Electrochemical Detection of Penicillin G Using Molecularly Imprinted Conductive Co-Polymer Sensor
Abstract
:1. Introduction
2. Materials and Methods
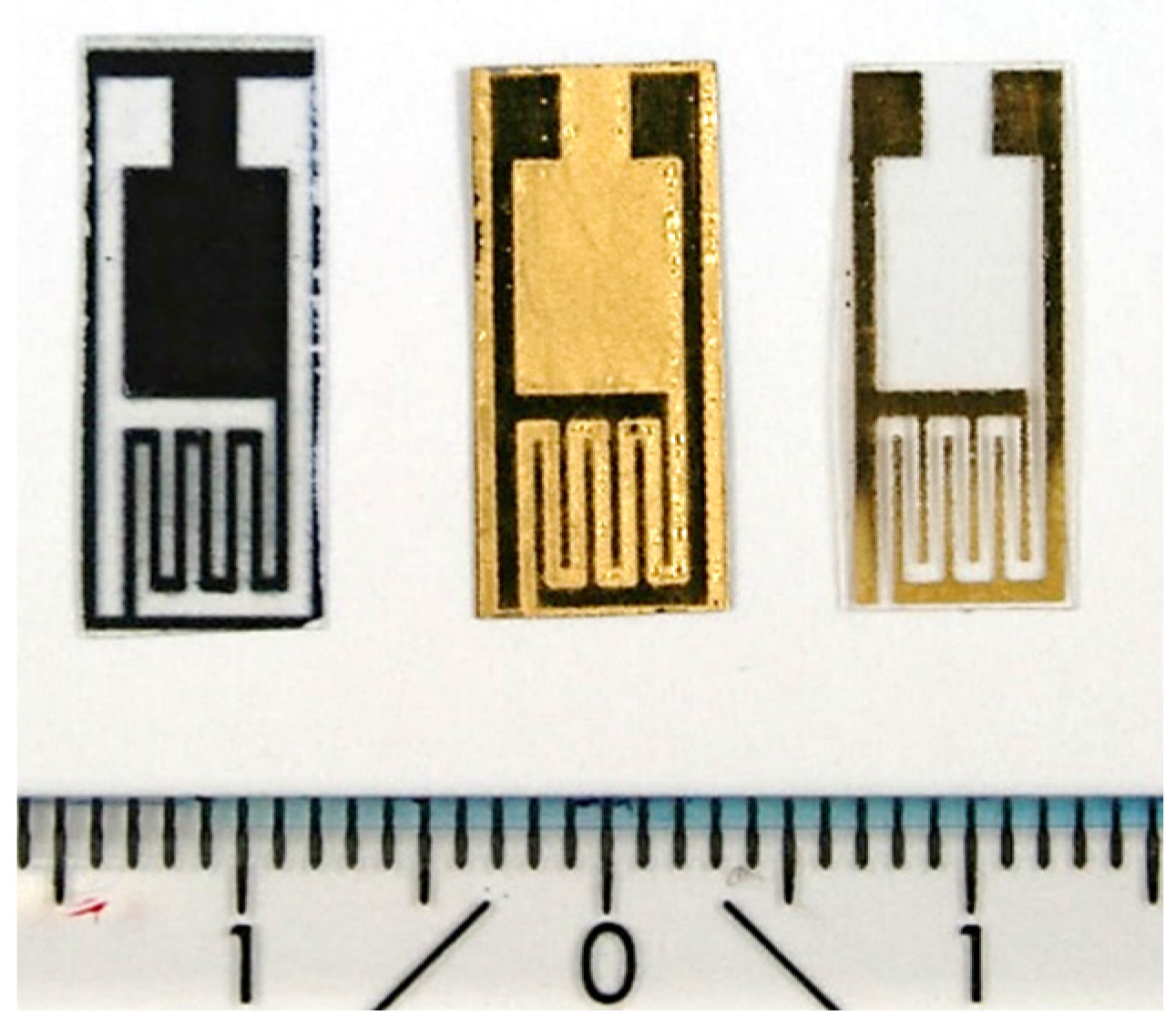
2.1. Interdigitated Electrodes on PET Substrates
2.2. Chemicals
- Deionized water as solvent and rinsing liquid.
- Penicillin G (PenG) from Merck™: CAS 69-57-8 (C16H17N2NaO4S), (>96 wt.%), 356.38 g/mol.
- Pyrrole-3-carboxylic acid (PyCOOH) from Merck™: CAS 931-03-3 (C5H5NO2) (96 wt.%), 111.10 g/mol.
- Pyrrole (Py) from Merck™: CAS 109-97-7 (C4H5N) (98 wt.%), 67.09 g/mol.
- Sulfuric Acid from Merck™: CAS 7664-93-9 (H2SO4) (95 wt.%), 98.08 g/mol.
- Ammonium persulfate (APS) from Merck™: CAS 7727-54-0 ((NH4)2S2O8) (>98 wt.%), 228.2 g/mol.
- Methanol (MeOH) from M Supelco™: CAS 67-56-1 (CH3OH) (>99 wt.%), 32.04 g/mol.
- Hydrochloric acid from VWR Chemicals™: CAS 7647-01-0 (HCl) (>37 wt.%), 36.46 g/mol.
2.3. Equipment
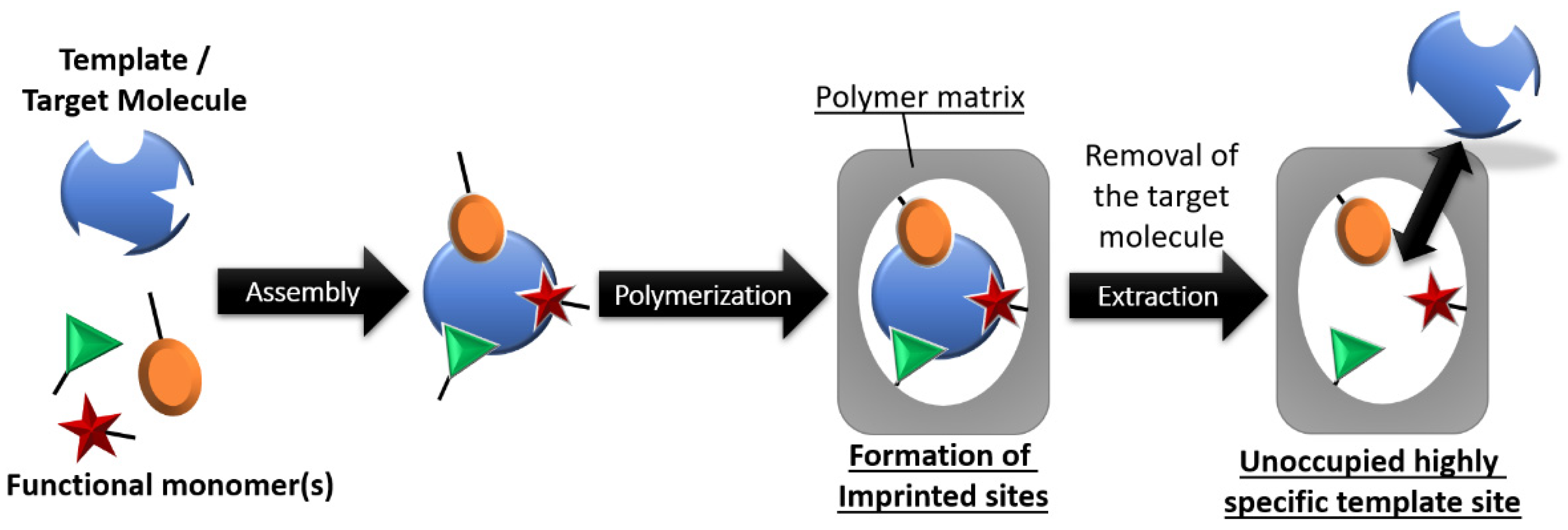
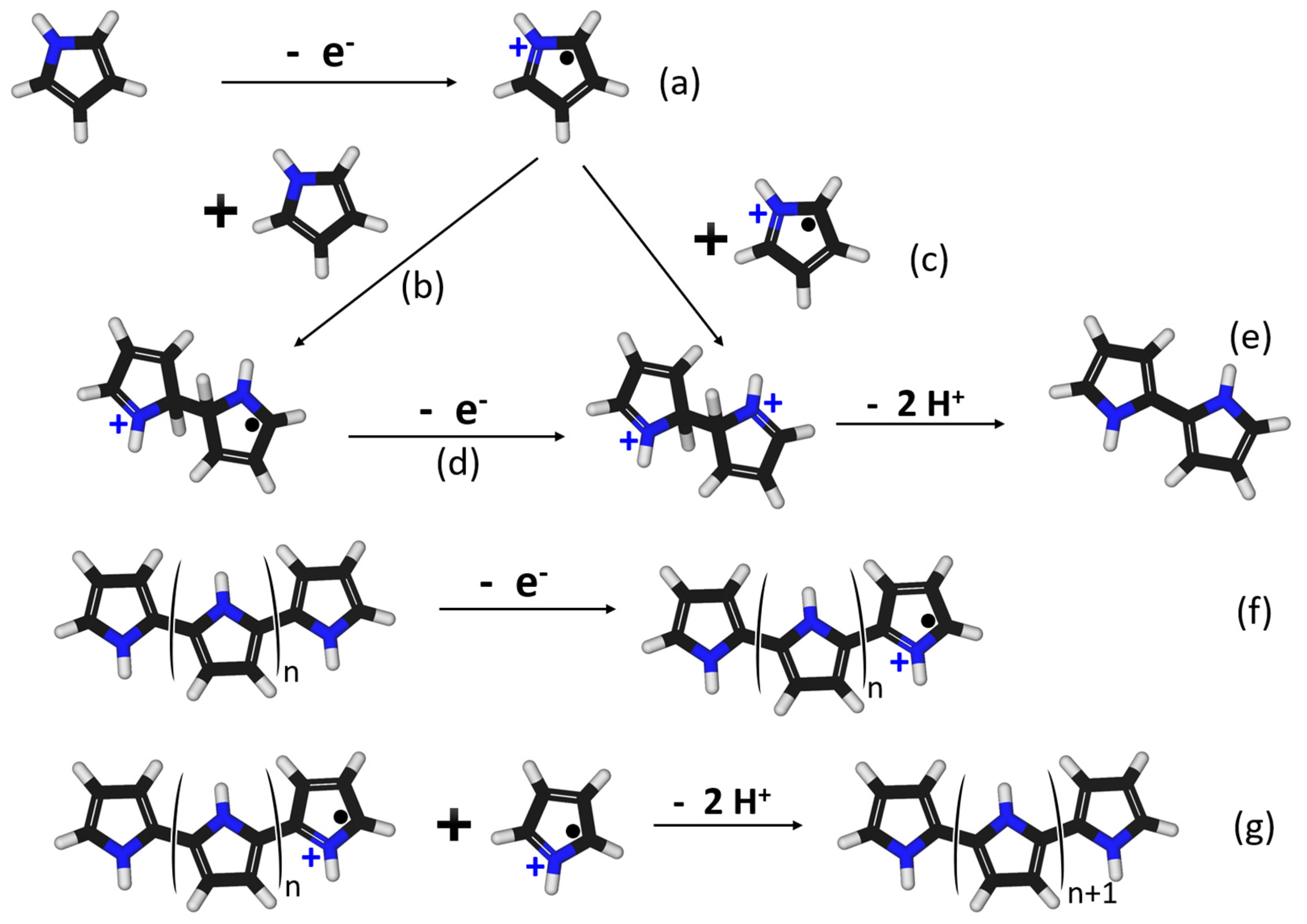
2.4. Synthesis of the MICP
3. Results and Discussion
3.1. Characterization
3.1.1. Extraction Efficiency
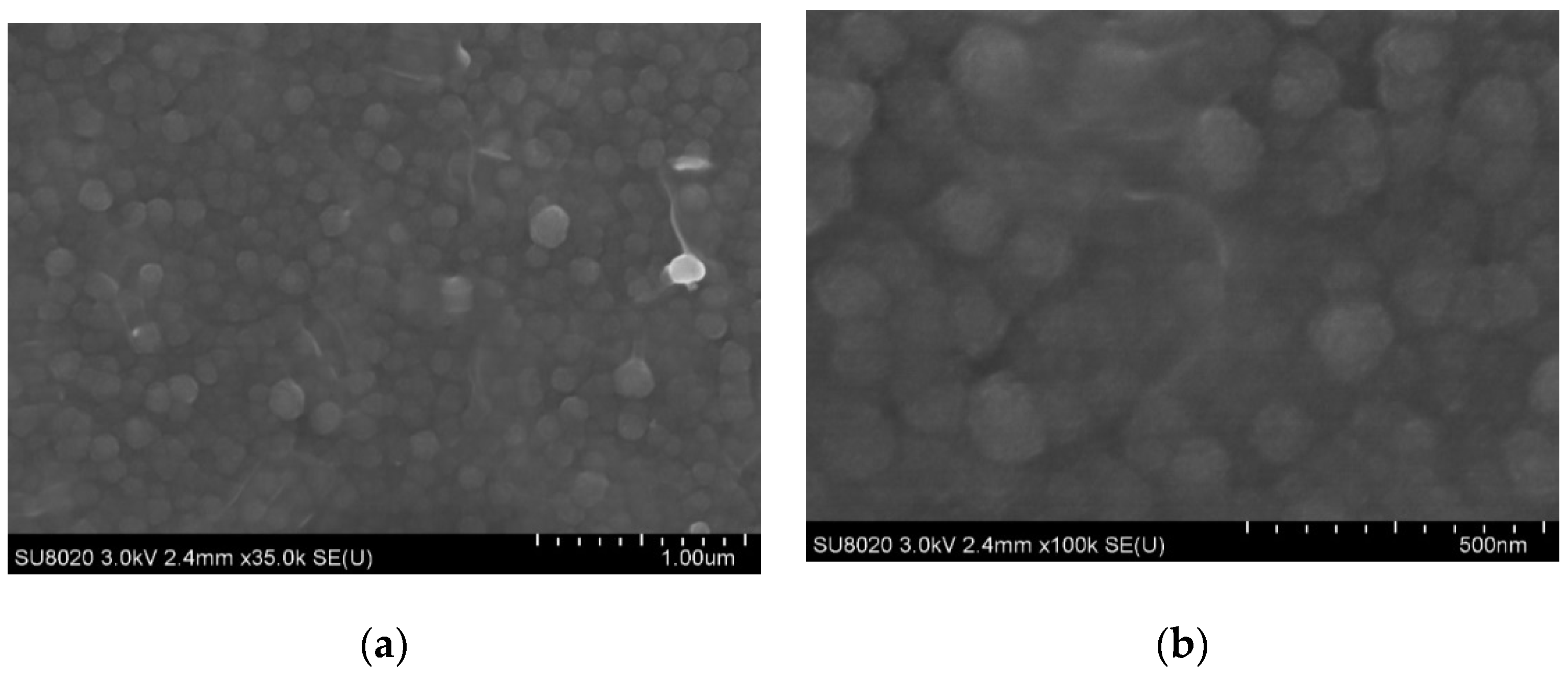
3.1.2. SEM Observation
3.1.3. Thickness Measurements
3.2. Sensor’s Performances
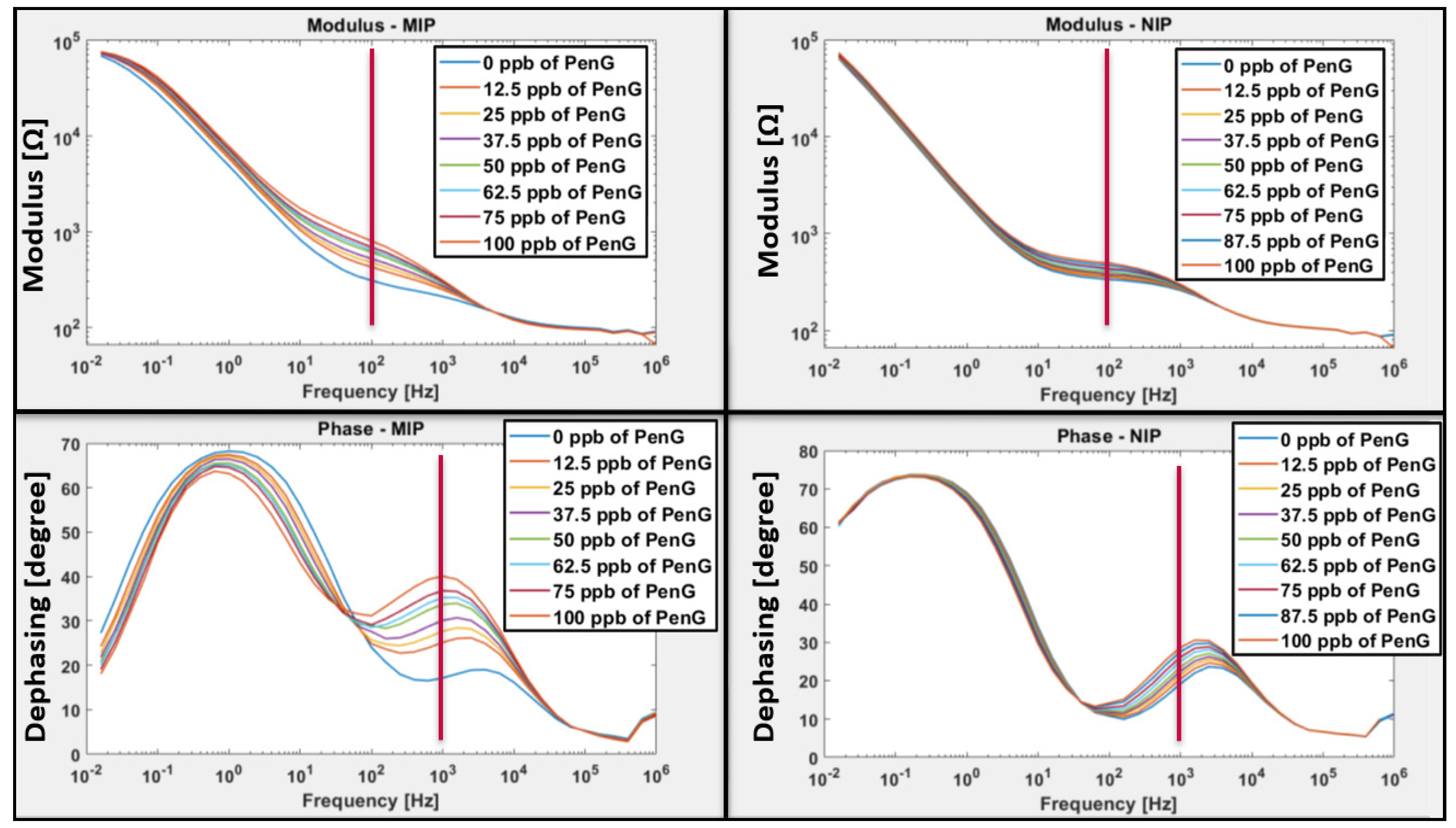
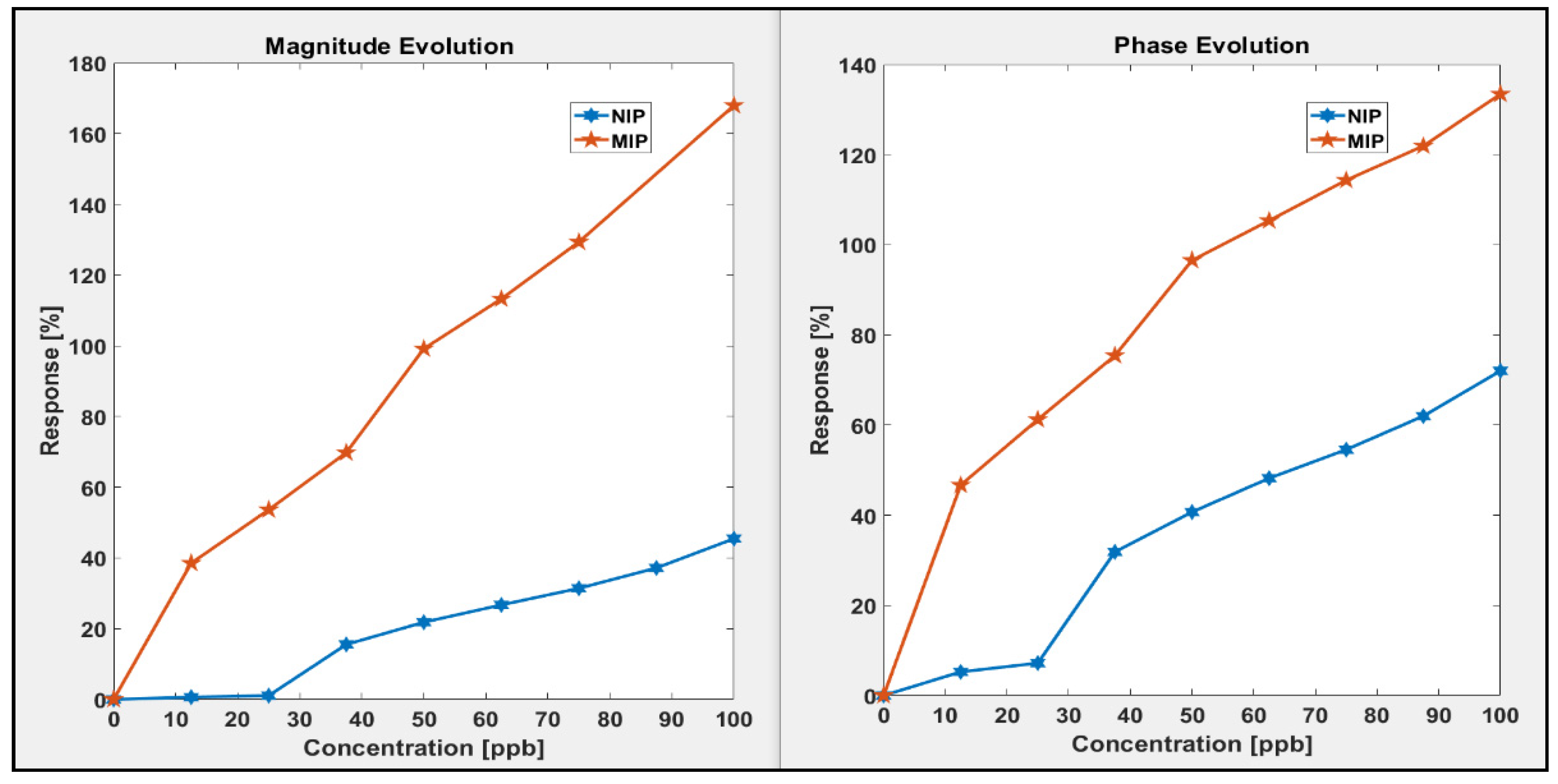
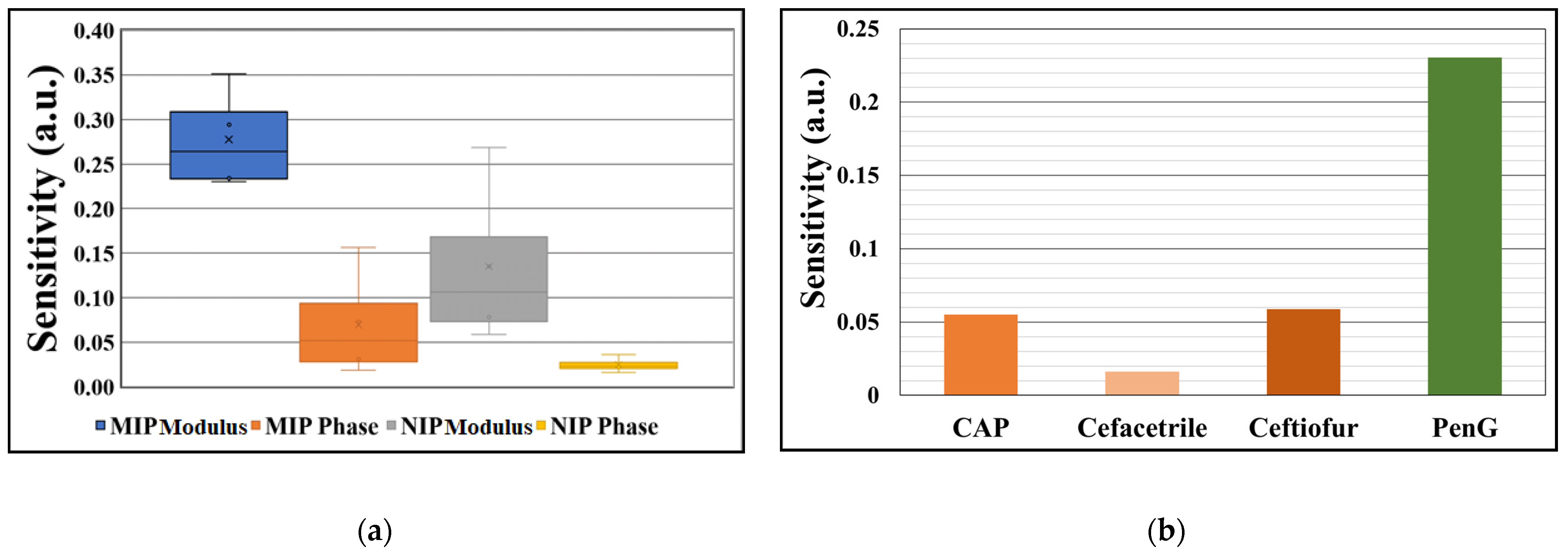
3.2.1. Sensitivity
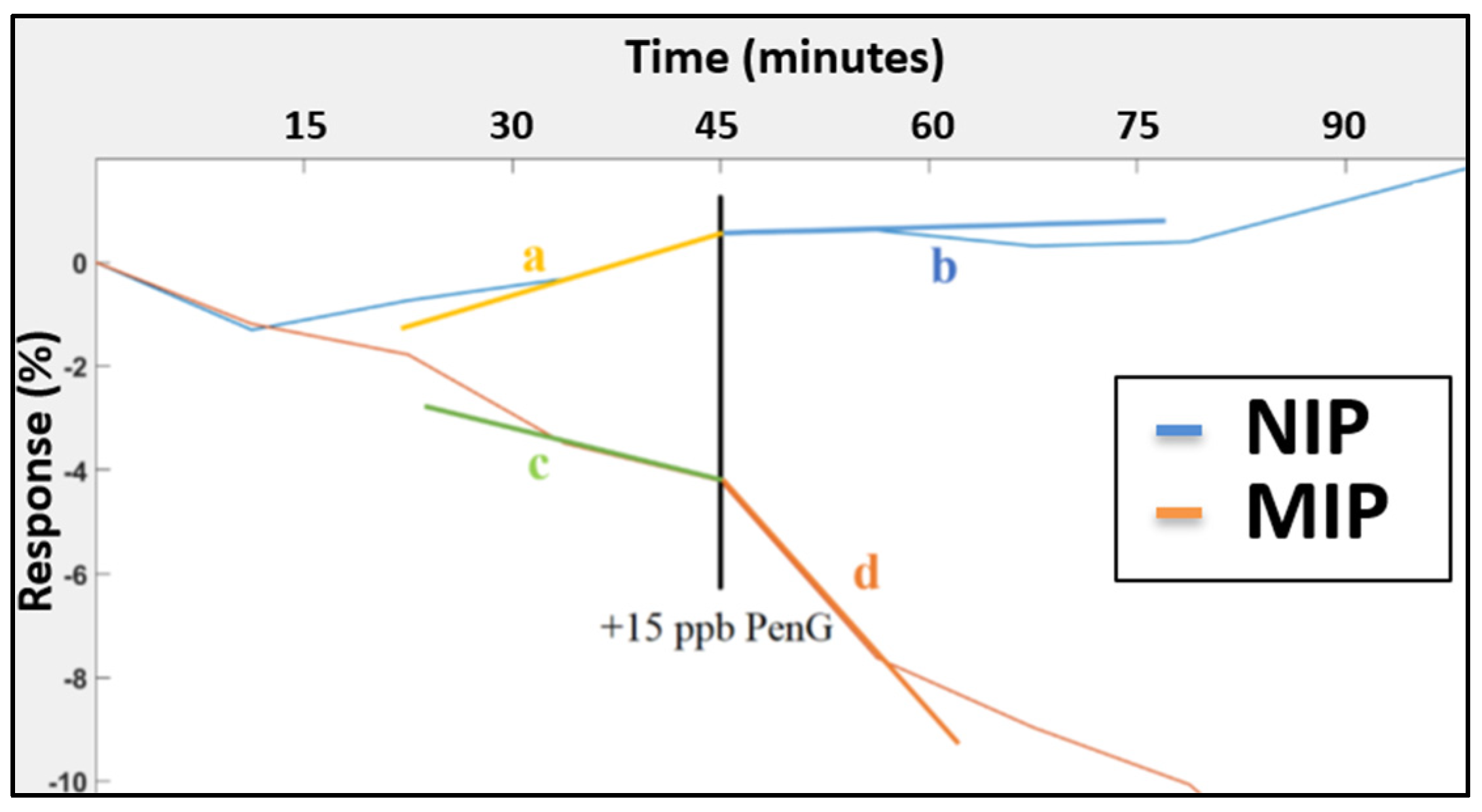
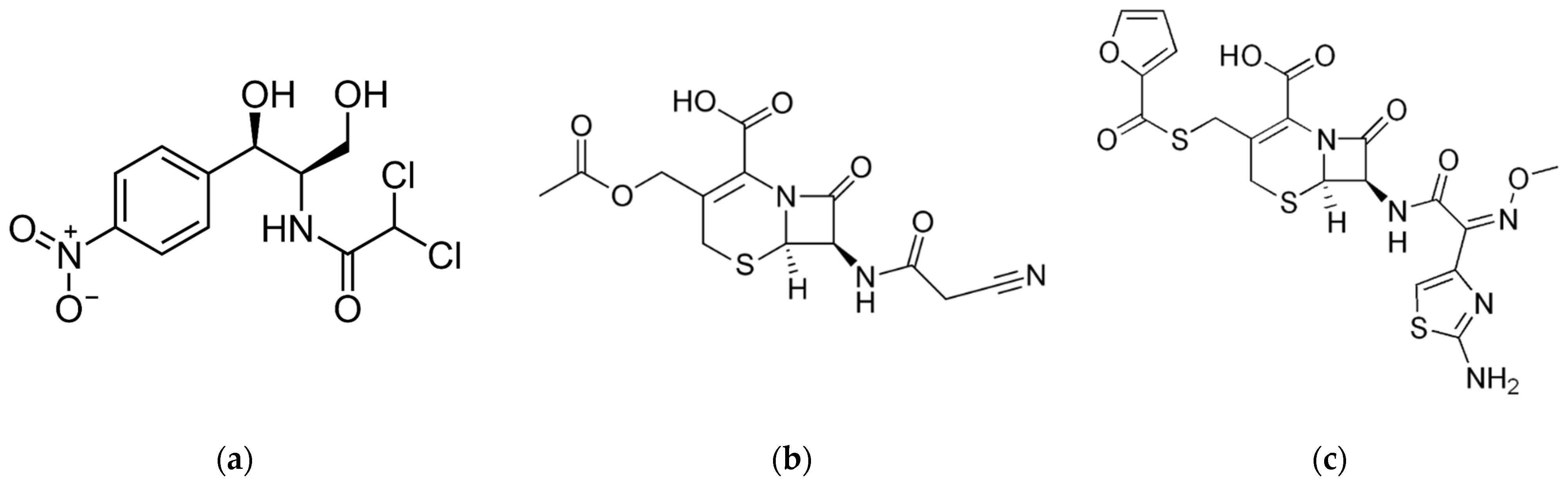
3.2.2. Reproducibility and Selectivity
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rehman, M.S.U.; Rashid, N.; Ashfaq, M.; Saif, A.; Ahmad, N.; Han, J.I. Global risk of pharmaceutical contamination from highly populated developing countries. Chemosphere 2015, 138, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Sachi, S.; Ferdous, J.; Sikder, M.H.; Hussani, S.M.A.K. Antibiotic residues in milk: Past, present, and future. J. Adv. Vet. Anim. Res. 2019, 6, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, A.C.; Wang, W.; Koteva, K.; Barton, H.A.; McArthur, A.G.; Wright, G.D. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moro, G.; Bottari, F.; Sleegers, N.; Florea, A.; Cowen, T.; Moretto, L.M.; Piletsky, S.; De Wael, K. Conductive imprinted polymers for the direct electrochemical detection of β-lactam antibiotics: The case of cefquinome. Sens. Actuators B Chem. 2019, 297, 126786. [Google Scholar] [CrossRef]
- Smith, R. Commission Regulation (EU) No 330/2010. Official Journal of the European Union, 2010. Available online: https://ec.europa.eu/health/sites/health/files/files/eudralex/vol-5/reg_2010_37/reg_2010_37_en.pdf (accessed on 7 April 2022).
- Babington, R.; Matas, S.; Marco, M.P.; Galve, R. Current bioanalytical methods for detection of penicillins. Anal. Bioanal. Chem. 2012, 403, 1549–1566. [Google Scholar] [CrossRef]
- Cháfer-Pericás, C.; Maquieira, Á.; Puchades, R. Fast screening methods to detect antibiotic residues in food samples. TrAC Trends Anal. Chem. 2010, 29, 1038–1049. [Google Scholar] [CrossRef]
- Beltrán, M.C.; Berruga, M.I.; Molina, A.; Althaus, R.L.; Molina, M.P. Performance of current microbial tests for screening antibiotics in sheep and goat milk. Int. Dairy J. 2015, 41, 13–15. [Google Scholar] [CrossRef]
- Kantiani, L.; Farré, M.; Barceló, D. Analytical methodologies for the detection of β-lactam antibiotics in milk and feed samples. TrAC-Trends Anal. Chem. 2009, 28, 729–744. [Google Scholar] [CrossRef]
- Pietschmann, J.; Dittmann, D.; Spiegel, H.; Krause, H.J.; Schröper, F. A Novel Method for Antibiotic Detection in Milk Based on Competitive Magnetic Immunodetection. Foods 2020, 9, 1773. [Google Scholar] [CrossRef]
- Gustavsson, E.; Degelaen, J.; Bjurling, P.; Sternesjö, Å. Determination of β-Lactams in Milk Using a Surface Plasmon Resonance-Based Biosensor. J. Agric. Food Chem. 2004, 52, 2791–2796. [Google Scholar] [CrossRef]
- Kivirand, K.; Kagan, M.; Rinken, T. Biosensors for the detection of antibiotic residues in milk. In Biosensors—Micro and Nanoscale Application; InTech Open: Rijeka, Croatia, 2015. [Google Scholar] [CrossRef] [Green Version]
- Lamar, J.; Petz, M. Development of a receptor-based microplate assay for the detection of beta-lactam antibiotics in different food matrices. Anal. Chim. Acta 2007, 586, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Song, K.M.; Jeong, E.; Jeon, W.; Cho, M.; Ban, C. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence-colorimetric methods. Anal. Bioanal. Chem. 2012, 402, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Luo, L.; Suryoprabowo, S.; Peng, J.; Kuang, H.; Xu, C. Development of an Immunochromatographic Strip Test for Rapid Detection of Ciprofloxacin in Milk Samples. Sensors 2014, 14, 16785–16798. [Google Scholar] [CrossRef] [PubMed]
- Alidoust, M.; Baharfar, M.; Manouchehri, M.; Yamini, Y.; Tajik, M.; Seidi, S. Emergence of microfluidic devices in sample extraction; an overview of diverse methodologies, principals, and recent advancements. TrAC Trends Anal. Chem. 2021, 143, 116352. [Google Scholar] [CrossRef]
- Weber, P.; Riegger, B.R.; Niedergall, K.; Tovar, G.E.M.; Bach, M.; Gauglitz, G. Nano-MIP based sensor for penicillin G: Sensitive layer and analytical validation. Sens. Actuators B Chem. 2018, 267, 26–33. [Google Scholar] [CrossRef]
- Sapurina, I.Y.; Shishov, M.A. Oxidative Polymerization of Aniline Molecular Synthesis of Polyaniline and the Formation of Supramolecular Structures. N. Polym. Spec. Appl. 2012, 740, 251–312. [Google Scholar]
- Holthoff, E. Molecularly imprinted polymers enhance detection capabilities. SPIE Newsroom 2013, 2–4. [Google Scholar] [CrossRef] [Green Version]
- Alemán, C.; Casanovas, J.; Torras, J.; Bertrán, O.; Armelin, E.; Oliver, R.; Estrany, F. Cross-linking in polypyrrole and poly(N-methylpyrrole): Comparative experimental and theoretical studies. Polymer 2008, 49, 1066–1075. [Google Scholar] [CrossRef]
- Gonzalez-Vila, A.; Delius, M.; Lahem, D.; Zhang, C.; Mégret, P.; Caucheteur, C. Molecularly imprinted electropolymerization on a metal-coated optical fiber for gas sensing applications. Sens. Actuators B Chem. 2017, 244, 1145–1151. [Google Scholar] [CrossRef]
- Charlier, H. Chemical Sensors Based on Molecularly Imprinted Conductive Polymers for the Detection of Antibiotics; University of Mons (UMONS)-Faculty of Engineering: Mons, Belgium, 2021. [Google Scholar]
- Diaz, A.; Bargon, J. Handbook of Conducting Polymers; TA Skothei.: New York, NY, USA, 1986; Volume 1. [Google Scholar]
- Andrieux, C.P.; Audebert, P.; Hapiot, P.; Savéant, J.M. Identification of the first steps of the electrochemical polymerization of pyrroles by means of fast potential step techniques. J. Phys. Chem. 1991, 95, 10158–10164. [Google Scholar] [CrossRef]
- Tan, Y.; Ghandi, K. Kinetics and mechanism of pyrrole chemical polymerization. Synth. Met. 2013, 175, 183–191. [Google Scholar] [CrossRef]
- Clochard, M.-C.; Baudin, C.; Betz, N.; Le Moël, A.; Bittencourt, C.; Houssiau, L.; Pireaux, J.-J.; Caldemaison, D. New sulfonated pyrrole and pyrrole 3-carboxylic acid copolymer membranes via track-etched templates. React. Funct. Polym. 2006, 66, 1296–1305. [Google Scholar] [CrossRef]
- Khung, Y.L.; Narducci, D. Synergizing nucleic acid aptamers with 1-dimensional nanostructures as label-free field-effect transistor biosensors. Biosens. Bioelectron. 2013, 50, 278–293. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Kin, J.H.; Lee, N.; Kim, B.G.; Jang, J. A novel sensor platform based on aptamer-conjugated polypyrrole nanotubes for label-free electrochemical protein detection. ChemBioChem 2008, 9, 634–641. [Google Scholar] [CrossRef]
- Urraca, J.L.; Hall, A.J.; Moreno-Bondi, M.C.; Sellergren, B. A Stoichiometric Molecularly Imprinted Polymer for the Class-Selective Recognition of Antibiotics in Aqueous Media. Angew. Chem. 2006, 118, 5282–5285. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charlier, H.; David, M.; Lahem, D.; Debliquy, M. Electrochemical Detection of Penicillin G Using Molecularly Imprinted Conductive Co-Polymer Sensor. Appl. Sci. 2022, 12, 7914. https://doi.org/10.3390/app12157914
Charlier H, David M, Lahem D, Debliquy M. Electrochemical Detection of Penicillin G Using Molecularly Imprinted Conductive Co-Polymer Sensor. Applied Sciences. 2022; 12(15):7914. https://doi.org/10.3390/app12157914
Chicago/Turabian StyleCharlier, Hugues, Mariel David, Driss Lahem, and Marc Debliquy. 2022. "Electrochemical Detection of Penicillin G Using Molecularly Imprinted Conductive Co-Polymer Sensor" Applied Sciences 12, no. 15: 7914. https://doi.org/10.3390/app12157914
APA StyleCharlier, H., David, M., Lahem, D., & Debliquy, M. (2022). Electrochemical Detection of Penicillin G Using Molecularly Imprinted Conductive Co-Polymer Sensor. Applied Sciences, 12(15), 7914. https://doi.org/10.3390/app12157914





