping their own default settings in each of the five software packages, Mimics showed the smallest final number of triangles on average over the 15 models, i.e., around half that of the other software packages, which resulted eventually in the smallest and most consistent file size in terms of standard deviation (Table 2). At the end, a very similar ratio of triangles per MByte was observed, approximately 20 thousand, apart from Mimics, where this value was halved. In addition, the corresponding standard deviations reported in % of the mean values reveal a value of approximately 20% for Mimics, and less than 1% in the other software packages.
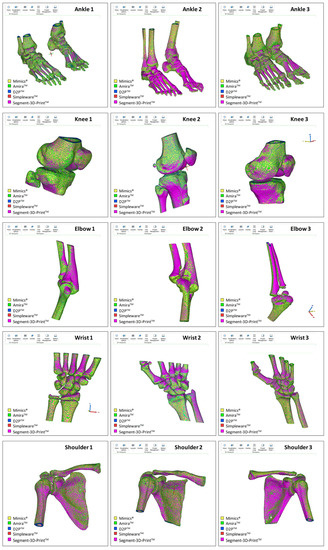
Figure 2.
Superimposition of the resulting meshes from the five commercial software packages investigated; this is shown for each of the 15 DICOM files (three subjects for each of the five different anatomical areas). The STL models shown here for rough comparison were imported into Mimics for the present representation.
4. Discussion
For the first time, the main features of five commercial software packages (Mimics®, AmiraTM, D2PTM, SimplewareTM, and Segment 3D PrintTM) for the generation of STL bone models were investigated. Relevant performance in bone model reconstruction was analyzed by the same operator, using a single computer workstation, and compared accurately in fifteen different CT scans, from five anatomical areas with distinct morphological complexity, by the same operator and using the same computer hardware to avoid possible bias. Amongst the scope, there was also the quantitative comparison of specific features with those from four freeware software packages (3DSlicer, ITK-SNAP, InVesalius, and VuePACS3D—the latter being accessible free-of-charge in our radiological unit), analyzed recently by the same authors [25]. To our knowledge, these nine software packages are amongst the most popular for medical imaging segmentation of the musculo-skeletal system [22,27,28,35]. To make rational and objective comparisons, both the CT scans and the operator were the same in the two studies, and manual intervention was limited as much as possible also in the present analysis. Additionally, all software packages were used on the same computer to avoid the situation wherein segmentation performance is affected by the hardware specifications.
The present work did not seek to investigate either the segmentation algorithms, tools, and features of these commercial software packages, clearly very different from one another, or the degree of automation in 3D model reconstruction, but rather to assess only the main quantitative features and the gross results for comparison. Therefore, apart from a few basic final refinements, only the standard segmentation tools, such as thresholding, were used in each software package for bone shape reconstruction.
The present work obviously has limitations. To limit the cost of the present exercise, access to these commercial software packages was sought in the geographical area of the authors; five amongst the most popular were found and tested with no additional charges. Clearly, many other software packages are available on the market, but it was not possible to further enlarge this exercise, also because the condition of a single operator and a single computer was pursued. In this respect, the operator could not become very familiar with each software package in a short time; therefore, local users, already familiar with these, provided some support, only that necessary to obtain the final results. Threshold values for optimal mask visualization in bone segmentation were set by the operator in each software program, according to the specific density of each bone under analysis but generally in the range of 130–226 Hounsfield unit values, as in the previous work [25]. Of course, these commercial software packages feature many additional tools for manual editing and refinements, particularly with regard to the final number and the density of mesh triangles, but these were not exploited, to maintain the comparison of the initial basic performance. Finally, given the scope of this work, there was no need to distinguish between the different bones within a model, so all bones in each scan were segmented as a single object.
Our findings compare well with recent similar studies in the literature in terms of tested scan resolution, threshold, and accuracy of 3D bone model reconstruction [23,36]. With respect to similar previous studies where features and reconstruction techniques from different software packages are investigated and compared [22,27,28,35], the present work offers quantitative objective outcomes also in terms of the number of triangles, file size, and relevant ratio. In addition, the present analysis was not biased by single anatomical areas or limited scans, but involved full morphological reconstructions from five different anatomical areas with different complexity, overall from fifteen different subjects.
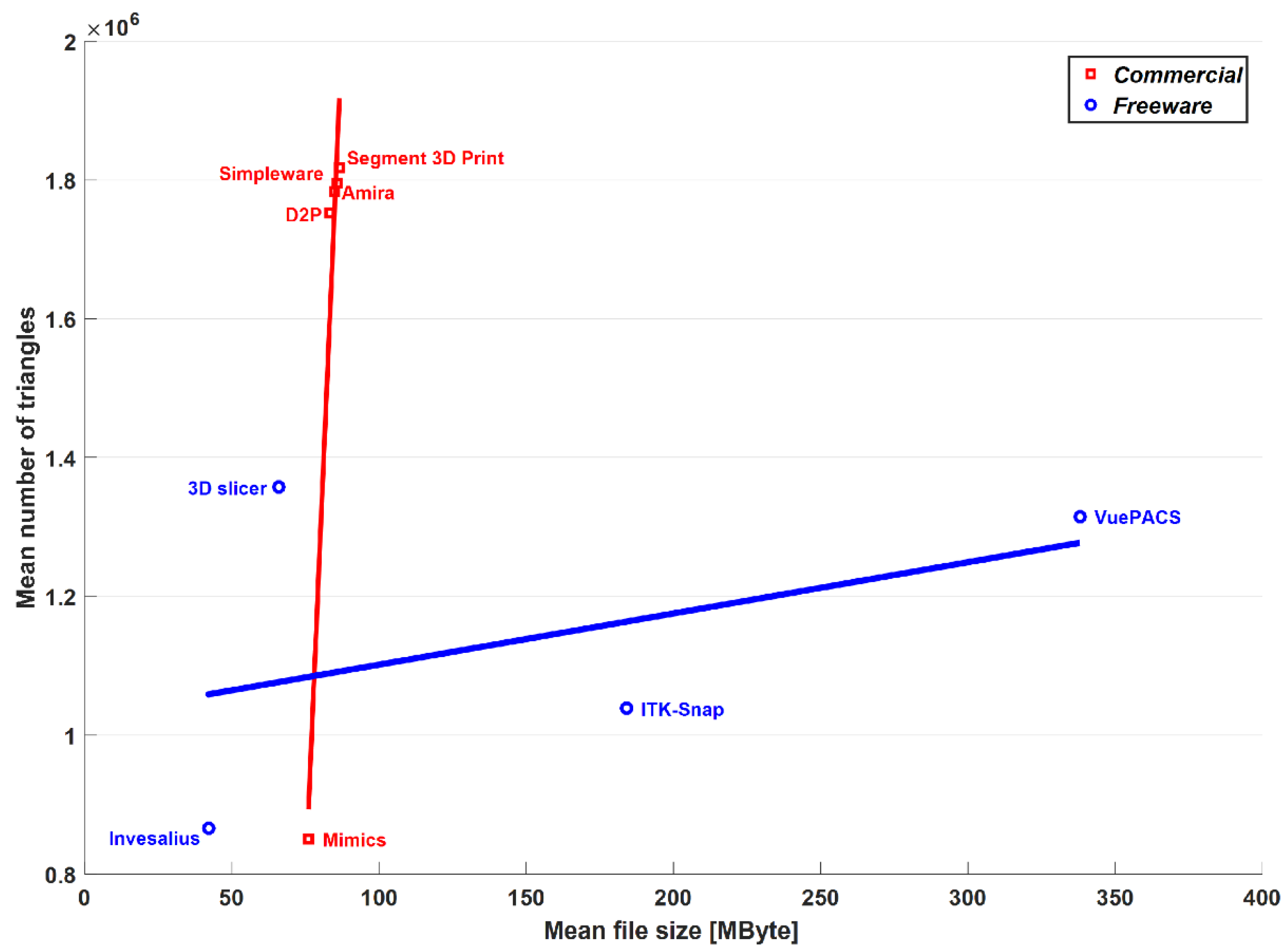
With respect to the freeware software analysis by the present authors [25], the segmentation thresholds and reconstruction algorithms were of course very different. In terms of the mean number of triangles, the four freeware programs were within the range of the commercial software packages, with Mimics® as the minimum (around 850,000) and Segment 3D PrintTM as the maximum (around 1,750,000). The mean file size was found compatible with these differences, as expressed well in the triangles-per-MByte ratio (Table 2), which, apart from MimicsTM, was approximately 20 thousand in the present commercial software packages and in the free 3D SlicerTM (Brigham and Women’s Hospital, Boston, MA, USA) and InVesaliusTM (Renato Archer Information Technology Center, Campinas, Brazil) of the previous paper; in ITK-SNAPTM (PICSL University of Pennsylvania, Philadelphia, PA, USA) and VuePACS3D (CarestreamTM, Rochester, NY, USA), this ratio was, respectively, 5.7 and 3.9 thousand, relatively closer to MimicsTM. However, the ability to export the models both in American Standard Code for Information Interchange (ASCII) and binary STL files gives the option of more readable data for debugging and coding, or less space to store the same amount of data, respectively. Furthermore, it is very interesting to note (Figure 3) that for both the commercial and freeware software packages providing binary code for STL export, there is an overall linear trend between the file size and the number of triangles of the output mesh.
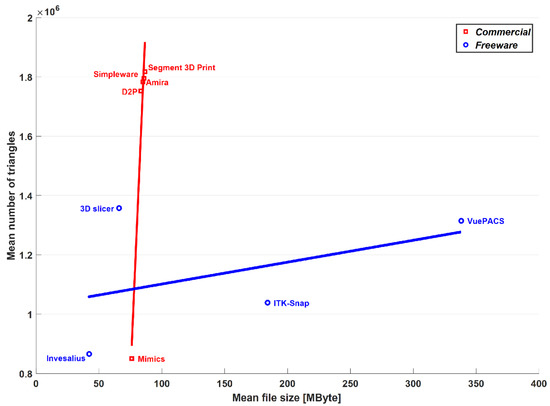
Figure 3.
Graphical representation in terms of mean number of triangles and mean file size of the output meshes over the 15 anatomical models. Results obtained in the present study with commercial software are superimposed to the corresponding results previously obtained by these authors with freeware software on the same models [25]. Corresponding linear regression lines are superimposed for comparison.
Although this behavior is detectable in both the software types, only for the commercial ones is this statistically significant, the correlation coefficient being 0.98, with an associated p-value equal to 0.004. However, despite the features and the good performance of the freeware software packages for 3D bone model reconstruction reported recently by these authors [25], these software packages cannot currently be used worldwide in clinical practice because of the required certification as medical devices, according to the national regulations [34].
5. Conclusions
The present analysis has assessed five commercial software packages and found that the main features are better than those of freeware software packages, as expected. Clearly, only the basic features of these commercial packages were evaluated in the present analysis, whereas their ancillary utility would be a matter for future studies. However, these features shall be assessed also together with other aspects, such as license conditions, costs, accessibility, ease-of-use, etc. Moreover, the intended use of the final 3D bone models should be considered, e.g., whether they are for finite element or musculo-skeletal modeling, prosthesis or custom jig design, clinical research, or medical education. Future relevant work shall compare the results obtained from traditional manual or semi-automatic segmentation tools with modern automatic segmentation software packages, as performed recently for other software packages [37,38].
Author Contributions
Conceptualization: C.B., E.M. and A.L.; methodology: C.B., S.D. and L.C.; software: M.O., K.M. and B.B.; validation: M.O., K.M. and B.B.; formal analysis: C.B., S.D. and L.C.; investigation: C.B., E.M. and A.L.; resources: A.L., S.D. and E.M.; writing—original draft preparation: C.B. and A.L.; writing—review and editing: C.B., M.O., E.M., B.B., K.M., S.D., L.C. and A.L.; visualization: M.O., K.M., B.B. and L.C.; supervision: C.B. and A.L.; project administration: A.L.; funding acquisition: A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Italian Ministry of Economy and Finance, program “5 per mille”.
Informed Consent Statement
Patient consent was waived since DICOM data came from a local DICOM repository and were provided in fully anonymized form.
Data Availability Statement
The authors confirm that the data supporting the results of this study are available within the article.
Conflicts of Interest
All authors declare that there are no personal or commercial relationships related to this work that would lead to a conflict of interest.
References
- Nolte, D.; Tsang, C.K.; Zhang, K.Y.; Ding, Z.; Kedgley, A.E.; Bull, A.M.J. Non-linear scaling of a musculoskeletal model of the lower limb using statistical shape models. J. Biomech. 2016, 49, 3576–3581. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Besier, T.F. Accuracy of femur reconstruction from sparse geometric data using a statistical shape model. Comput. Methods Biomech. Biomed. Eng. 2017, 20, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Nardini, F.; Belvedere, C.; Sancisi, N.; Conconi, M.; Leardini, A.; Durante, S.; Parenti Castelli, V. An Anatomical-Based Subject-Specific Model of In-Vivo Knee Joint 3D Kinematics From Medical Imaging. Appl. Sci. 2020, 10, 2100. [Google Scholar] [CrossRef] [Green Version]
- Osti, F.; Santi, G.M.; Neri, M.; Liverani, A.; Frizziero, L.; Stilli, S.; Maredi, E.; Zarantonello, P.; Gallone, G.; Stallone, S.; et al. CT Conversion Workflow for Intraoperative Usage of Bony Models: From DICOM Data to 3D Printed Models. Appl. Sci. 2019, 9, 708. [Google Scholar] [CrossRef] [Green Version]
- Belvedere, C.; Siegler, S.; Fortunato, A.; Caravaggi, P.; Liverani, E.; Durante, S.; Ensini, A.; Konow, T.; Leardini, A. New comprehensive procedure for custom-made total ankle replacements: Medical imaging, joint modeling, prosthesis design, and 3D printing. J. Orthop. Res. 2019, 37, 760–768. [Google Scholar] [CrossRef]
- Xia, R.Z.; Zhai, Z.J.; Chang, Y.Y.; Li, H.W. Clinical Applications of 3-Dimensional Printing Technology in Hip Joint. Orthop. Surg. 2019, 11, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Galvez, M.; Asahi, T.; Baar, A.; Carcuro, G.; Cuchacovich, N.; Fuentes, J.A.; Mardones, R.; Montoya, C.E.; Negrin, R.; Otayza, F.; et al. Use of Three-dimensional Printing in Orthopaedic Surgical Planning. J. Am. Acad. Orthop. Surg. Glob. Res. Rev. 2018, 2, e071. [Google Scholar] [CrossRef]
- Parthasarathy, J. 3D modeling, custom implants and its future perspectives in craniofacial surgery. Ann. Maxillofac. Surg. 2014, 4, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Malik, H.H.; Darwood, A.R.; Shaunak, S.; Kulatilake, P.; El-Hilly, A.A.; Mulki, O.; Baskaradas, A. Three-dimensional printing in surgery: A review of current surgical applications. J. Surg. Res. 2015, 199, 512–522. [Google Scholar] [CrossRef]
- Martelli, N.; Serrano, C.; van den Brink, H.; Pineau, J.; Prognon, P.; Borget, I.; El Batti, S. Advantages and disadvantages of 3-dimensional printing in surgery: A systematic review. Surgery 2016, 159, 1485–1500. [Google Scholar] [CrossRef]
- Auricchio, F.; Marconi, S. 3D printing: Clinical applications in orthopaedics and traumatology. EFORT Open Rev. 2016, 1, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Belvedere, C.; Cadossi, M.; Mazzotti, A.; Giannini, S.; Leardini, A. Fluoroscopic and Gait Analyses for the Functional Performance of a Custom-Made Total Talonavicular Replacement. J. Foot Ankle Surg. 2017, 56, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S.; Badiali, G.; Cercenelli, L.; Bortolani, B.; Marcelli, E.; Cipriani, R.; Contedini, F.; Marchetti, C.; Tarsitano, A. Combination of CAD/CAM and Augmented Reality in Free Fibula Bone Harvest. Plast. Reconstr. Surg. Glob. Open 2019, 7, e2510. [Google Scholar] [CrossRef] [PubMed]
- Bahraminasab, M. Challenges on optimization of 3D-printed bone scaffolds. Biomed. Eng. Online 2020, 19, 69. [Google Scholar] [CrossRef]
- Van Eijnatten, M.; van Dijk, R.; Dobbe, J.; Streekstra, G.; Koivisto, J.; Wolff, J. CT image segmentation methods for bone used in medical additive manufacturing. Med. Eng. Phys. 2018, 51, 6–16. [Google Scholar] [CrossRef]
- King, A.I. A review of biomechanical models. J. Biomech. Eng. 1984, 106, 97–104. [Google Scholar] [CrossRef]
- Leardini, A.; Belvedere, C.; Nardini, F.; Sancisi, N.; Conconi, M.; Parenti-Castelli, V. Kinematic models of lower limb joints for musculo-skeletal modelling and optimization in gait analysis. J. Biomech. 2017, 62, 77–86. [Google Scholar] [CrossRef]
- Galbusera, F.; Cina, A.; Panico, M.; Albano, D.; Messina, C. Image-based biomechanical models of the musculoskeletal system. Eur. Radiol. Exp. 2020, 4, 49. [Google Scholar] [CrossRef]
- An, G.; Hong, L.; Zhou, X.B.; Yang, Q.; Li, M.Q.; Tang, X.Y. Accuracy and efficiency of computer-aided anatomical analysis using 3D visualization software based on semi-automated and automated segmentations. Ann. Anat. 2017, 210, 76–83. [Google Scholar] [CrossRef]
- Bucking, T.M.; Hill, E.R.; Robertson, J.L.; Maneas, E.; Plumb, A.A.; Nikitichev, D.I. From medical imaging data to 3D printed anatomical models. PLoS ONE 2017, 12, e0178540. [Google Scholar] [CrossRef] [Green Version]
- Durastanti, G.; Leardini, A.; Siegler, S.; Durante, S.; Bazzocchi, A.; Belvedere, C. Comparison of cartilage and bone morphological models of the ankle joint derived from different medical imaging technologies. Quant. Imaging Med. Surg. 2019, 9, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Kresanova, Z.; Kostolny, J. Comparison of Software for Medical Segmentation. Cent. Eur. Res. J. 2018, 4, 66–80. [Google Scholar]
- Tan, C.J.; Parr, W.C.H.; Walsh, W.R.; Makara, M.; Johnson, K.A. Influence of Scan Resolution, Thresholding, and Reconstruction Algorithm on Computed Tomography-Based Kinematic Measurements. J. Biomech. Eng. 2017, 139, 104503. [Google Scholar] [CrossRef] [PubMed]
- Huotilainen, E.; Jaanimets, R.; Valasek, J.; Marcian, P.; Salmi, M.; Tuomi, J.; Makitie, A.; Wolff, J. Inaccuracies in additive manufactured medical skull models caused by the DICOM to STL conversion process. J. Craniomaxillofac. Surg. 2014, 42, e259–e265. [Google Scholar] [CrossRef]
- Matsiushevich, K.; Belvedere, C.; Leardini, A.; Durante, S. Quantitative comparison of freeware software for bone mesh from DICOM files. J. Biomech. 2019, 84, 247–251. [Google Scholar] [CrossRef]
- Lee, L.; Liew, S. A survey of medical image processing tools. In Proceedings of the 4th International Conference on Software Engineering and Computer Systems (ICSECS), Kuantan, Malaysia, 27–29 June 2011; pp. 171–176. [Google Scholar]
- Argüello, D.; Sánchez Acevedo, H.G.; González-Estrada, O.A. Comparison of segmentation tools for structural analysis of bone tissues by finite elements. J. Phys. 2019, 1386, 012113. [Google Scholar] [CrossRef]
- Virzi, A.; Muller, C.O.; Marret, J.B.; Mille, E.; Berteloot, L.; Grevent, D.; Boddaert, N.; Gori, P.; Sarnacki, S.; Bloch, I. Comprehensive Review of 3D Segmentation Software Tools for MRI Usable for Pelvic Surgery Planning. J. Digit. Imaging 2020, 33, 99–110. [Google Scholar] [CrossRef]
- Fourie, Z.; Damstra, J.; Schepers, R.H.; Gerrits, P.O.; Ren, Y. Segmentation process significantly influences the accuracy of 3D surface models derived from cone beam computed tomography. Eur. J. Radiol. 2012, 81, e524–e530. [Google Scholar] [CrossRef]
- Kamio, T.; Suzuki, M.; Asaumi, R.; Kawai, T. DICOM segmentation and STL creation for 3D printing: A process and software package comparison for osseous anatomy. 3D Print Med. 2020, 6, 17. [Google Scholar] [CrossRef]
- Ahn, C.; Bui, T.D.; Lee, Y.W.; Shin, J.; Park, H. Fully automated, level set-based segmentation for knee MRIs using an adaptive force function and template: Data from the osteoarthritis initiative. Biomed. Eng. Online 2016, 15, 99. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Jian, F.; Wu, H.; Li, H. An improved level set method for vertebra CT image segmentation. Biomed. Eng. Online 2013, 12, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sander, I.M.; McGoldrick, M.T.; Helms, M.N.; Betts, A.; van Avermaete, A.; Owers, E.; Doney, E.; Liepert, T.; Niebur, G.; Liepert, D.; et al. Three-dimensional printing of X-ray computed tomography datasets with multiple materials using open-source data processing. Anat. Sci. Educ. 2017, 10, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Lipprandt, M.; Röhrig, R.; Neumuth, T. Digital health—Software as a medical device in focus of the medical device regulation (MDR). IT Inf. Technol. 2019, 61, 211–218. [Google Scholar] [CrossRef]
- Wallner, J.; Schwaiger, M.; Hochegger, K.; Gsaxner, C.; Zemann, W.; Egger, J. A review on multiplatform evaluations of semi-automatic open-source based image segmentation for cranio-maxillofacial surgery. Comput. Methods Programs Biomed. 2019, 182, 105102. [Google Scholar] [CrossRef]
- Soodmand, E.; Kluess, D.; Varady, P.A.; Cichon, R.; Schwarze, M.; Gehweiler, D.; Niemeyer, F.; Pahr, D.; Woiczinski, M. Interlaboratory comparison of femur surface reconstruction from CT data compared to reference optical 3D scan. Biomed. Eng. Online 2018, 17, 29. [Google Scholar] [CrossRef] [Green Version]
- Ortolani, M.; Leardini, A.; Pavani, C.; Scicolone, S.; Girolami, M.; Bevoni, R.; Lullini, G.; Durante, S.; Berti, L.; Belvedere, C. Angular and linear measurements of adult flexible flatfoot via weight-bearing CT scans and 3D bone reconstruction tools. Sci. Rep. 2021, 11, 16139. [Google Scholar] [CrossRef]
- De Carvalho, K.A.M.; Walt, J.S.; Ehret, A.; Tazegul, T.E.; Dibbern, K.; Mansur, N.S.B.; Lalevee, M.; de Cesar Netto, C. Comparison between Weightbearing-CT semiautomatic and manual measurements in Hallux Valgus. Foot Ankle Surg. 2022, 28, 518–525. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).