PNMAVis: Visual Analysis Tool of Protein Normal Mode for Understanding Cavity Dynamics
Abstract
:1. Introduction
- Quickly obtain the possible movements of cavity-lining amino acids and potential key amino acids through the coarse-grained NMA model.
- Give the characterization form of cavity-lining amino acids of the dynamic cavity, introduce the structural and biochemical properties of the cavity, and enrich the data characteristics of the cavity.
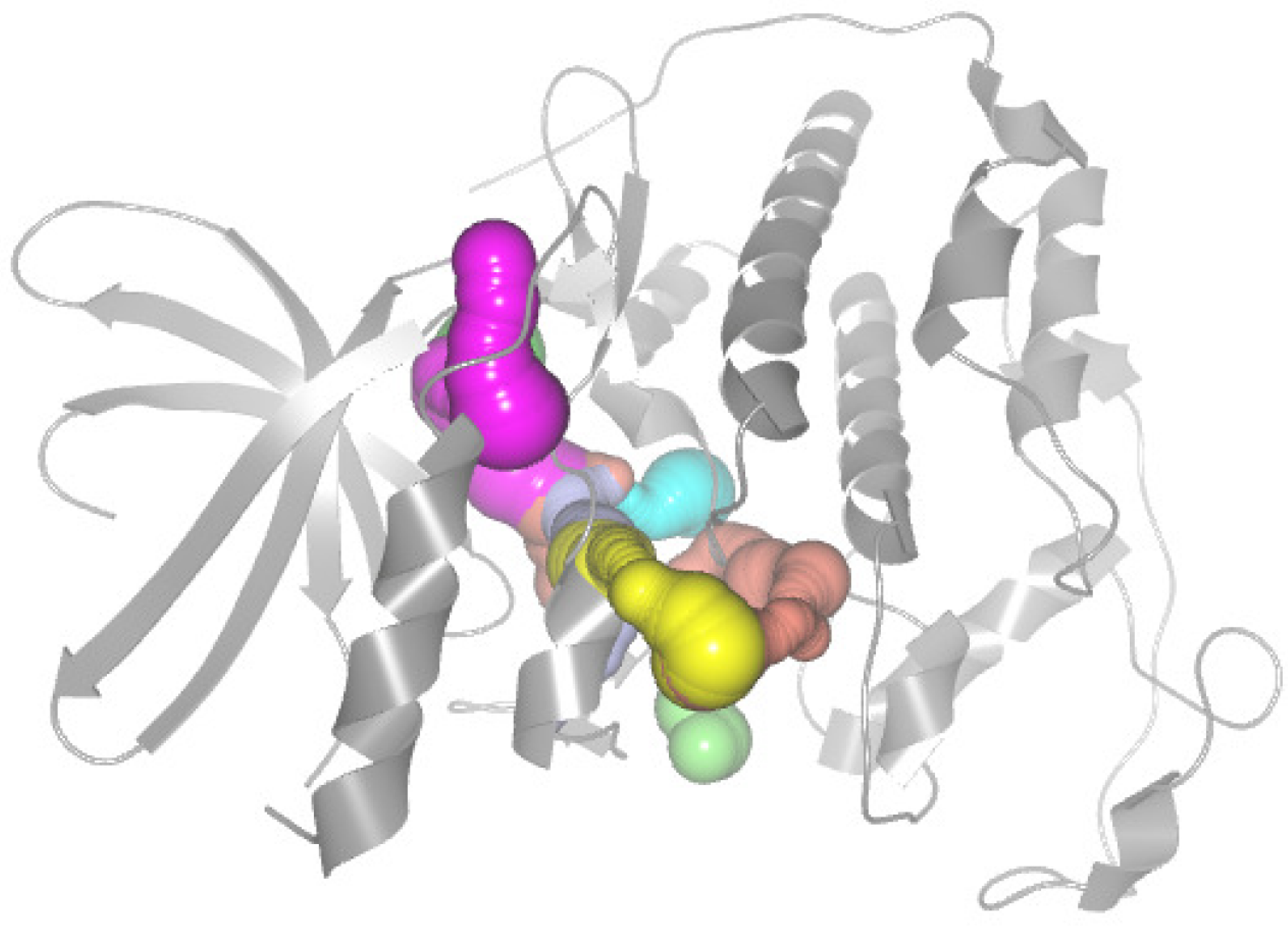
- Elaborate design 2D and 3D views to explore the direct and indirect factors that affect the dynamic characteristics of the cavity.
2. Data and Task Abstraction
- Q1
- Is it the cavity-lining amino acids or other key amino acids that cause the drastic change of the protein cavity?
- Q2
- Which cavity-lining amino acids of the protein cause drastic changes in the cavity?
- Q3
- Which key amino acid atoms of the protein affect the movement of cavity-lining amino acids, resulting in drastic changes in the cavity?
- Q4
- How to explore the motion correlation between cavity-lining amino acids and key amino acids?
- Q5
- What is the relationship between cavity-lining amino acids, key amino acids, and cavity motion in three-dimensional space?
- T1: Observe the cavity dynamics in different modes, and compare and analyze the distribution and fluctuation of amino acid atoms (Q1–Q4).
- T2: Observe the key chemical and physical properties (Q1, Q2) represented by cavity-lining amino acid atoms under different modes.
- T3: It can easily reveal the relationship between cavity-lining amino acid atoms and key amino acid atoms (Q3).
- T4: The possibility of identifying key amino acids by clustering amino acids with motions similarity (Q3, Q4).
- T5: Enhance the understanding of the effect of amino acids on cavity dynamics by studying 3D structures (Q5).
- T6: Analyze the possibility of influencing the cavity-lining amino acid atoms and key amino acid atoms with drastic changes in the cavity from the biological field cases and public data sets (Q1–Q5).
3. Related Work
3.1. Normal Mode Analysis
3.2. Molecular Cavity Calculation and Extraction
3.3. Visualization Method of Molecular Cavity Dynamics
4. Data Preparation
4.1. Normal Mode Calculation
4.2. Cavity Processing
4.3. Dimension Reduction & Clustering
5. Visualization
5.1. Protein Cavity Detail View and Amino Acid Fluctuation HeatMap View
5.2. Amino Acid Fluctuation Similarity Clustering View and Detailed Clustering View
5.3. Protein Cavity Dynamics Influencing Factors View
5.4. Protein Cavity Dynamics 3D View
6. Interactive Exploration
7. Results
7.1. Case Study
7.2. Feedback
8. Discussion and Conclusions
- (1)
- Our work realizes a set of linked 2D and 3D Views, which can be explored interactively to quickly locate the direct factors (cavity-lining amino acid atoms) and indirect factors (key atoms) that affect the dynamic characteristics of the cavity.
- (2)
- Using normal mode data to establish the relationship between cavity-lining amino acid atom movement, key amino acid atom movement, and cavity shape changes, so as to help users find potential allosteric sites.
- (3)
- The system utilizes multiple views from multiple perspectives for the user to explore the stability of the cavity, including cavity characteristics and the properties of the amino acids surrounding the cavity.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koudelakova, T.; Chaloupkova, R.; Brezovsky, J.; Prokop, Z.; Sebestova, E.; Hesseler, M.; Khabiri, M.; Plevaka, M.; Kulik, D.; Smatanova, I.K.; et al. Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel. Angew. Chem. 2013, 125, 2013–2017. [Google Scholar] [CrossRef]
- Pavlova, M.; Klvana, M.; Prokop, Z.; Chaloupkova, R.; Banas, P.; Otyepka, M.; Wade, R.C.; Tsuda, M.; Nagata, Y.; Damborsky, J. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat. Chem. Biol. 2009, 5, 727–733. [Google Scholar] [CrossRef]
- Klvana, M.; Pavlova, M.; Koudelakova, T.; Chaloupkova, R.; Dvorak, P.; Prokop, Z.; Stsiapanava, A.; Kuty, M.; Kuta-Smatanova, I.; Dohnalek, J.; et al. Pathways and mechanisms for product release in the engineered haloalkane dehalogenases explored using classical and random acceleration molecular dynamics simulations. J. Mol. Biol. 2009, 392, 1339–1356. [Google Scholar] [CrossRef]
- Hensen, U.; Meyer, T.; Haas, J.; Rex, R.; Vriend, G.; Grubmüller, H. Exploring protein dynamics space: The dynasome as the missing link between protein structure and function. PLoS ONE 2012, 7, e33931. [Google Scholar] [CrossRef] [Green Version]
- Sykora, J.; Brezovsky, J.; Koudelakova, T.; Lahoda, M.; Fortova, A.; Chernovets, T.; Chaloupkova, R.; Stepankova, V.; Prokop, Z.; Smatanova, I.K.; et al. Dynamics and hydration explain failed functional transformation in dehalogenase design. Nat. Chem. Biol. 2014, 10, 428–430. [Google Scholar] [CrossRef]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-level characterization of the structural dynamics of proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef] [Green Version]
- Chovancova, E.; Pavelka, A.; Benes, P.; Strnad, O.; Brezovsky, J.; Kozlikova, B.; Gora, A.; Sustr, V.; Klvana, M.; Medek, P.; et al. Caver 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012, 8, e1002708. [Google Scholar] [CrossRef] [Green Version]
- Byška, J.; Muzic, M.L.; Groller, M.E.; Viola, I.; Kozlikova, B. Animoaminominer: Exploration of protein tunnels and their properties in molecular dynamics. IEEE Trans. Vis. Comput. Graph. 2015, 22, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shi, J.; Liu, J.; Zhao, J.; Zhou, F.; Zhang, W.; Chen, K.; Zhao, X.; Zhu, C.; Chen, W. Evaluating effects of background stories on graph perception. IEEE Trans. Vis. Comput. Graph. 2021. [Google Scholar] [CrossRef]
- Bedoucha, P.; Reuter, N.; Hauser, H.; Byška, J. Visual exploration of large normal mode spaces to study protein flexibility. Comput. Graph. 2020, 90, 73–83. [Google Scholar] [CrossRef]
- Doruker, P.; Atilgan, A.R.; Bahar, I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: Application to α-amylase inhibitor. Proteins Struct. Funct. Bioinform. 2000, 40, 512–524. [Google Scholar] [CrossRef]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. imods: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Sanejouand, Y.H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001, 14, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Liu, X.; Huang, W.; Lu, S.; Shen, Q.; Zhang, L.; Zhang, J. Improved method for the identification and validation of allosteric sites. J. Chem. Inf. Model. 2017, 57, 2358–2363. [Google Scholar] [CrossRef]
- Levitt, D.G.; Banaszak, L.J. Pocket: A computer graphies method for identifying and displaying protein cavities and their surrounding amino acids. J. Mol. Graph. 1992, 10, 229–234. [Google Scholar] [CrossRef]
- Feng, L.; Wang, F.; Zhang, J.; Tang, Y.; Zhao, J.; Zhou, L.; Wang, J.; Guo, D.; Singh, A.K. Particle-based calculation and visualization of protein cavities using SES models. IEEE J. Biomed. Health Inform. 2021, 26, 2447–2457. [Google Scholar] [CrossRef]
- Hendlich, M.; Rippmann, F.; Barnickel, G. Ligsite: Automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph. Model. 1997, 15, 359–363. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, X.; Peng, C.; Wang, J.; Xu, Z.; Chen, K.; Shi, J.; Zhu, W. D3pockets: A method and web server for systematic analysis of protein pocket dynamics. J. Chem. Inf. Model. 2019, 59, 3353–3358. [Google Scholar] [CrossRef]
- Schmidtke, P.; Guilloux, V.L.; Maupetit, J.; Tufféry, P. Fpocket: Online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 2010, 38 (Suppl. S2), W582–W589. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Pisabarro, M.T. Mspocket: An orientation-independent algorithm for the detection of ligand binding pockets. Bioinformatics 2011, 27, 351–358. [Google Scholar] [CrossRef] [Green Version]
- Brady, G.P.; Stouten, P.F. Fast prediction and visualization of protein binding pockets with pass. J. Comput.-Aided Mol. Des. 2000, 14, 383–401. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhou, Y.; Tanaka, I.; Yao, M. Roll: A new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 2010, 26, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, S.H.; Ferraz, F.A.; Honorato, R.V.; Xavier-Neto, J.; Sobreira, T.J.; de Oliveira, P.S. Kvfinder: Steered identification of protein cavities as a pymol plugin. BMC Bioinform. 2014, 15, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões, T.M.; Gomes, A.J. Cavvis—A field-of-view geometric algorithm for protein cavity detection. J. Chem. Inf. Model. 2019, 59, 786–796. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Sehnal, D.; Deshpande, M.; Vařeková, R.S.; Mir, S.; Berka, K.; Midlik, A.; Pravda, L.; Velankar, S.; Koča, J. Litemol suite: Interactive web-based visualization of large-scale macromolecular structure data. Nat. Methods 2017, 14, 1121–1122. [Google Scholar] [CrossRef]
- Byška, J.; Trautner, T.; Marques, S.M.; Damborský, J.; Kozlíková, B.; Waldner, M. Analysis of long molecular dynamics simulations using interactive focus+context visualization. Comput. Graph. Forum J. Eur. Assoc. Comput. Graph. 2019, 38, 441–453. [Google Scholar] [CrossRef]
- Patro, R.; Ip, C.Y.; Bista, S.; Thirumalai, D.; Cho, S.S.; Varshney, A. Mdmap: A system for data-driven layout and exploration of molecular dynamics simulations. In Proceedings of the 2011 IEEE Symposium on Biological Data Visualization (BioVis), Providence, RI, USA, 23–24 October 2011; pp. 111–118. [Google Scholar] [CrossRef] [Green Version]
- Lindow, N.; Baum, D.; Bondar, A.N.; Hege, H.C. Exploring cavity dynamics in biomolecular systems. BMC Bioinform. 2013, 14 (Suppl. 19), S5. [Google Scholar] [CrossRef] [Green Version]
- Furmanová, K.; Jareová, M.; Byka, J.; Jurík, A.; Parulek, J.; Hauser, H.; Kozlíková, B. Interactive exploration of ligand transportation through protein tunnels. BMC Bioinform. 2017, 18 (Suppl. S2), 22. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Han, D.; Xu, X.; Ye, K.; Nie, J. Spatiotemporal multiscale molecular cavity visualization and visual analysis. J. Vis. 2020, 23, 661–676. [Google Scholar] [CrossRef]
- Guo, D.; Wang, Q.; Liang, M.; Liu, W.; Nie, J. Molecular cavity topological representation for pattern analysis: A nlp analogy-based word2vec method. Int. J. Mol. Sci. 2019, 20, 6019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burch, M.; Ten Brinke, K.B.; Castella, A.; Peters, G.K.S.; Shteriyanov, V.; Vlasvinkel, R. Dynamic graph exploration by interactively linked node-link diagrams and matrix visualizations. Vis. Comput. Ind. Biomed. Art 2021, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.P.; Fuglebakk, E.; Hollup, S.M.; Skjærven, L.; Cragnolini, T.; Grindhaug, S.H.; Tekle, K.M.; Reuter, N. Webnm@ v2.0: Web server and services for comparing protein flexibility. BMC Bioinform. 2014, 15, 427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinsen, K.; Thomas, A.; Field, M. Analysis of domain motion in large proteins. Proteins Struct. Funct. Bioinform. 1999, 34, 369–382. [Google Scholar] [CrossRef]
- Lukman, S.; Grant, G. A network of dynamically conserved residues deciphers the motions of maltose transporter. Proteins 2009, 76, 588–597. [Google Scholar] [CrossRef]
- Wei, Y.; Mei, H.; Zhao, Y.; Zhou, S.; Lin, B.; Jiang, H.; Chen, W. Evaluating Perceptual Bias During Geometric Scaling of Scatterplots. IEEE Trans. Vis. Comput. Graph. 2020, 26, 100–111. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Luo, F.; Chen, M.; Wang, Y.; Xia, J.; Zhou, F.; Wang, Y.; Chen, Y.; Chen, W. Evaluating Multi-Dimensional Visualizations for Understanding Fuzzy Clusters. IEEE Trans. Vis. Comput. Graph. 2019, 25, 12–21. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, H.C. The classic: Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Clin. Orthop. Relat. Res. 2007, 462, 3–5. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein data bank (PDB): The single global macromolecular structure archive. In Protein Crystallography; Humana Press: New York, NY, USA, 2017; pp. 627–641. [Google Scholar]
- Adam, J.; David, B.; Jan, B.; Marques, S.M.; Katarina, F.; Lukas, D.; Piia, K.; Jan, B.; Ondrej, S.; Jan, S. Caver analyst 2.0: Analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 2018, 20, 3586–3588. [Google Scholar]
















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, D.; Feng, L.; Zhang, T.; Guo, Y.; Wang, Y.; Xu, X. PNMAVis: Visual Analysis Tool of Protein Normal Mode for Understanding Cavity Dynamics. Appl. Sci. 2022, 12, 7919. https://doi.org/10.3390/app12157919
Guo D, Feng L, Zhang T, Guo Y, Wang Y, Xu X. PNMAVis: Visual Analysis Tool of Protein Normal Mode for Understanding Cavity Dynamics. Applied Sciences. 2022; 12(15):7919. https://doi.org/10.3390/app12157919
Chicago/Turabian StyleGuo, Dongliang, Li Feng, Taoxiang Zhang, Yaoyao Guo, Yanfen Wang, and Ximing Xu. 2022. "PNMAVis: Visual Analysis Tool of Protein Normal Mode for Understanding Cavity Dynamics" Applied Sciences 12, no. 15: 7919. https://doi.org/10.3390/app12157919
APA StyleGuo, D., Feng, L., Zhang, T., Guo, Y., Wang, Y., & Xu, X. (2022). PNMAVis: Visual Analysis Tool of Protein Normal Mode for Understanding Cavity Dynamics. Applied Sciences, 12(15), 7919. https://doi.org/10.3390/app12157919






