Performance Analysis for COVID-19 Diagnosis Using Custom and State-of-the-Art Deep Learning Models
Abstract
:1. Introduction
- To the best of our knowledge, this study is the first that benchmarks deep learning models on the largest COVID-19 dataset.
- In this study, we developed a custom CNN model for benchmarking on the subject dataset.
- The custom CNN model outperformed MobileNetV2 in terms of precision, which is of high scientific value.
- The current study includes a detailed and comprehensive evaluation of the performance involving several performance metrics to observe the impact of increasing the COVID-19 dataset on the research work.
2. Related Work
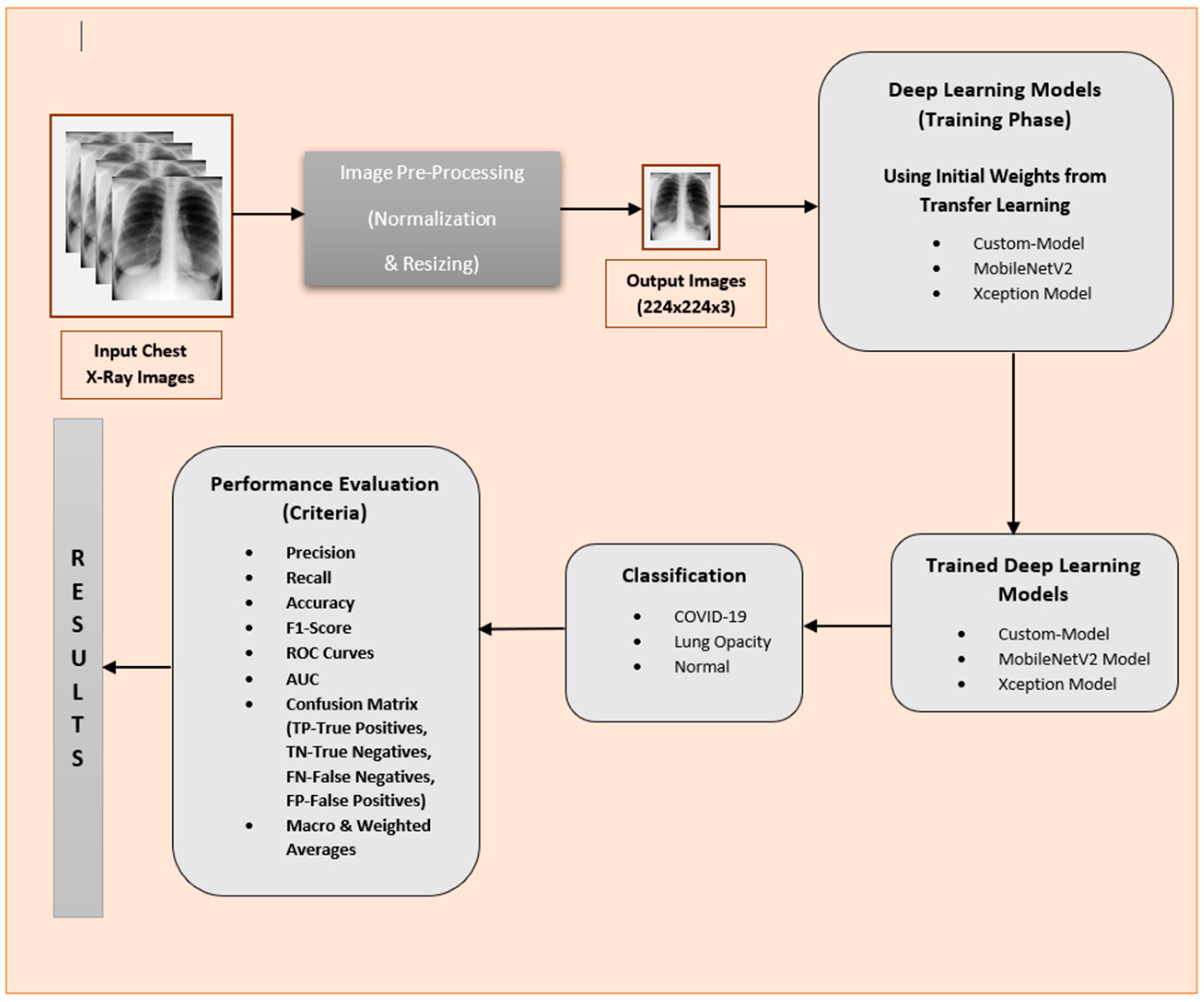
3. Methodology
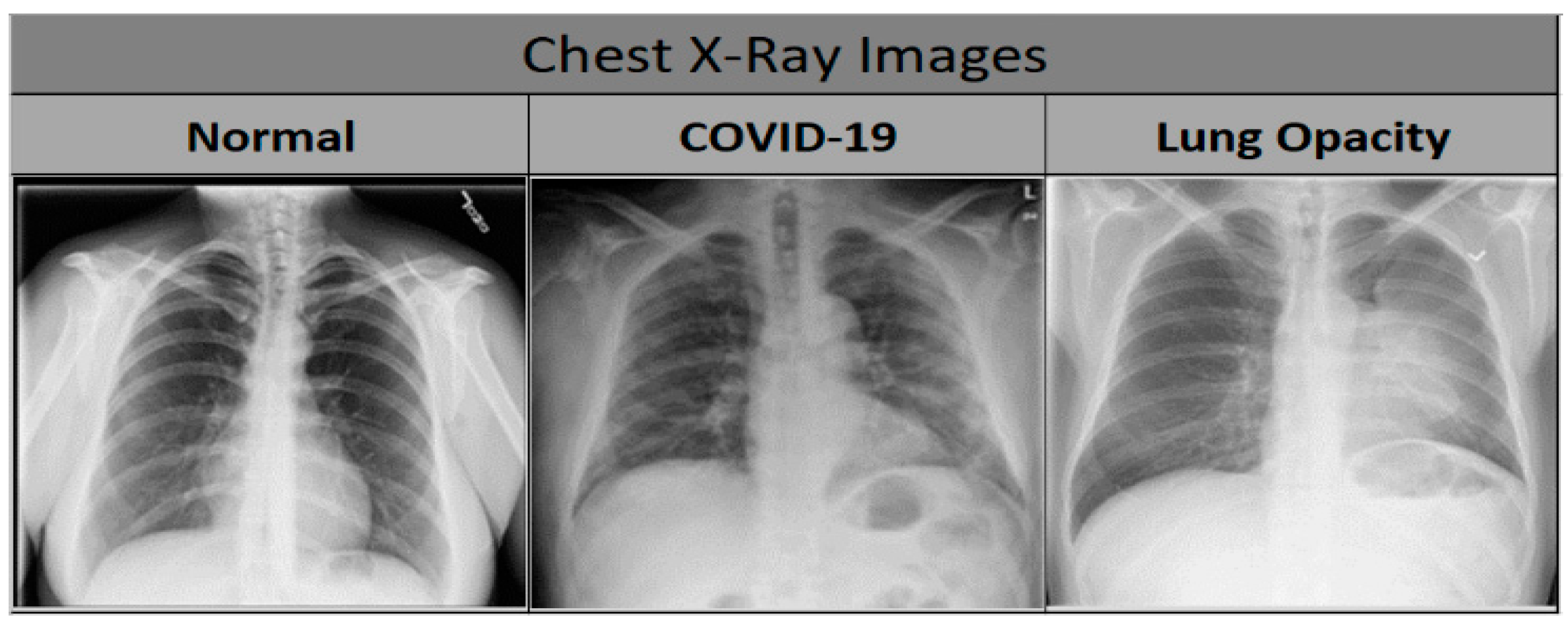
3.1. Dataset
3.2. Approach
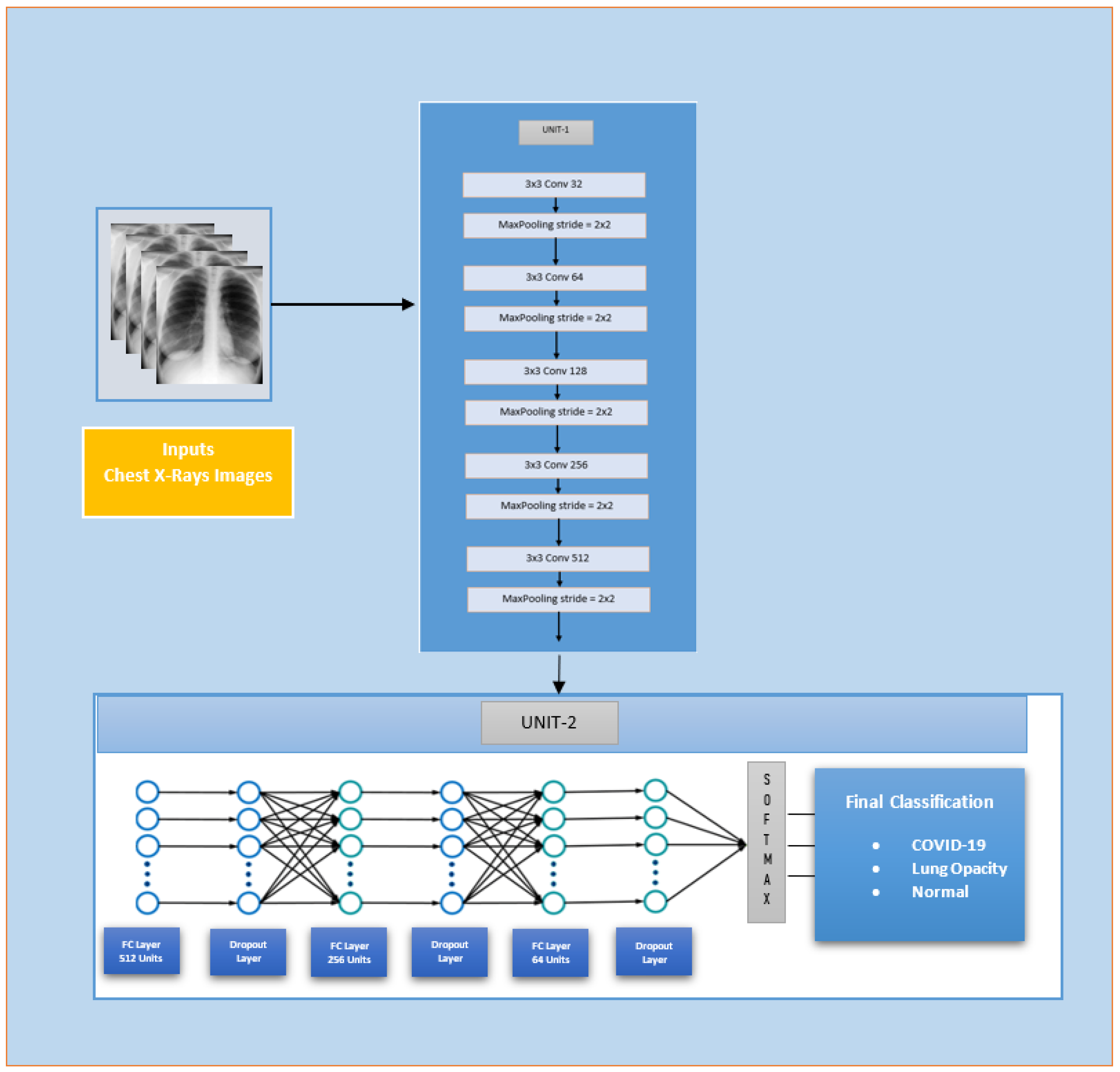
3.2.1. Custom-Model
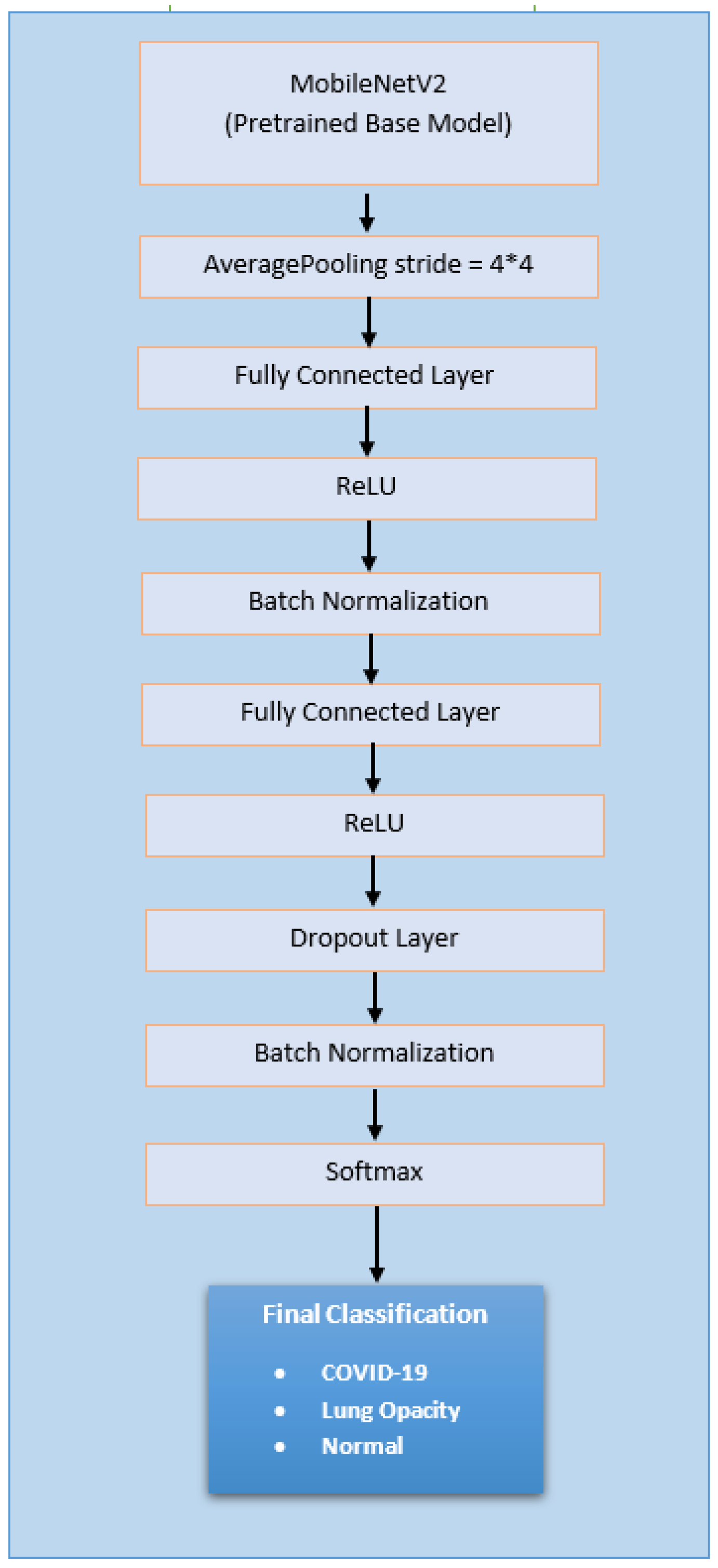
3.2.2. Deep Learning Model-1 (Extended Mobilenetv2 Model)
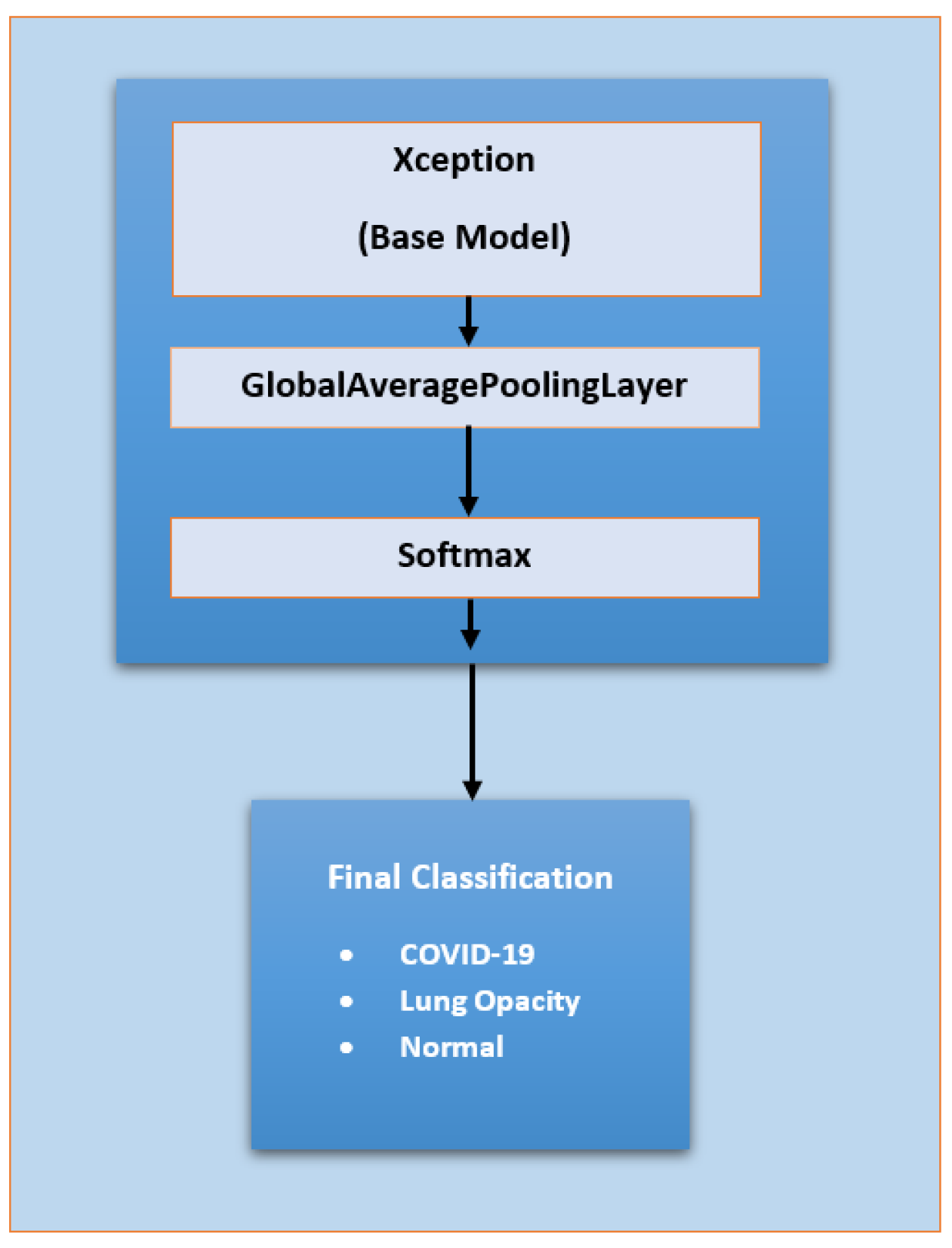
3.2.3. Deep Learning Model-2 (Extended Xception Model)
3.3. Experimental Setup
3.3.1. System and Software Setup
3.3.2. Input Setup
3.3.3. Hyperparameter Choice
3.3.4. Train–Test Ratio
3.3.5. Evaluation Metrics and Tools
- Precision
- 2.
- Recall
- 3.
- F1 Score
- 4.
- Accuracy
- 5.
- ROC (Receiver Operating Characteristic) Curve
- 6.
- Confusion Matrix
4. Results
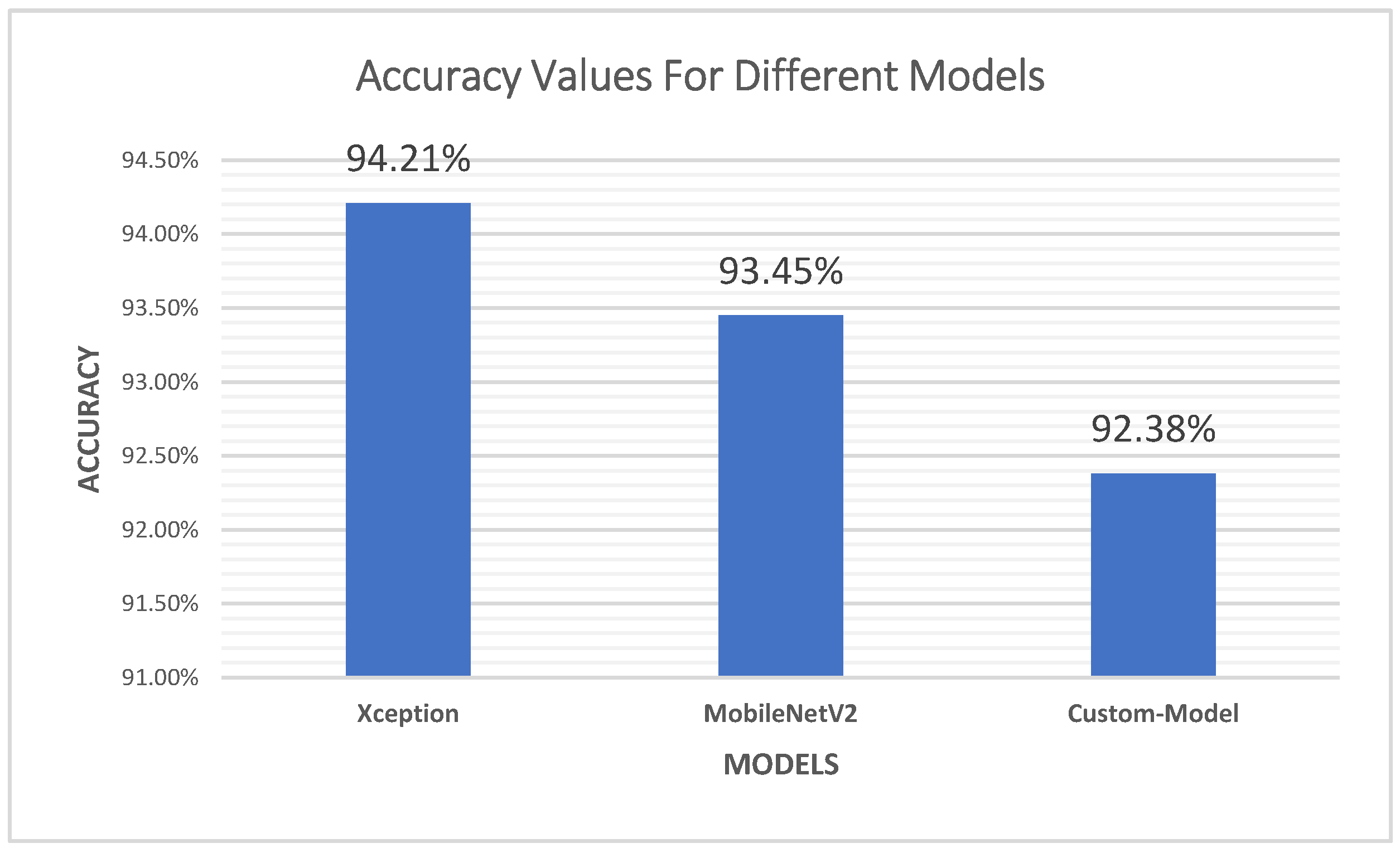
4.1. Accuracy
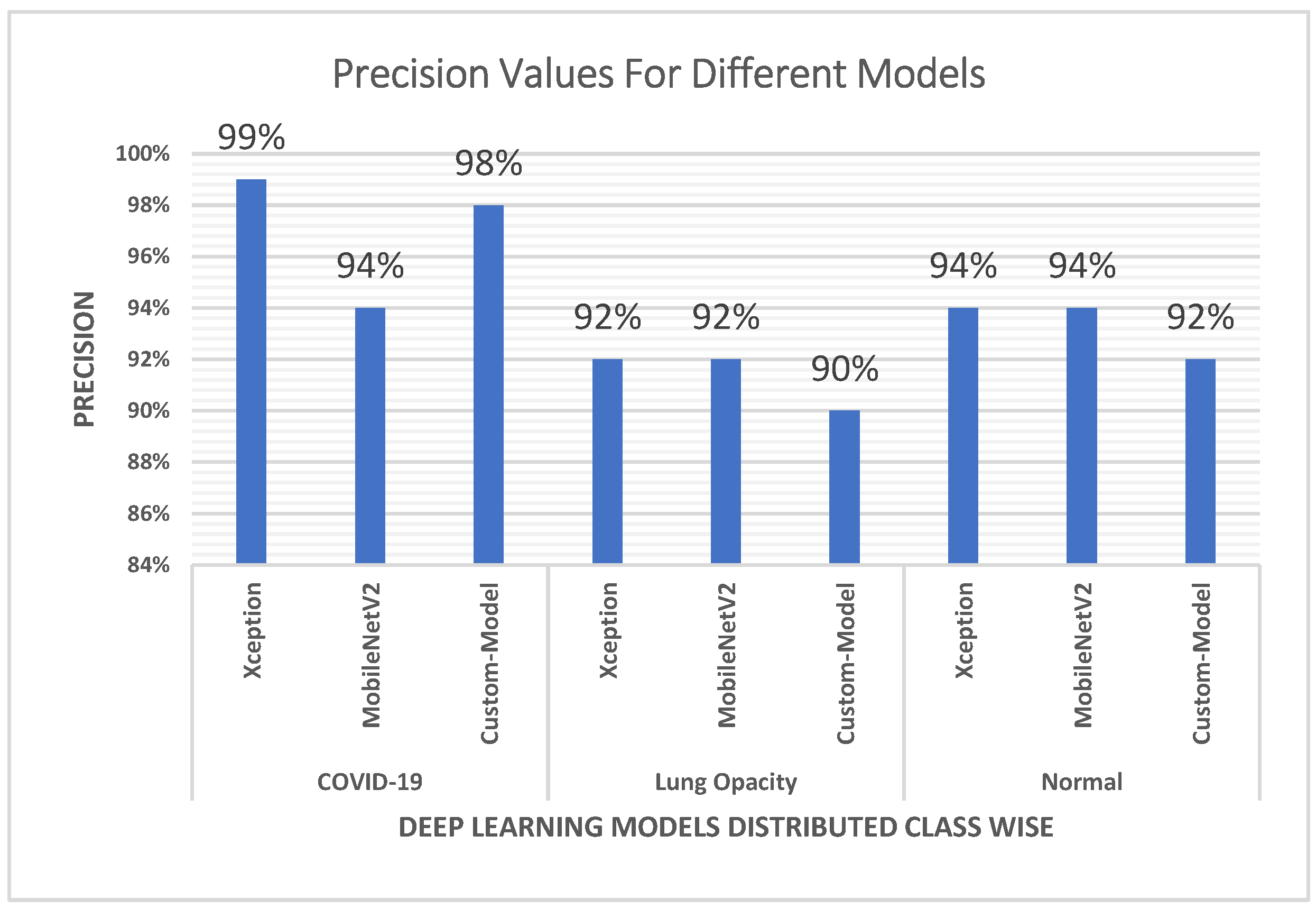
4.2. Precision
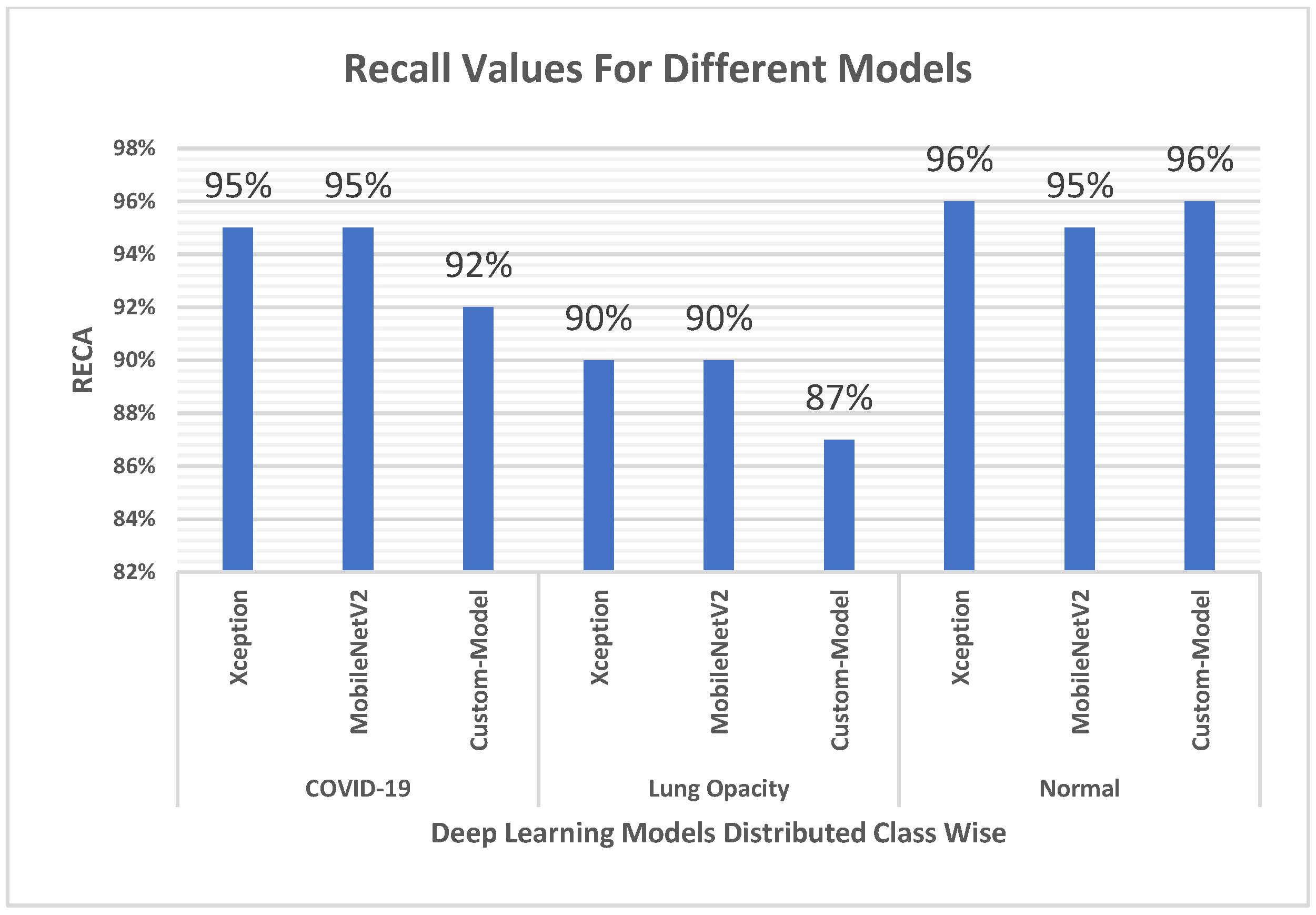
4.3. Recall
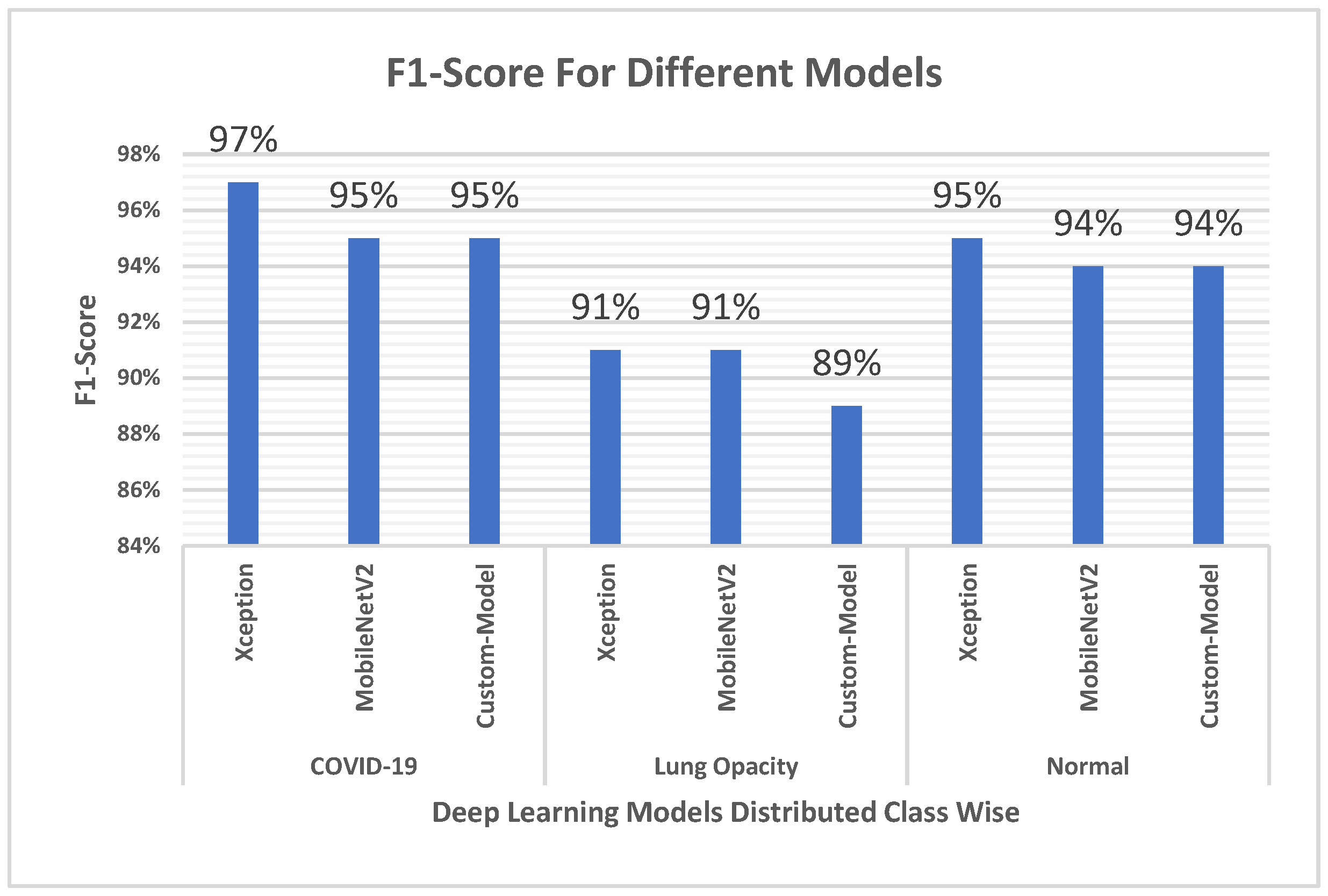
4.4. F1 Score
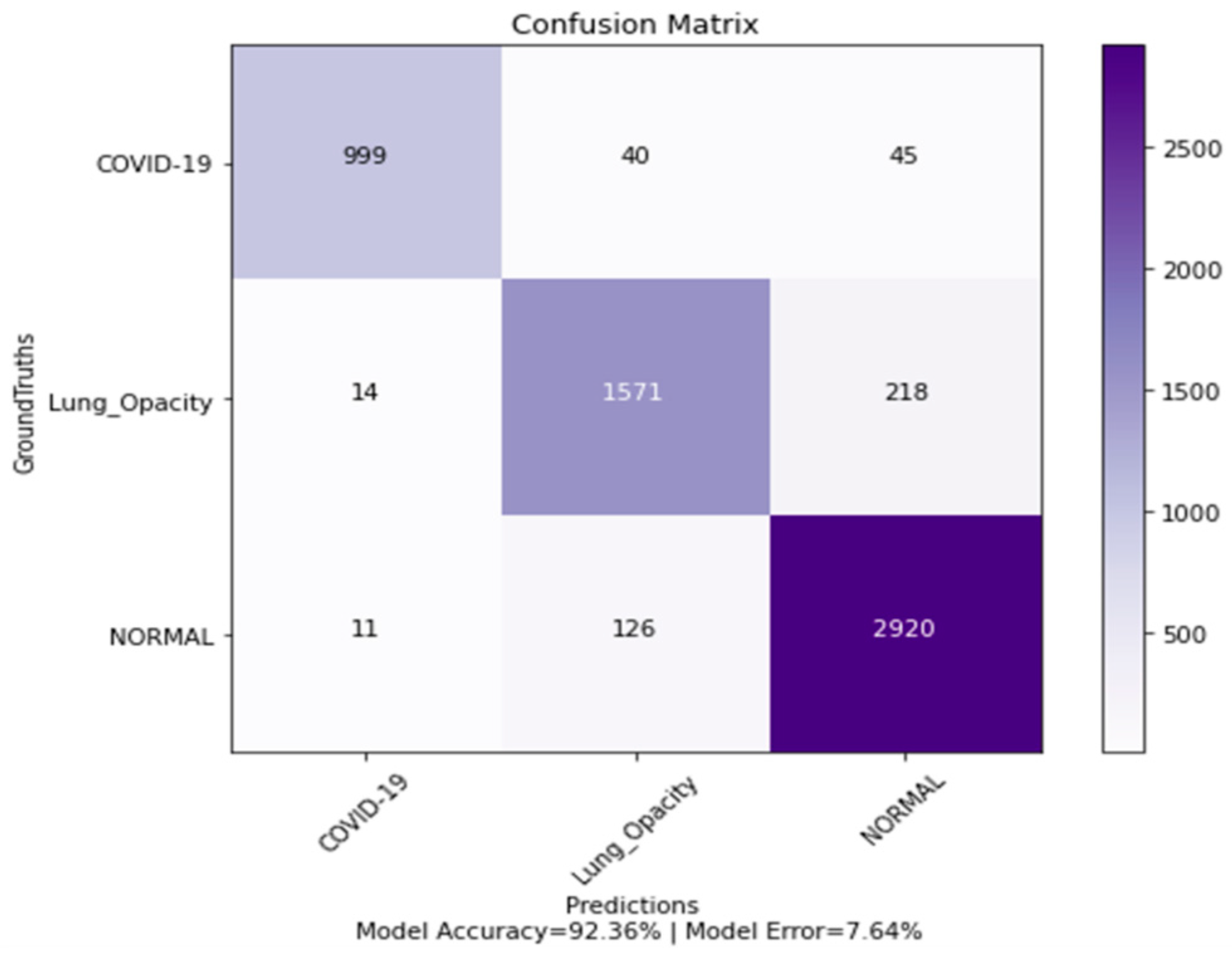
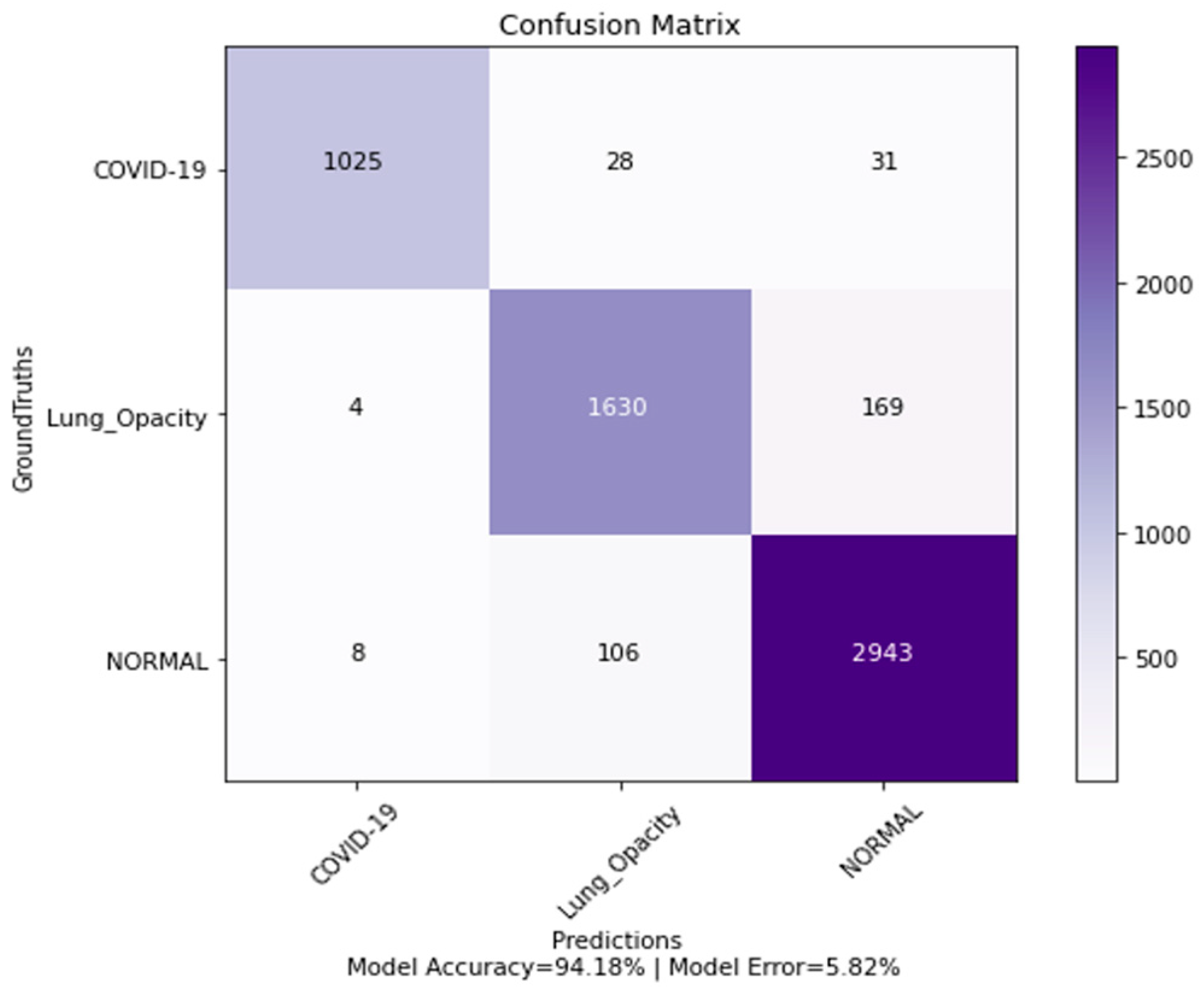
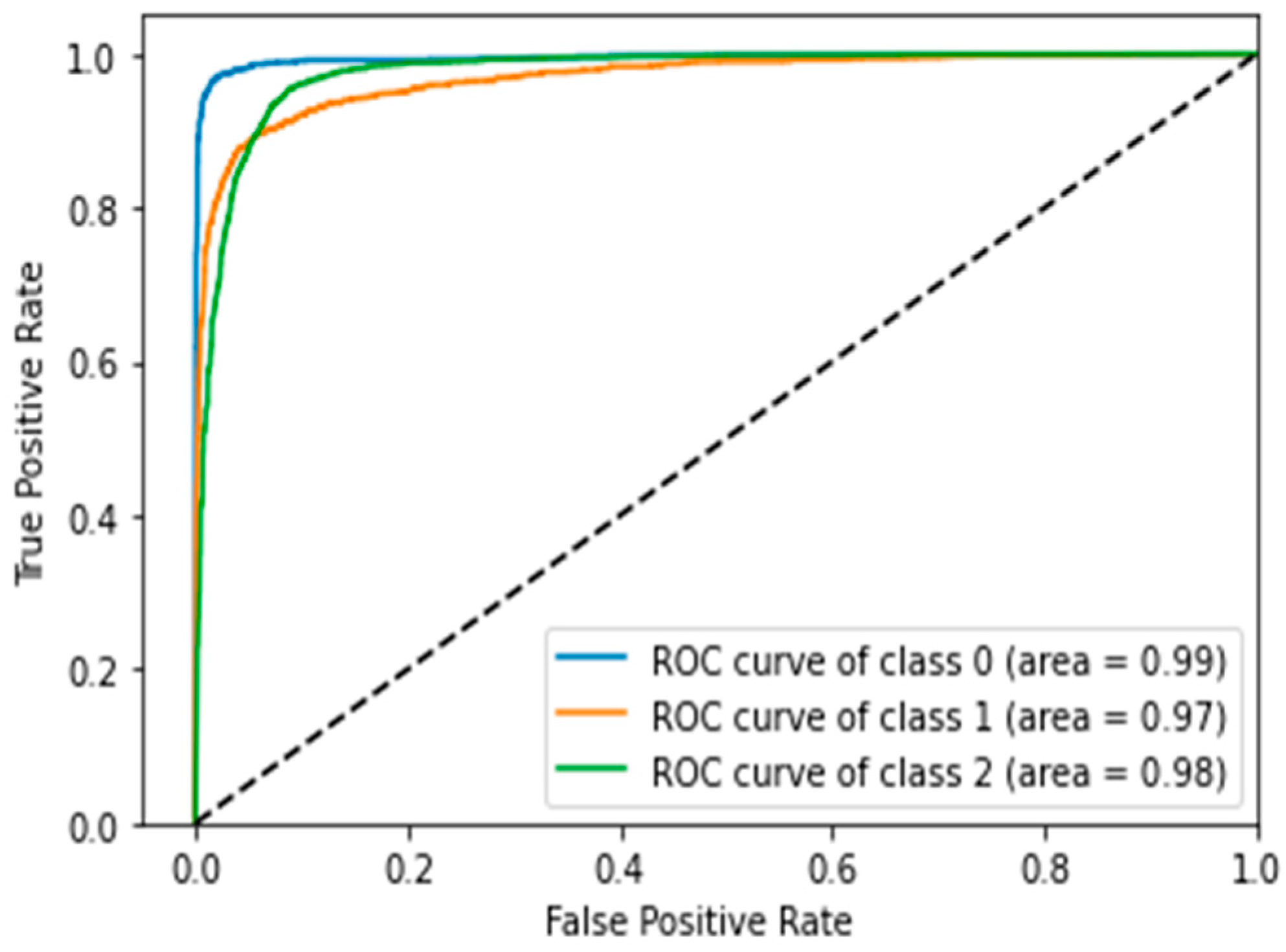
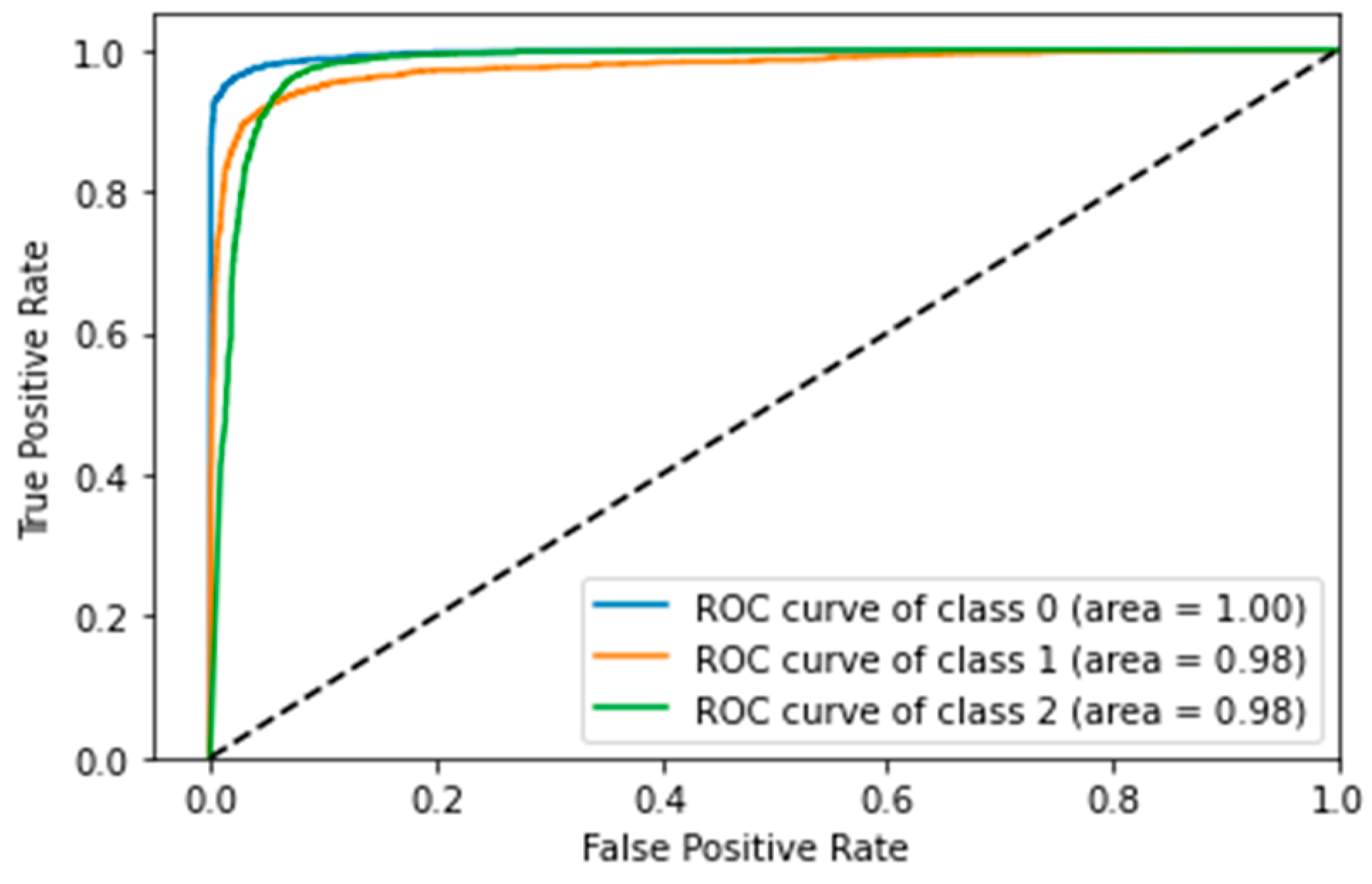
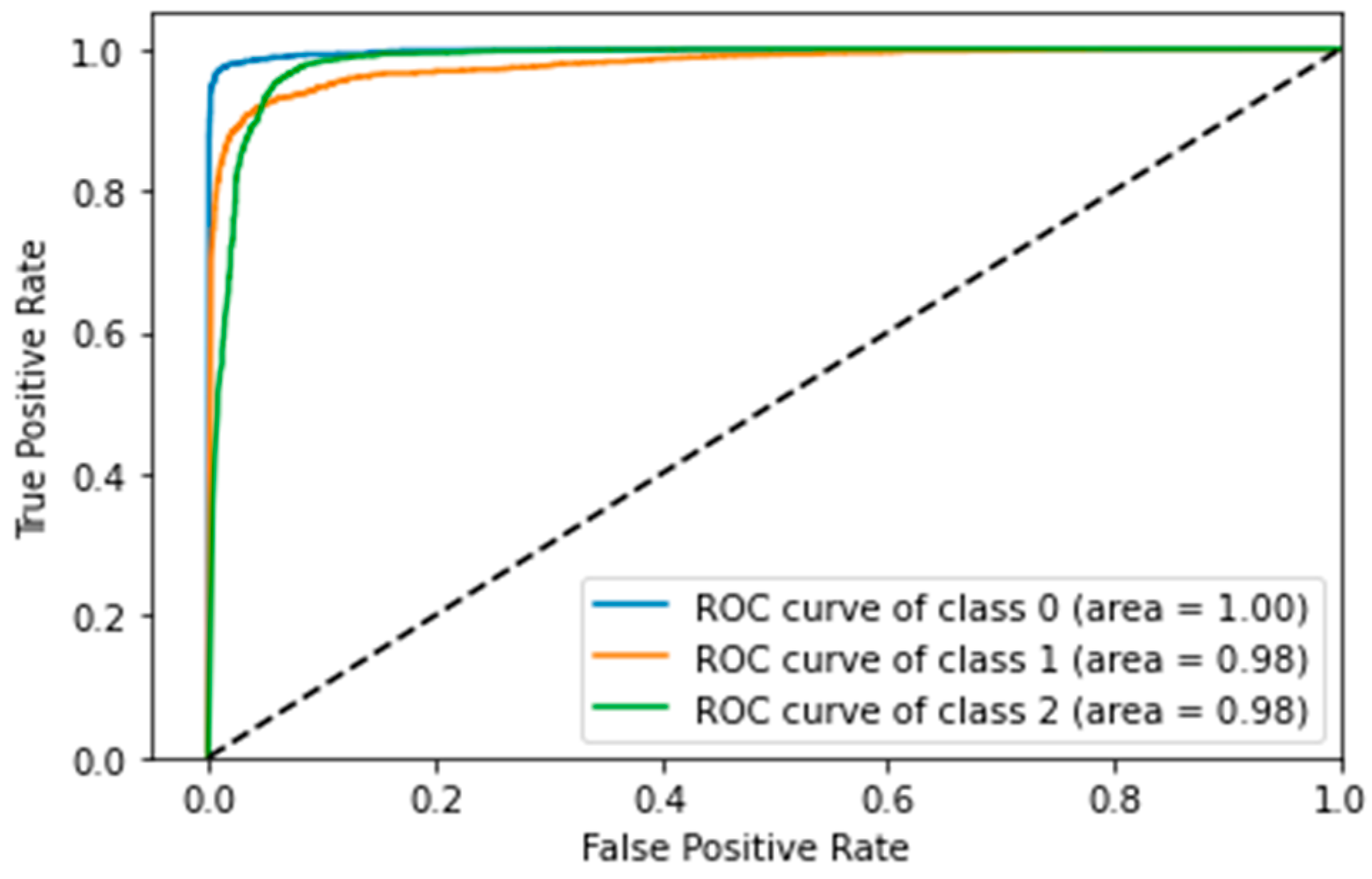
4.5. Confusion Matrix, ROC Curves and Area under Curve
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hasoon, J.N.; Fadel, A.H.; Hameed, R.S.; Mostafa, S.A.; Khalaf, B.A.; Mohammed, M.A.; Nedoma, J. COVID-19 anomaly detection and classification method based on supervised machine learning of chest X-ray images. Results Phys. 2021, 31, 105045. [Google Scholar] [CrossRef] [PubMed]
- Awan, M.J.; Bilal, M.H.; Yasin, A.; Nobanee, H.; Khan, N.S.; Zain, A.M. Detection of COVID-19 in Chest X-ray Images: A Big Data Enabled Deep Learning Approach. Int. J. Environ. Res. Public Health 2021, 18, 10147. [Google Scholar] [CrossRef] [PubMed]
- Haafza, L.A.; Awan, M.J.; Abid, A.; Yasin, A.; Nobanee, H.; Farooq, M.S. Big Data COVID-19 Systematic Literature Review: Pandemic Crisis. Electronics 2021, 10, 3125. [Google Scholar] [CrossRef]
- Gupta, M.; Jain, R.; Arora, S.; Gupta, A.; Javed Awan, M.; Chaudhary, G.; Nobanee, H. AI-enabled COVID-19 Outbreak Analysis and Prediction: Indian States vs. Union Territories. Comput. Mater. Contin. 2021, 67, 933–950. [Google Scholar] [CrossRef]
- Awan, M.J.; Imtiaz, M.W.; Usama, M.; Rehman, A.; Ayesha, N.; Shehzad, H.M.F. COVID-19 Detection by using Deep learning-based Custom Convolution Neural Network (CNN). In Proceedings of the 2021 International Conference on Innovative Computing (ICIC), Lahore, Pakistan, 9–10 November 2021; pp. 1–7. [Google Scholar]
- Abdulkareem, K.H.; Mostafa, S.A.; Al-Qudsy, Z.N.; Mohammed, M.A.; Al-Waisy, A.S.; Kadry, S.; Lee, J.; Nam, Y. Automated System for Identifying COVID-19 Infections in Computed Tomography Images Using Deep Learning Models. J. Healthc. Eng. 2022, 2022, 5329014. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, R.; Benameur, N.; Mabrouk, R.; Mohammed, M.A.; Garcia-Zapirain, B.; Bedoui, M.H. A Deep Learning-Based Diagnosis System for COVID-19 Detection and Pneumonia Screening Using CT Imaging. Appl. Sci. 2022, 12, 4825. [Google Scholar] [CrossRef]
- Ibrahim, D.A.; Zebari, D.A.; Mohammed, H.J.; Mohammed, M.A. Effective hybrid deep learning model for COVID-19 patterns identification using CT images. Expert Syst. 2022, e13010. [Google Scholar] [CrossRef]
- Alyasseri, Z.A.A.; Al-Betar, M.A.; Doush, I.A.; Awadallah, M.A.; Abasi, A.K.; Makhadmeh, S.N.; Alomari, O.A.; Abdulkareem, K.H.; Adam, A.; Damasevicius, R.; et al. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2022, 39, e12759. [Google Scholar] [CrossRef]
- Awan, M.J.; Rahim, M.S.M.; Salim, N.; Rehman, A.; Nobanee, H.; Shabir, H. Improved Deep Convolutional Neural Network to Classify Osteoarthritis from Anterior Cruciate Ligament Tear Using Magnetic Resonance Imaging. J. Pers. Med. 2021, 11, 1163. [Google Scholar] [CrossRef]
- Ali, Y.; Farooq, A.; Alam, T.M.; Farooq, M.S.; Awan, M.J.; Baig, T.I. Detection of Schistosomiasis Factors Using Association Rule Mining. IEEE Access 2019, 7, 186108–186114. [Google Scholar] [CrossRef]
- Vaishnav, P.K.; Sharma, S.; Sharma, P. Analytical review analysis for screening COVID-19 disease. Int. J. Mod. Res. 2021, 1, 22–29. [Google Scholar]
- Tariq, A.; Awan, M.J.; Alshudukhi, J.; Alam, T.M.; Alhamazani, K.T.; Meraf, Z. Software Measurement by Using Artificial Intelligence. J. Nanomater. 2022, 2022, 7283171. [Google Scholar] [CrossRef]
- Nagi, A.T.; Wali, A.; Shahzada, A.; Ahmad, M.M. A Parellel two Stage Classifier for Breast Cancer Prediction and Comparison with Various Ensemble Techniques. VAWKUM Trans. Comput. Sci. 2018, 15, 121. [Google Scholar] [CrossRef]
- Naseem, U.; Rashid, J.; Ali, L.; Kim, J.; Emad Ul Haq, Q.; Awan, M.J.; Imran, M. An Automatic Detection of Breast Cancer Diagnosis and Prognosis based on Machine Learning Using Ensemble of Classifiers. IEEE Access 2022. [Google Scholar] [CrossRef]
- Awan, M.J.; Mohd Rahim, M.S.; Salim, N.; Rehman, A.; Nobanee, H. Machine Learning-Based Performance Comparison to Diagnose Anterior Cruciate Ligament Tears. J. Healthc. Eng. 2022, 2022, 255012. [Google Scholar] [CrossRef]
- Sharma, T.; Nair, R.; Gomathi, S. Breast Cancer Image Classification using Transfer Learning and Convolutional Neural Network. Int. J. Mod. Res. 2022, 2, 8–16. [Google Scholar]
- Allioui, H.; Mohammed, M.A.; Benameur, N.; Al-Khateeb, B.; Abdulkareem, K.H.; Garcia-Zapirain, B.; Damaševičius, R.; Maskeliūnas, R. A multi-agent deep reinforcement learning approach for enhancement of COVID-19 CT image segmentation. J. Pers. Med. 2022, 12, 309. [Google Scholar] [CrossRef]
- Shamim, S.; Awan, M.J.; Mohd Zain, A.; Naseem, U.; Mohammed, M.A.; Garcia-Zapirain, B. Automatic COVID-19 Lung Infection Segmentation through Modified Unet Model. J. Healthc. Eng. 2022, 2022, 6566982. [Google Scholar] [CrossRef]
- Albahli, A.S.; Algsham, A.; Aeraj, S.; Alsaeed, M.; Alrashed, M.; Rauf, H.T.; Arif, M.; Mohammed, M.A. COVID-19 public sentiment insights: A text mining approach to the Gulf countries. CMC Comput. Mater. Contin. 2021, 67, 1613–1627. [Google Scholar] [CrossRef]
- Obaid, O.I.; Mohammed, M.A.; Mostafa, S.A. Long Short-Term Memory Approach for Coronavirus Disease Predicti. J. Inf. Technol. Manag. 2020, 12, 11–21. [Google Scholar]
- Rousan, L.A.; Elobeid, E.; Karrar, M.; Khader, Y. Chest X-ray findings and temporal lung changes in patients with COVID-19 pneumonia. BMC Pulm. Med. 2020, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Ben Jabra, M.; Koubaa, A.; Benjdira, B.; Ammar, A.; Hamam, H. COVID-19 diagnosis in chest X-rays using deep learning and majority voting. Appl. Sci. 2021, 11, 2884. [Google Scholar] [CrossRef]
- Chandra, T.B.; Verma, K.; Singh, B.K.; Jain, D.; Netam, S.S. Coronavirus disease (COVID-19) detection in chest X-ray images using majority voting based classifier ensemble. Expert Syst. Appl. 2021, 165, 113909. [Google Scholar] [CrossRef] [PubMed]
- Heidari, M.; Mirniaharikandehei, S.; Khuzani, A.Z.; Danala, G.; Qiu, Y.; Zheng, B. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inform. 2020, 144, 104284. [Google Scholar] [CrossRef]
- Al-Waisy, A.S.; Al-Fahdawi, S.; Mohammed, M.A.; Abdulkareem, K.H.; Mostafa, S.A.; Maashi, M.S.; Arif, M.; Garcia-Zapirain, B. COVID-CheXNet: Hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020. [Google Scholar] [CrossRef]
- Awan, M.J.; Rahim, M.S.M.; Salim, N.; Mohammed, M.A.; Garcia-Zapirain, B.; Abdulkareem, K.H. Efficient Detection of Knee Anterior Cruciate Ligament from Magnetic Resonance Imaging Using Deep Learning Approach. Diagnostics 2021, 11, 105. [Google Scholar] [CrossRef]
- Vaid, S.; Kalantar, R.; Bhandari, M. Deep learning COVID-19 detection bias: Accuracy through artificial intelligence. Int. Orthop. 2020, 44, 1539–1542. [Google Scholar] [CrossRef]
- Albahli, S.; Albattah, W. Detection of coronavirus disease from X-ray images using deep learning and transfer learning algorithms. J. X-ray Sci. Technol. 2020, 28, 841–850. [Google Scholar] [CrossRef]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Ismael, A.M.; Şengür, A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021, 164, 114054. [Google Scholar] [CrossRef]
- Awan, M.J.; Masood, O.A.; Mohammed, M.A.; Yasin, A.; Zain, A.M.; Damaševičius, R.; Abdulkareem, K.H. Image-Based Malware Classification Using VGG19 Network and Spatial Convolutional Attention. Electronics 2021, 10, 2444. [Google Scholar] [CrossRef]
- Das, A.K.; Ghosh, S.; Thunder, S.; Dutta, R.; Agarwal, S.; Chakrabarti, A. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal. Appl. 2021, 24, 1111–1124. [Google Scholar] [CrossRef]
- Gianchandani, N.; Jaiswal, A.; Singh, D.; Kumar, V.; Kaur, M. Rapid COVID-19 diagnosis using ensemble deep transfer learning models from chest radiographic images. J. Ambient. Intell. Humaniz. Comput. 2020. [Google Scholar] [CrossRef] [PubMed]
- Marques, G.; Agarwal, D.; Díez, I.D.L.T. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020, 96, 106691. [Google Scholar] [CrossRef]
- Maghdid, H.S.; Asaad, A.T.; Ghafoor, K.Z.; Sadiq, A.S.; Mirjalili, S.; Khan, M.K. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. Multimodal Image Exploit. Learn. 2021, 11734, 99–110. [Google Scholar]
- Hall, L.O.; Paul, R.; Goldgof, D.B.; Goldgof, G.M. Finding COVID-19 from Chest X-rays using Deep Learning on a Small Dataset. arXiv 2020, arXiv:2004.02060. [Google Scholar]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020, 65, 101794. [Google Scholar] [CrossRef]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef]
- Pandit, M.K.; Banday, S.A. SARS n-CoV2-19 detection from chest X-ray images using deep neural networks. Int. J. Pervasive Comput. Commun. 2020, 16, 419–427. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, D.M.; Khandakar, A. COVID-19 Radiography Database. 2021. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database (accessed on 19 March 2022).
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al Emadi, N. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 2020, 8, 132665–132676. [Google Scholar] [CrossRef]
- Mujahid, A.; Awan, M.J.; Yasin, A.; Mohammed, M.A.; Damaševičius, R.; Maskeliūnas, R.; Abdulkareem, K.H. Real-Time Hand Gesture Recognition Based on Deep Learning YOLOv3 Model. Appl. Sci. 2021, 11, 4164. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Wu, J. Introduction to convolutional neural networks. Natl. Key Lab. Nov. Softw. Technol. Nanjing Univ. China 2017, 5, 495. [Google Scholar]
- Albawi, S.; Bayat, O.; Al-Azawi, S.; Ucan, O.N. Social touch gesture recognition using convolutional neural network. Comput. Intell. Neurosci. 2018, 2018, 697310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awan, M.; Rahim, M.; Salim, N.; Ismail, A.; Shabbir, H. Acceleration of knee MRI cancellous bone classification on google colaboratory using convolutional neural network. Int. J. Adv. Trends Comput. Sci. 2019, 8, 83–88. [Google Scholar] [CrossRef]
- Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258.
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ’16), Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Bengio, Y. Practical recommendations for gradient-based training of deep architectures. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Dhont, J.; Wolfs, C.; Verhaegen, F. Automatic COVID-19 diagnosis based on chest radiography and deep learning—Success story or dataset bias? Med. Phys. 2021, 49, 978–987. [Google Scholar] [CrossRef]
- López-Cabrera, J.D.; Orozco-Morales, R.; Portal-Diaz, J.A.; Lovelle-Enríquez, O.; Pérez-Díaz, M. Current limitations to identify COVID-19 using artificial intelligence with chest X-ray imaging. Health Technol. 2021, 11, 411–424. [Google Scholar] [CrossRef]
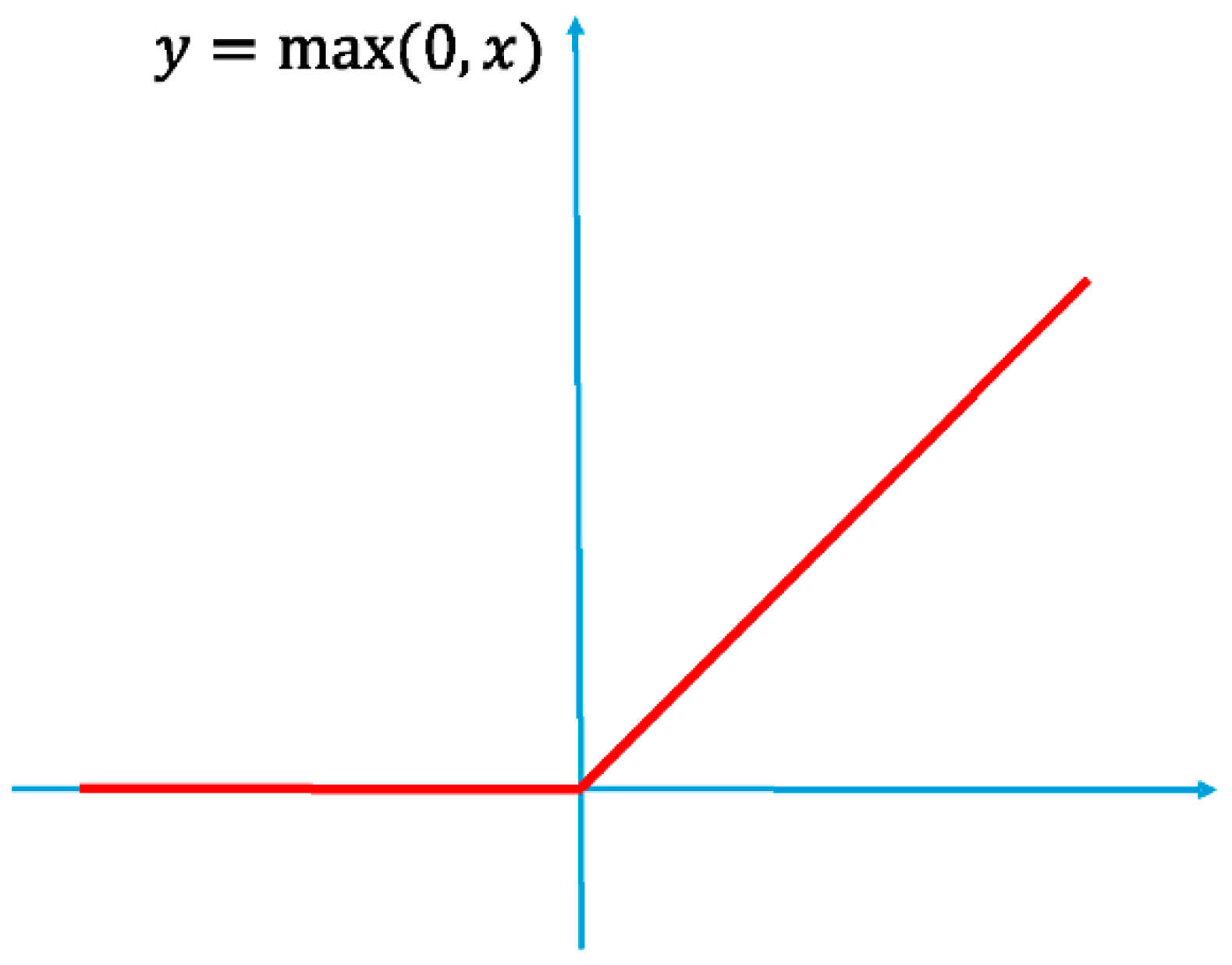

| Architecture | Optimizer | Learning Rate | Batch Size | Image Shuffling in Batches | Number of Epochs for Convergence | Patience | Number of Epochs |
|---|---|---|---|---|---|---|---|
| Custom-Model | Adam | 1.00 × 10−3 | 32 | Yes | N/A | N/A | 200 |
| MobileNetV2 | Adam | 1.00 × 10−5 | 200 | Yes | 573 | 100 | 1000 |
| Xception | Adam | Initial LR = 1.00 × 10−3, Final LR = 1.00 × 10−5 | 32 | Yes | 126 (Initial Convergence), 146 (Final Convergence) | 50 (Initial Patience), 50 (Final Patience) | 250 |
| Architecture | Average Scheme | Precision | Recall | F1 Score |
|---|---|---|---|---|
| Custom-Model | Macro Average | 0.93 | 0.92 | 0.92 |
| MobileNetV2 Model | 0.94 | 0.93 | 0.93 | |
| Xception Model | 0.95 | 0.94 | 0.94 | |
| Custom-Model | Weighted Average | 0.92 | 0.92 | 0.92 |
| MobileNetV2 Model | 0.93 | 0.94 | 0.93 | |
| Xception Model | 0.94 | 0.94 | 0.94 |
| Studies | Cases Number X-ray Datasets | Method Utilized | Accuracy (%) Binary Classification | Accuracy (%) Multi-Class Classification | ||
|---|---|---|---|---|---|---|
| COVID-19 | Pneumonia | Normal | ||||
| [23] | 237 | 1336 | 1338 | Extended MobileNetV2 architecture | N/A | 99.66% |
| [26] | 400 | 400 | COVID-CheXNet architecture | 99.90% | ||
| [25] | 415 | 5179 | 2880 | VGG16 CNN | 93.9 | |
| [29] | 850 | 500 | 915 | Inception ResNetV2, InceptionNetV3 and NASNetLarge | N/A | 97.87%, 97.87% and 96.24% |
| [31] | 250 | 2753 | 3520 | VGG16 with AveragePooling2D, Flatten, Dense, Dropout and a Dense layer using Softmax function | N/A | 97%. |
| [32] | 180 | 200 | ResNet18, ResNet50, ResNet101, VGG16, and VGG19 with different SVM kernel functions (Linear, Quadratic, Cubic, and Gaussian) and a customized CNN model | 92.6% (Pre-Trained Resnet 50 Model) 91.6% (Proposed CNN Model, Average accuracy using different SVM kernels) | ||
| [33] | 538 | 468 | Ensemble of three models | 95.70% | ||
| [34] | 401 | 401 | 401 | Ensemble of two state-of-the-art classifiers with additional CNN layers | 96.15% | 99.21% |
| [27] | 181 | 364 | Modified VGG-19 architecture | 96.30% | ||
| [35] | 404 | 404 | 404 | Extended EfficientNetB4 architecture | 99.62% | 97.11% |
| [39] | 127 | 500 | 500 | Extended Xception architecture | N/A | 94.59% |
| [39] | 157 | 500 | 500 | Extended Xception architecture | N/A | 90.21% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagi, A.T.; Awan, M.J.; Mohammed, M.A.; Mahmoud, A.; Majumdar, A.; Thinnukool, O. Performance Analysis for COVID-19 Diagnosis Using Custom and State-of-the-Art Deep Learning Models. Appl. Sci. 2022, 12, 6364. https://doi.org/10.3390/app12136364
Nagi AT, Awan MJ, Mohammed MA, Mahmoud A, Majumdar A, Thinnukool O. Performance Analysis for COVID-19 Diagnosis Using Custom and State-of-the-Art Deep Learning Models. Applied Sciences. 2022; 12(13):6364. https://doi.org/10.3390/app12136364
Chicago/Turabian StyleNagi, Ali Tariq, Mazhar Javed Awan, Mazin Abed Mohammed, Amena Mahmoud, Arnab Majumdar, and Orawit Thinnukool. 2022. "Performance Analysis for COVID-19 Diagnosis Using Custom and State-of-the-Art Deep Learning Models" Applied Sciences 12, no. 13: 6364. https://doi.org/10.3390/app12136364
APA StyleNagi, A. T., Awan, M. J., Mohammed, M. A., Mahmoud, A., Majumdar, A., & Thinnukool, O. (2022). Performance Analysis for COVID-19 Diagnosis Using Custom and State-of-the-Art Deep Learning Models. Applied Sciences, 12(13), 6364. https://doi.org/10.3390/app12136364










