CoroNet: Deep Neural Network-Based End-to-End Training for Breast Cancer Diagnosis
Abstract
:1. Introduction
2. Materials and Procedures
2.1. Materials
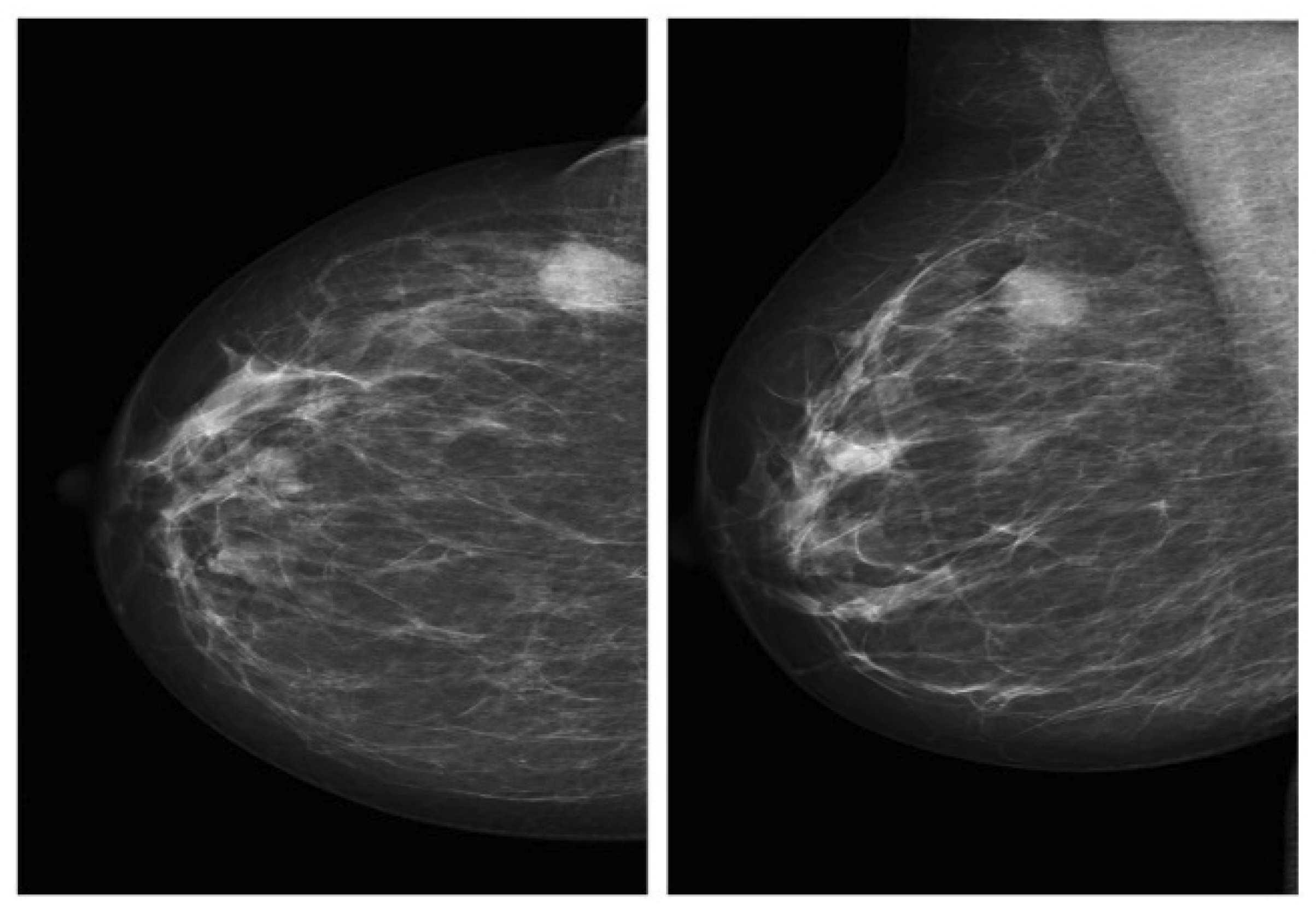
2.1.1. Mammography Datasets
2.1.2. Data Pre-Processing
- Import the training and testing data as numpy arrays from shared npy files.
- When the baseline patches are not needed, remove them and the corresponding labels from the arrays (even indices).
- Remap the labels, depending on how many, and which classes are involved in the specific classification. If the task is to only distinguish between the masses and calcification, only two labels (0–1) are needed. Conversely, four labels (03) are required when it is also important to discriminate benign abnormalities from malignant ones.
- Normalize the pixel values to be in a range that is compatible with the chosen model. Scratch CNN models using input in the range (0, 1) floating point, while VGGNet and other pretrained models are designed to work with images in (0, 255) that are further pre-processed with custom transformations (channel swapping, mean subtraction,…).
- Shuffle the training set and corresponding labels accordingly.
- Distribute the training data to “validation” and “training” subsets. The former will be used to compute the loss function exploited by the optimizer, where the actual performance is monitored on an independent group during training, using a validation set.
- Instantiate Keras generators as data sources for the network. Data augmentation settings can be specified at this stage.
- The data are formatted as expected (size, range…)
- The images content is still meaningful and was not accidentally corrupted during the process.
2.2. Methodology
2.2.1. Pre-Trained Models
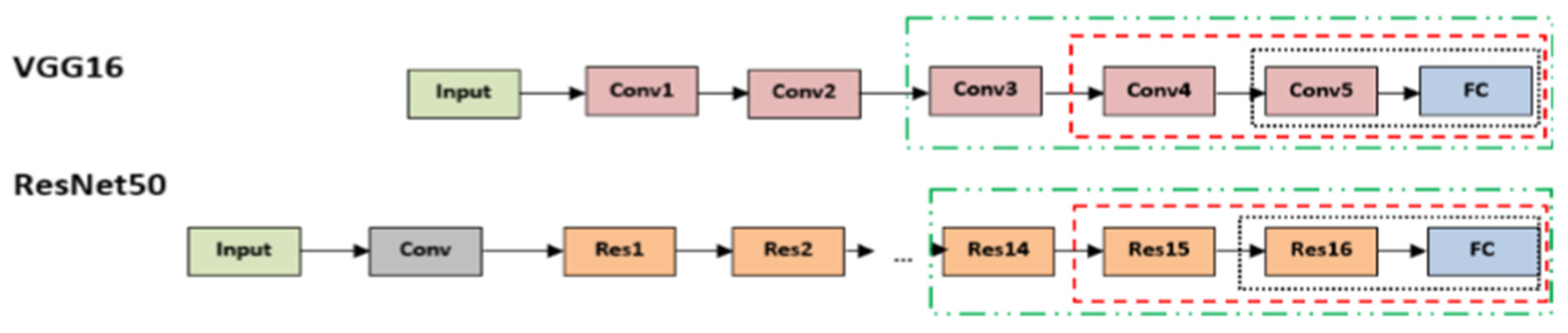
2.2.2. Pre-Trained VGG Architecture
2.2.3. Pre-Trained ResNet50 Architecture
2.2.4. Pre-Trained MobileNet Architecture
3. New Method
3.1. Convolutional Neural Network (CNN)
- ✓
- Convolution layers;
- ✓
- Pooling layers;
- ✓
- Fully connected layers.
3.2. Architecture and Development of the Model
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hassan, N.M.; Hamad, S.; Mahar, K. Mammogram breast cancer CAD systems for mass detection and classification: A review. Multimed. Tools Appl. 2022, 81, 20043–20075. [Google Scholar] [CrossRef]
- Ponraj, D.N.; Jenifer, M.E.; Poongodi, P.; Manoharan, J.S. A survey on the preprocessing techniques of mammogram for the detection of breast cancer. J. Emerg. Trends Comput. Inf. Sci. 2011, 2, 656–664. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Saira Charan, S.; Khan, M.J.; Khurshid, K. Breast Cancer Detection in Mammograms using Convolution Neural Network. In Proceedings of the 2018 International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), Sukkur, Pakistan, 3–4 March 2018; Volume 1, pp. 2–6. [Google Scholar]
- Omonigho, E.L.; David, M.; Adejo, A.; Aliyu, S. Breast Cancer: Tumor Detection in Mammogram Images Using Modified AlexNet Deep Convolution Neural Network. In Proceedings of the 2020 International Conference in Mathematics, Computer Engineering and Computer Science (ICMCECS), Ayobo, Nigeria, 18–21 March 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Elmore, J.G.; Jackson, S.L.; Abraham, L.; Miglioretti, D.L.; Carney, P.A.; Geller, B.M.; Yankaskas, B.C.; Kerlikowske, K.; Onega, T.; Rosenberg, R.D.; et al. Variability in Interpretive Performance at Screening Mammography and Radiologists’ Characteristics Associated with Accuracy. Radiology 2009, 253, 641–651. [Google Scholar] [CrossRef]
- Lehman, C.D.; Wellman, R.D.; Buist, D.S.M.; Kerlikowske, K.; Tosteson, A.N.A.; Miglioretti, D.L. Diagnostic Accuracy of Digital Screening Mammography With and Without Computer-Aided Detection. JAMA Intern. Med. 2015, 175, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Tosteson, A.N.A.; Fryback, D.G.; Hammond, C.S.; Hanna, L.G.; Grove, M.R.; Brown, M.; Wang, Q.; Lindfors, K.; Pisano, E.D. Consequences of False-Positive Screening Mammograms. JAMA Intern. Med. 2014, 174, 954–961. [Google Scholar] [CrossRef]
- Houssami, N.; Hunter, K. The epidemiology, radiology and biological characteristics of interval breast cancers in population mammography screening. NPJ Breast Cancer 2017, 3, 12 . [Google Scholar] [CrossRef]
- Gilbert, F.J.; Astley, S.M.; Gillan, M.G.; Agbaje, O.F.; Wallis, M.G.; James, J.; Boggis, C.R.; Duffy, S.W. Single Reading with Computer-Aided Detection for Screening Mammography. N. Engl. J. Med. 2008, 359, 1675–1684. [Google Scholar] [CrossRef]
- Giger, M.L.; Chan, H.-P.; Boone, J. Anniversary paper: History and status of CAD and quantitative image analysis: The role of Medical Physics and AAPM. Med. Phys. 2008, 35, 5799–5820. [Google Scholar] [CrossRef]
- Fenton, J.J.; Taplin, S.H.; Carney, P.A.; Abraham, L.; Sickles, E.A.; D’Orsi, C.; Berns, E.A.; Cutter, G.; Hendrick, R.E.; Barlow, W.E.; et al. Influence of Computer-Aided Detection on Performance of Screening Mammography. N. Engl. J. Med. 2007, 356, 1399–1409. [Google Scholar] [CrossRef]
- Kohli, A.; Jha, S. Why CAD Failed in Mammography. J. Am. Coll. Radiol. 2018, 15, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, K.; Acharya, U.R.; Chua, C.K.; Min, L.C.; Abraham, K.T.; Ng, K.-H. Computer-Aided Breast Cancer Detection Using Mammograms: A Review. IEEE Rev. Biomed. Eng. 2012, 6, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Heath, M.; Bowyer, K.; Kopans, D.; Moore, R.; Kegelmeyer, W.P. The digital database for screening mammography. In Proceedings of the 5th International Workshop on Digital Mammography, Toronto, Canada, 1–14 June 2000; pp. 212–218. [Google Scholar]
- Khan, S.; Hussain, M.; Aboalsamh, H.; Bebis, G. A comparison of different Gabor feature extraction approaches for mass classification in mammography. Multimed. Tools Appl. 2015, 76, 33–57. [Google Scholar] [CrossRef] [Green Version]
- Narváez, F.; Alvarez, J.; Garcia-Arteaga, J.D.; Tarquino, J.; Romero, E. Characterizing Architectural Distortion in Mammograms by Linear Saliency. J. Med. Syst. 2016, 41, 26. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Rao, R.V.; Chen, P.; Zhang, Y.; Liu, A.; Wei, L. Abnormal Breast Detection in Mammogram Images by Feed-forward Neural Network Trained by Jaya Algorithm. Fundam. Inform. 2017, 151, 191–211. [Google Scholar] [CrossRef]
- Nithya, R.; Santhi, B. Classification of Normal and Abnormal Patterns in Digital Mammograms for Diagnosis of Breast Cancer. Int. J. Comput. Appl. 2011, 28, 21–25. [Google Scholar] [CrossRef]
- Zhu, W.; Lou, Q.; Vang, Y.S.; Xie, X. Deep multi-instance networks with sparse label assignment for whole mammogram classification. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017; pp. 603–611. [Google Scholar]
- Sampaio, W.B.; Diniz, E.M.; Silva, A.C.; de Paiva, A.C.; Gattass, M. Detection of masses in mammogram images using CNN, geostatistic functions and SVM. Comput. Biol. Med. 2011, 41, 653–664. [Google Scholar] [CrossRef]
- Lévy, D.; Jain, A. Breast mass classification from mammograms using deep convolutional neural networks. arXiv 2016, arXiv:1612.00542. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. NIPS 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Guan, S.; Loew, M. Breast cancer detection using synthetic mammograms from generative adversarial networks in convolutional neural networks. J. Med. Imaging 2019, 6, 31411. [Google Scholar] [CrossRef]
- Xi, P.; Shu, C.; Goubran, R. Abnormality detection in mammography using deep convolutional neural networks. In Proceedings of the 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Rome, Italy, 11–13 June 2018; pp. 1–6. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ragab, D.A.; Sharkas, M.; Marshall, S.; Ren, J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ 2019, 7, e6201. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Margolies, L.R.; Rothstein, J.H.; Fluder, E.; McBride, R.; Sieh, W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci. Rep. 2019, 9, 12495. [Google Scholar] [CrossRef] [PubMed]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef]
- Hussain, Z.; Gimenez, F.; Yi, D.; Rubin, D. Differential Data Augmentation Techniques for Medical Imaging Classification Tasks. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: Bethesda, MD, USA, 2018; Volume 2017, pp. 979–984. [Google Scholar]
- Costa, A.C.; Oliveira, H.C.R.; Catani, J.H.; de Barros, N.; Melo, C.F.E.; Vieira, M.A.C. Data augmentation for detection of architectural distortion in digital mammography using deep learning approach. arXiv 2018, arXiv:1807.03167. [Google Scholar]
- Elmore, J.G.; Armstrong, K.; Lehman, C.D.; Fletcher, S.W. Screening for Breast Cancer. JAMA J. Am. Med. Assoc. 2005, 293, 1245–1256. [Google Scholar] [CrossRef]
- Friedewald, S.M.; Rafferty, E.A.; Rose, S.L.; Durand, M.A.; Plecha, D.M.; Greenberg, J.S.; Hayes, M.K.; Copit, D.S.; Carlson, K.L.; Cink, T.M.; et al. Breast Cancer Screening Using Tomosynthesis in Combination With Digital Mammography. JAMA J. Am. Med. Assoc. 2014, 311, 2499–2507. [Google Scholar] [CrossRef] [Green Version]
- Guan, S.; Loew, M. Breast Cancer Detection Using Transfer Learning in Convolutional Neural Networks. In Proceedings of the 2017 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 10–12 October 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Heath, M.; Bowyer, K.; Kopans, D.; Kegelmeyer, P.; Moore, R.; Chang, K.; Munishkumaran, S. Current Status of the Digital Database for Screening Mammography. In Digital Mammography; Springer: Berlin/Heidelberg, Germany, 1998; pp. 457–460. [Google Scholar] [CrossRef]
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef]
- Suckling, J. The Mammographic Image Analysis Society Digital Mammogram Database. Digit. Mammo 1994, 1069, 375–378. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.I.; Shah, J.L.; Bhat, M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020, 196, 105581. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]



| Location of Cancer | Number of New Cases (% of All Locations) | Number of New Deaths (% of All Locations) | ||
|---|---|---|---|---|
| Brain, nervous system | (1.6) | 308,102 | (2.5) | 251,329 |
| Colon | (6.0) | 1,148,515 | (5.8) | 576,858 |
| Female breast | (11.7) | 2,261,419 | (6.9) | 684,996 |
| Leukemia | (2.5) | 474,519 | (3.1) | 311,594 |
| Liver | (4.7) | 905,677 | (8.3) | 830,180 |
| Lung | (11.4) | 2,206,771 | (18.0) | 1,796,144 |
| Nonmelanoma of skin | (6.2) | 1,198,073 | (0.6) | 63,731 |
| Ovary | (1.6) | 313.959 | (2.1) | 207.252 |
| Prostate | (7.3) | 1,414,259 | (3.8) | 375,304 |
| Stomach | (5.6) | 1,089,103 | (7.7) | 768,793 |
| Dataset | Type | # of Images | View | Format | Classes | Resolution (Bit/Pixel) | Availability |
|---|---|---|---|---|---|---|---|
| DDSM [37] | Digital Mammogram (DM) | 10,480 | MLO/CC | LJPEG | Normal, benign and malignant | 8–16 | Publicly available |
| CIBS-DDSM [38] | Digital Mammogram (DM) | 10,239 | MLO/CC | DICOM | Benign and malignant | 10 | Publicly available |
| Type | Normal | Abnormal | Total | |
|---|---|---|---|---|
| Benign | Malignancy | |||
| Train | 1190 | 688 | 719 | 2597 |
| Test | 128 | 64 | 64 | 256 |
| Total | 1318 | 752 | 783 | 2853 |
| Model | Main Finding | Depth | Dataset | Error Rate | Input Size | Year |
|---|---|---|---|---|---|---|
| AlexNet | Utilizes Dropout and ReLU | 8 | ImageNet | 16.4 | 227 × 227 × 3 | 2012 |
| VGG | Increased depth, small filter size | 16, 19 | ImageNet | 7.3 | 224 × 224 × 3 | 2014 |
| ResNet | Robust against overfitting because of symmetry mapping-based skip links | 50,152 | ImageNet | 3.57 | 224 × 224 × 3 | 2016 |
| Xception | A depthwise convolution followed by a pointwise convolution | 71 | ImageNet | 0.055 | 229 × 229 × 3 | 2017 |
| MobileNet-v2 | Inverted residual structure | 53 | ImageNet | - | 224 × 224 × 3 | 2018 |
| Layer (Type) | Output Shape | No of Parameters |
|---|---|---|
| Xception (Model) | 5 × 5 × 2048 | 20,861,480 |
| flatten (Flatten) | 51,200 | 0 |
| dropout (Dropout) | 51,200 | 0 |
| dense (Dense) | 256 | 13,107,456 |
| dense_1 (Dense) | 4 | 1028 |
| Total parameters: 33,969,964 | ||
| Trainable parameters:33,915,436 | ||
| Non-trainable parameters: 54,528 | ||
| Models | Epoch Stop | Validation Accuracy | Training Accuracy | Validation Loss | Testing Loss |
|---|---|---|---|---|---|
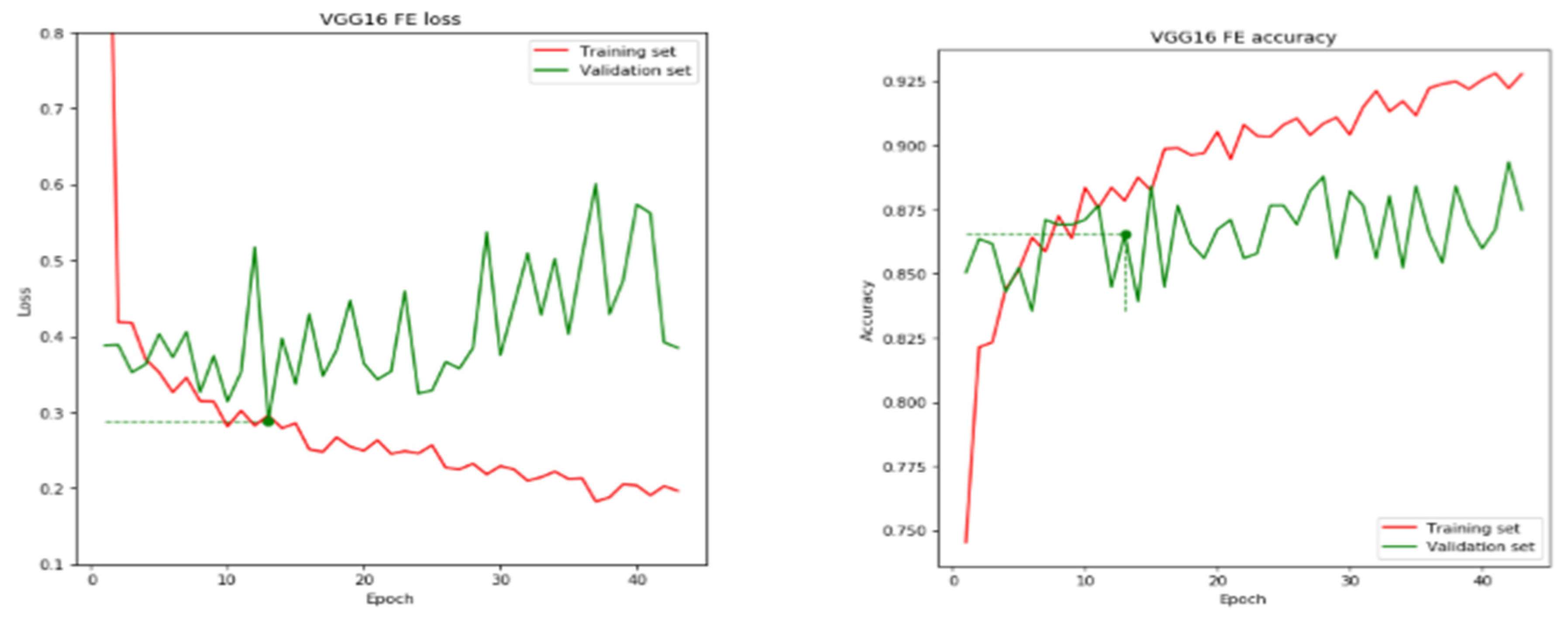
| VGG 16 | 13 | 86.54 | 68.90 | 0.2886 | 0.4320 |
| CoroNet | 84 | 94.73 | 99.73 | 0.6079 | 0.0069 |
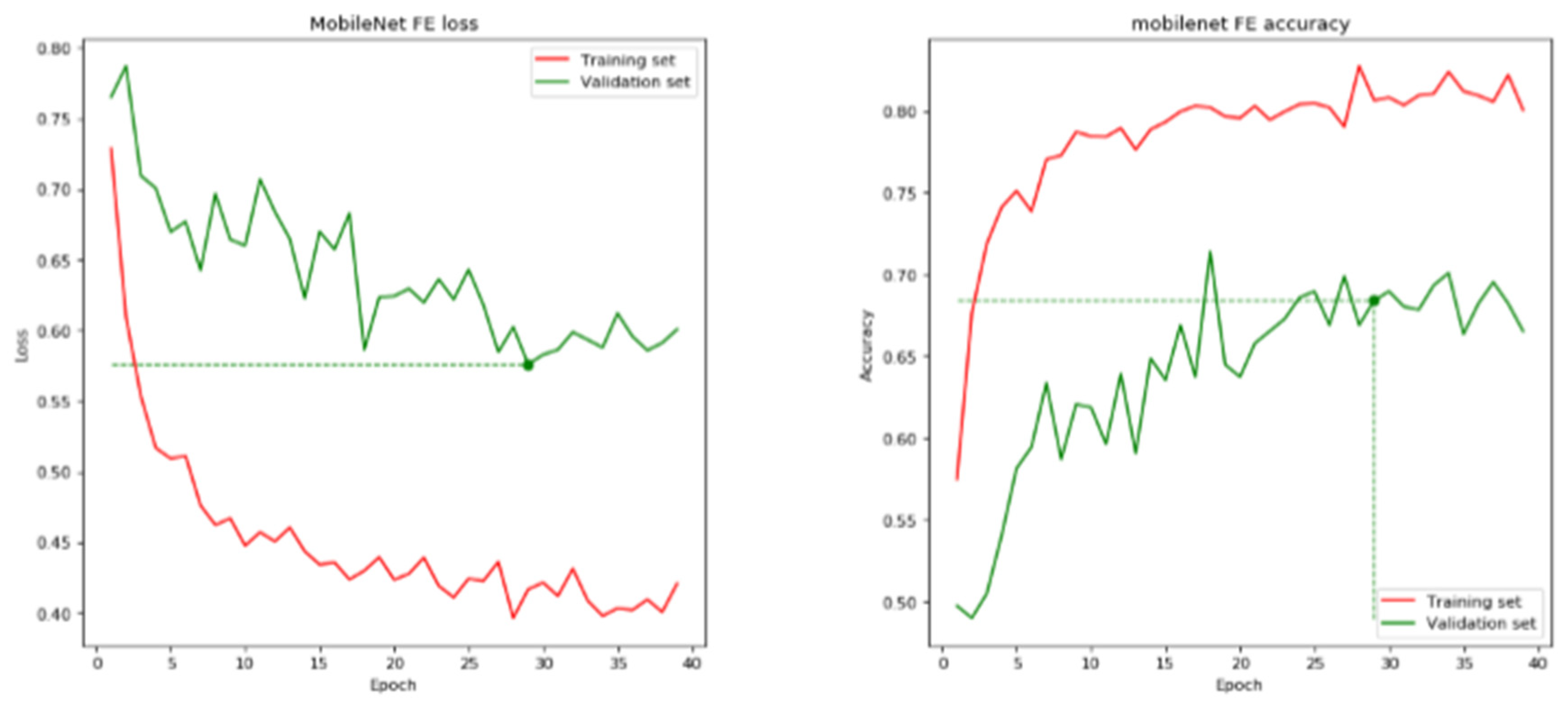
| MobileNet | 29 | 68.41 | 70.24 | 0.5759 | 0.6054 |
| ResNet50 | 12 | 72.15 | 74.40 | 0.5457 | 0.5948 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mobark, N.; Hamad, S.; Rida, S.Z. CoroNet: Deep Neural Network-Based End-to-End Training for Breast Cancer Diagnosis. Appl. Sci. 2022, 12, 7080. https://doi.org/10.3390/app12147080
Mobark N, Hamad S, Rida SZ. CoroNet: Deep Neural Network-Based End-to-End Training for Breast Cancer Diagnosis. Applied Sciences. 2022; 12(14):7080. https://doi.org/10.3390/app12147080
Chicago/Turabian StyleMobark, Nada, Safwat Hamad, and S. Z. Rida. 2022. "CoroNet: Deep Neural Network-Based End-to-End Training for Breast Cancer Diagnosis" Applied Sciences 12, no. 14: 7080. https://doi.org/10.3390/app12147080
APA StyleMobark, N., Hamad, S., & Rida, S. Z. (2022). CoroNet: Deep Neural Network-Based End-to-End Training for Breast Cancer Diagnosis. Applied Sciences, 12(14), 7080. https://doi.org/10.3390/app12147080






