Single Cell RNA-Sequence Analyses Reveal Uniquely Expressed Genes and Heterogeneous Immune Cell Involvement in the Rat Model of Intervertebral Disc Degeneration
Abstract
:1. Introduction
2. Materials and Methods
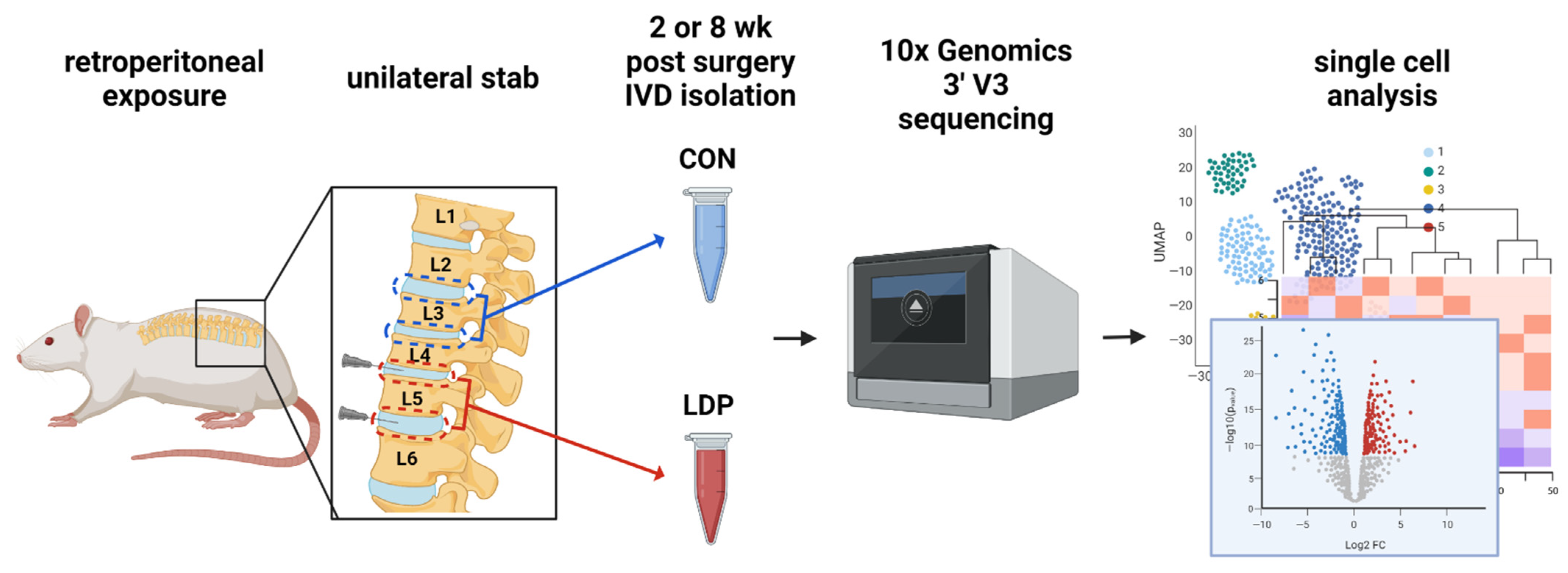
2.1. IVD Tissue Preparation and Single-Cell Isolation
2.2. cDNA Library Generation and Single-Cell RNA-Sequencing (scRNA-Seq) with Data Standardization
2.3. Dimensionality Reduction and Unsupervised Clustering
2.4. Cell Identification
2.5. Gene Specific Analysis
2.6. Gene Set Analyses
2.7. Statistical Analyses
3. Results
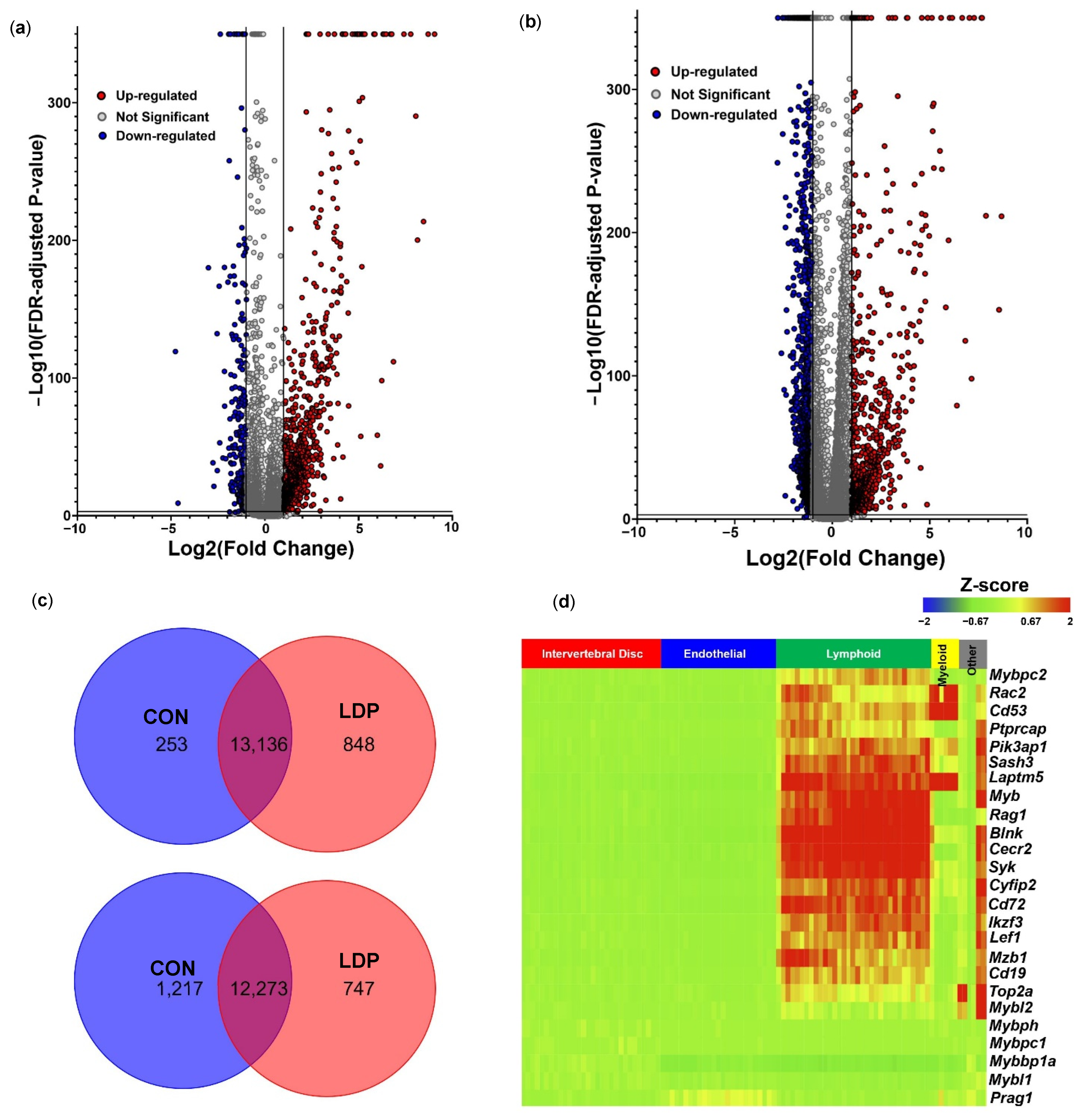
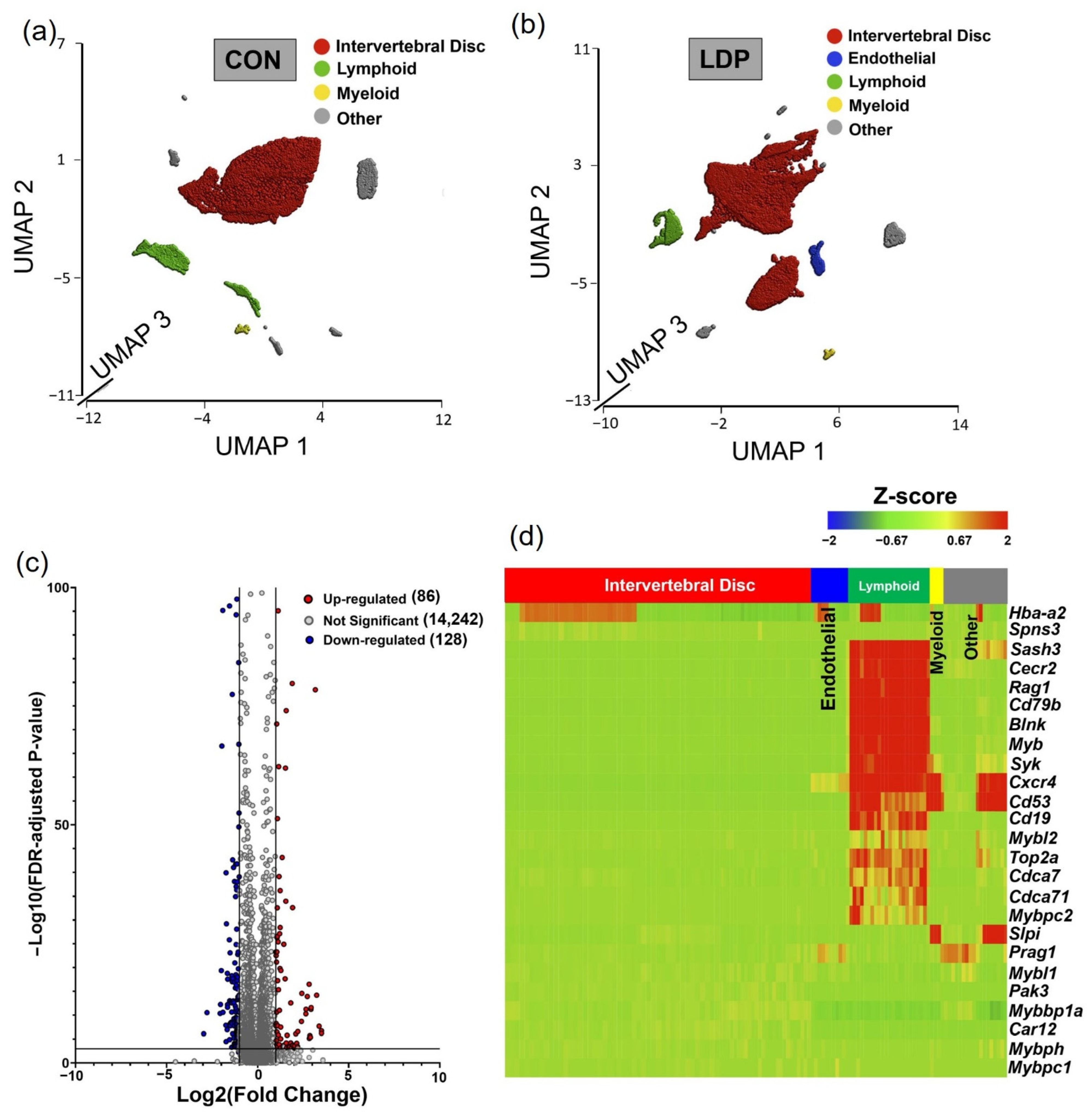
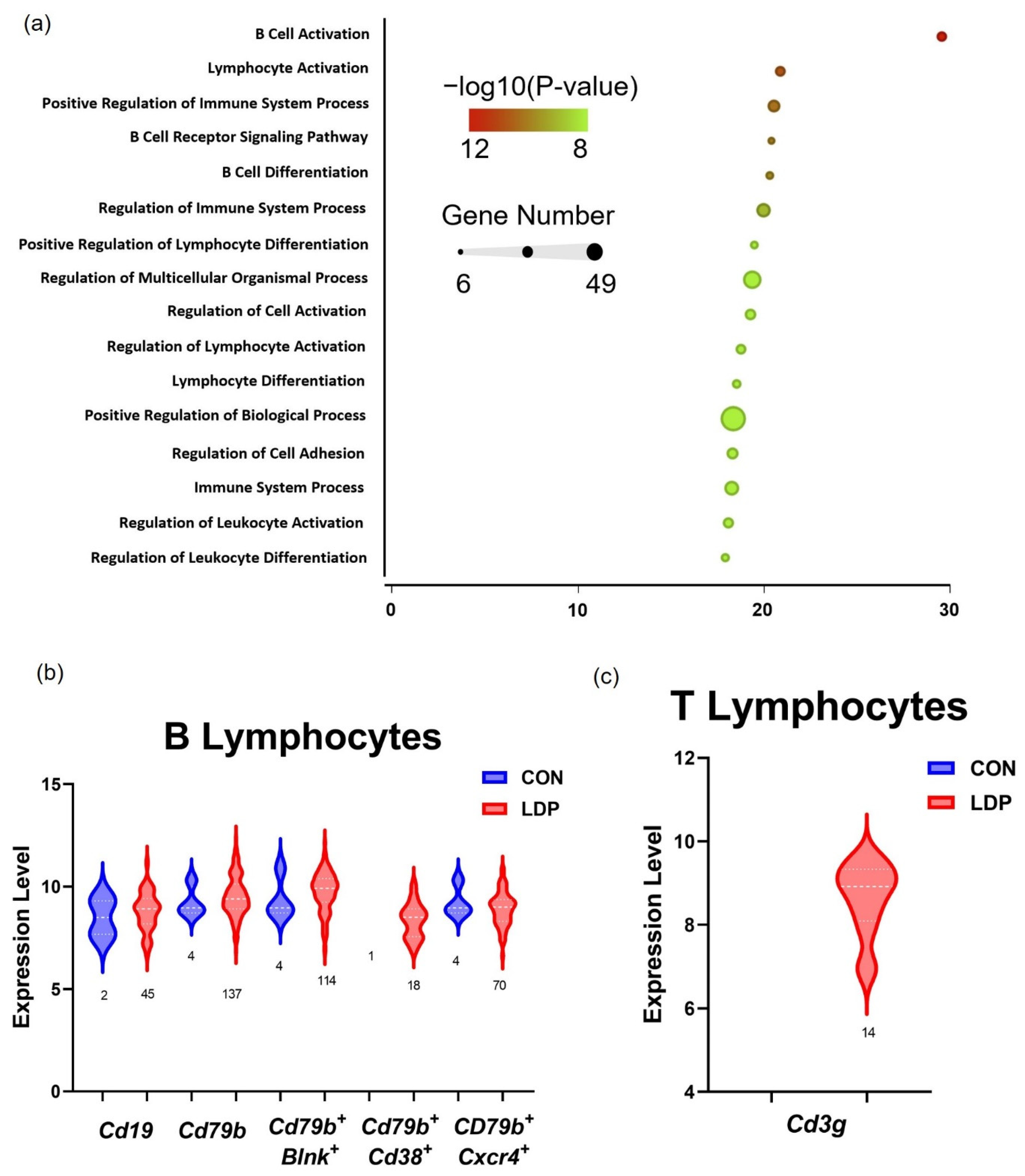
3.1. Cell Populations Identified in IVD Tissues from 2 Weeks Post-Surgery
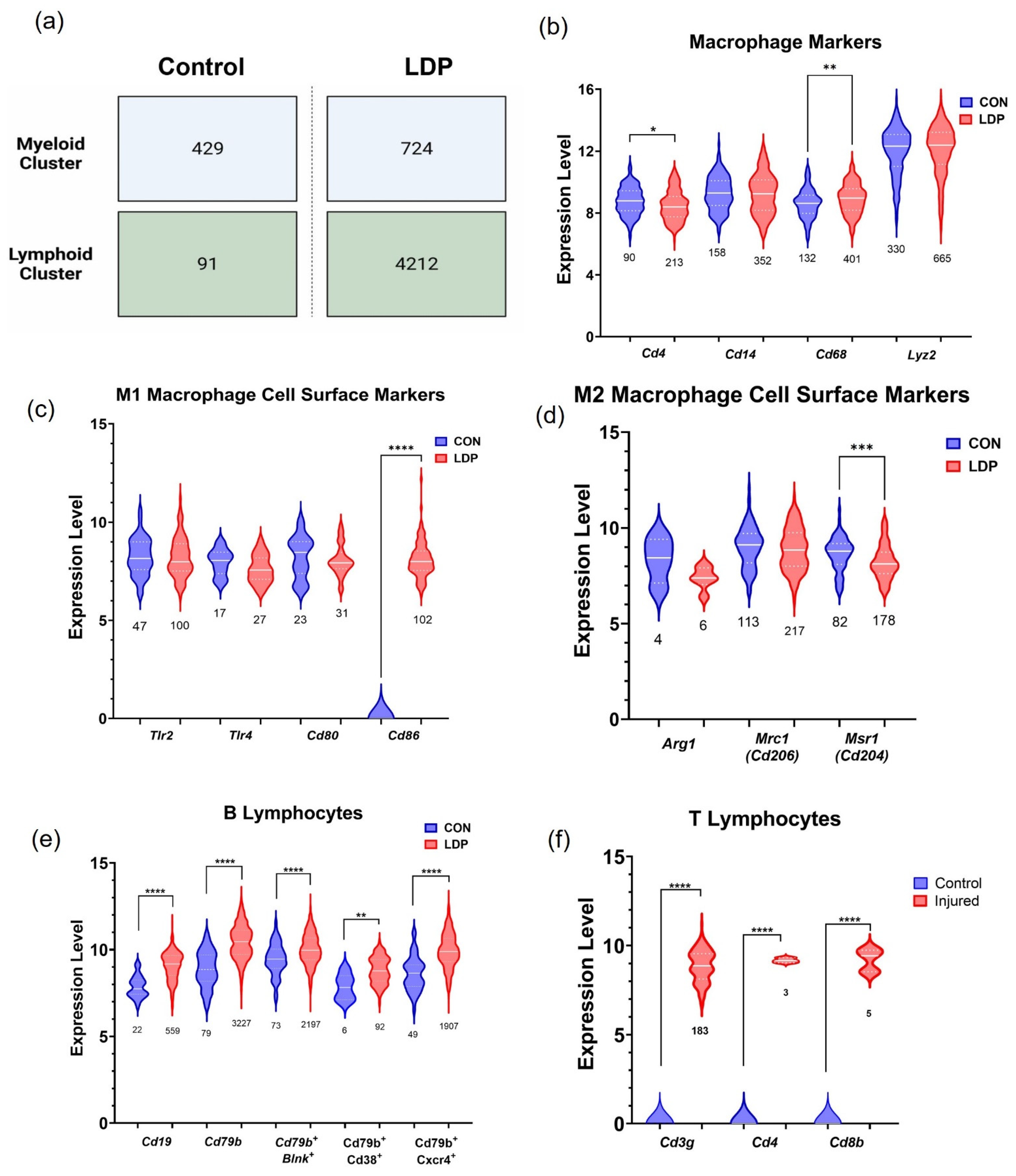
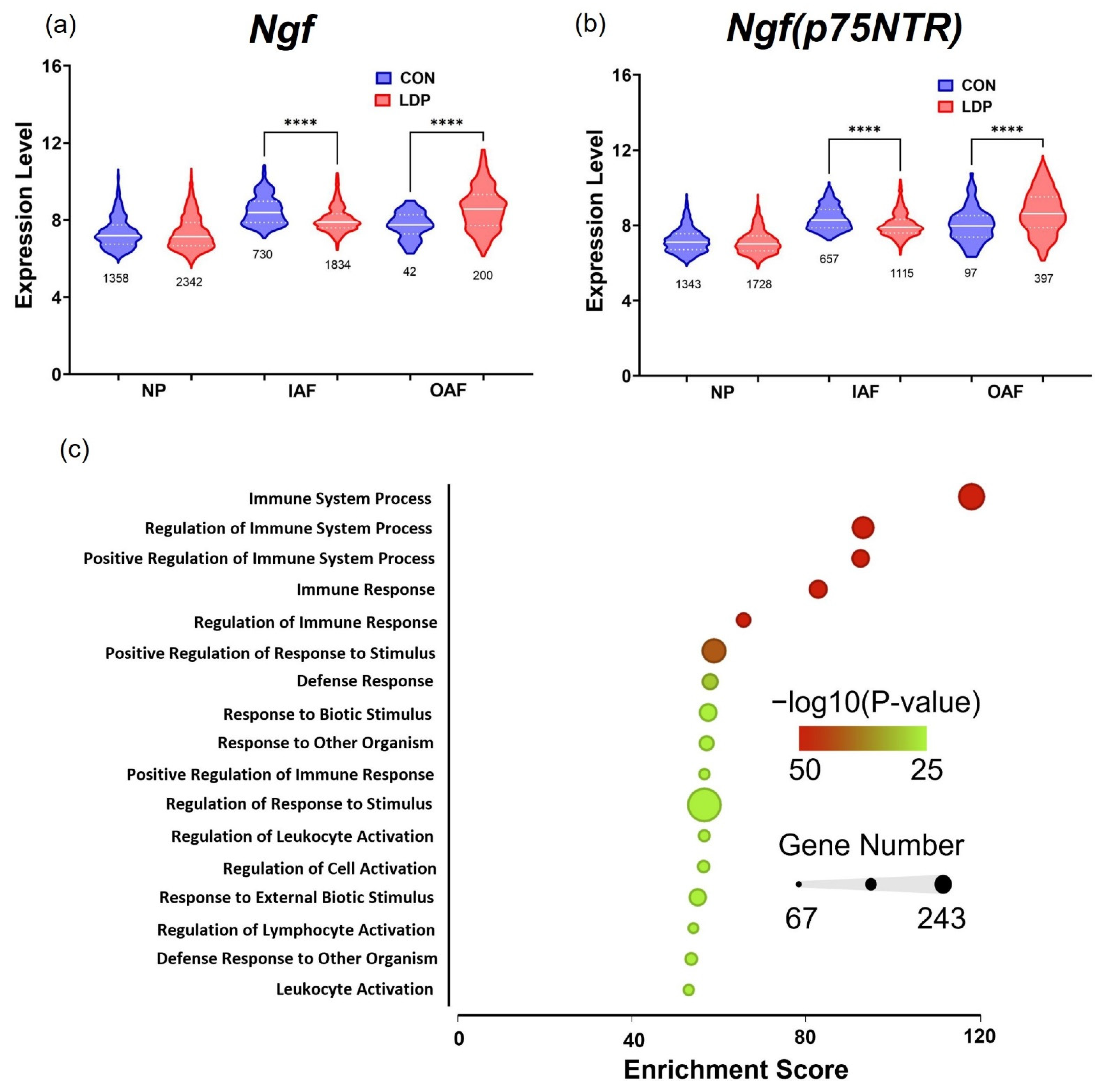
3.2. Cell Populations Identified in IVD Tissues from 8 Weeks Post-Surgery
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Pattappa, G.; Li, Z.; Peroglio, M.; Wismer, N.; Alini, M.; Grad, S. Diversity of intervertebral disc cells: Phenotype and function. J. Anat. 2012, 221, 480–496. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, I.M.; Karppinen, J.; Taimela, S.; Ott, J.; Barral, S.; Kaikkonen, K.; Heikkilä, O.; Mutanen, P.; Noponen, N.; Männikkö, M.; et al. Occupational and Genetic Risk Factors Associated with Intervertebral Disc Disease. Spine 2007, 31, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Sakai, D. Future perspectives of cell-based therapy for intervertebral disc disease. Eur. Spine J. 2008, 17, 452–458. [Google Scholar] [CrossRef]
- Noponen-Hietala, N.; Virtanen, I.; Karttunen, R.; Schwenke, S.; Jakkula, E.; Li, H.; Merikivi, R.; Barral, S.; Ott, J.; Karppinen, J.; et al. Genetic variations in IL6 associate with intervertebral disc disease characterized by sciatica. Pain 2005, 114, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Han, S.; Kong, M.; Tu, Q.; Zhang, L.; Ma, X. Single-cell RNA-seq analysis identifies unique chondrocyte subsets and reveals involvement of ferroptosis in human intervertebral disc degeneration. Osteoarthr. Cartil. 2021, 29, 1324–1334. [Google Scholar] [CrossRef]
- Sun, Y.; Lv, M.; Zhou, L.; Tam, V.; Lv, F.; Chan, D.; Wang, H.; Zheng, Z.; Cheung, K.; Leung, V. Enrichment of committed human nucleus pulposus cells expressing chondroitin sulfate proteoglycans under alginate encapsulation. Osteoarthr. Cartil. 2015, 23, 1194–1203. [Google Scholar] [CrossRef]
- Sive, J.I.; Baird, P.; Jeziorsk, M.; Watkins, A.; Hoyland, J.A.; Freemont, A.J. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol. Pathol. 2002, 55, 91–97. [Google Scholar] [CrossRef]
- Sakai, D.; Nakai, T.; Mochida, J.; Alini, M.; Grad, S. Differential Phenotype of Intervertebral Disc Cells: Microarray and Immunohistochemical Analysis of Canine Nucleus Pulposus and Anulus Fibrosus. Spine 2009, 34, 1448–1456. [Google Scholar] [CrossRef]
- Kim, K.-W.; Lim, T.-H.; Kim, J.G.; Jeong, S.-T.; Masuda, K.; An, H.S. The Origin of Chondrocytes in the Nucleus Pulposus and Histologic Findings Associated with the Transition of a Notochordal Nucleus Pulposus to a Fibrocartilaginous Nucleus Pulposus in Intact Rabbit Intervertebral Discs. Spine 2003, 28, 982–990. [Google Scholar] [CrossRef]
- Clouet, J.; Grimandi, G.; Pot-Vaucel, M.; Masson, M.; Fellah, H.B.; Guigand, L.; Cherel, Y.; Bord, E.; Rannou, F.; Weiss, P.; et al. Identification of phenotypic discriminating markers for intervertebral disc cells and articular chondrocytes. Rheumatology 2009, 48, 1447–1450. [Google Scholar] [CrossRef]
- Chen, J.; Yan, W.; Setton, L.A. Molecular phenotypes of notochordal cells purified from immature nucleus pulposus. Eur. Spine J. 2006, 15, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-S.; Cohn, M.J.; Harfe, B.D. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: Implications for disk degeneration and chordoma formation. Dev. Dyn. 2008, 237, 3953–3958. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Jing, L.; Chen, J. Changes in the Molecular Phenotype of Nucleus Pulposus Cells with Intervertebral Disc Aging. PLoS ONE 2012, 7, e52020. [Google Scholar] [CrossRef]
- Tam, V.; Chen, P.; Yee, A.; Solis, N.; Klein, T.; Kudelko, M.; Sharma, R.; Chan, W.C.; Overall, C.M.; Haglund, L.; et al. DIPPER, a spatiotemporal proteomics atlas of human intervertebral discs for exploring ageing and degeneration dynamics. eLife 2020, 9, e64940. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, Y.; Huang, L.; Shi, K.; Zhu, C.; Li, L.; Zhang, L.; Feng, G.; Song, Y. Novel biomarkers of intervertebral disc cells and evidence of stem cells in the intervertebral disc. Osteoarthr. Cartil. 2020, 29, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Calió, M.; Gantenbein, B.; Egli, M.; Poveda, L.; Ille, F. The Cellular Composition of Bovine Coccygeal Intervertebral Discs: A Comprehensive Single-Cell RNAseq Analysis. Int. J. Mol. Sci. 2021, 22, 4917. [Google Scholar] [CrossRef]
- Fernandes, L.M.; Khan, N.M.; Trochez, C.M.; Duan, M.; Diaz-Hernandez, M.E.; Presciutti, S.M.; Gibson, G.; Drissi, H. Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells. Sci. Rep. 2020, 10, 15263. [Google Scholar] [CrossRef]
- McCann, M.R.; Patel, P.; Frimpong, A.; Xiao, Y.; Siqueira, W.L.; Séguin, C.A. Proteomic Signature of the Murine In-tervertebral Disc. PLoS ONE 2015, 10, e0117807. [Google Scholar]
- Panebianco, C.J.; Dave, A.; Charytonowicz, D.; Sebra, R.; Iatridis, J.C. Single-cell RNA-sequencing atlas of bovine caudal intervertebral discs: Discovery of heterogeneous cell populations with distinct roles in homeostasis. FASEB J. 2021, 35, e21919. [Google Scholar] [CrossRef]
- Gan, Y.; He, J.; Zhu, J.; Xu, Z.; Wang, Z.; Yan, J.; Hu, O.; Bai, Z.; Chen, L.; Xie, Y.; et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus pro-genitors in human intervertebral discs. Bone Res. 2021, 9, 37. [Google Scholar] [CrossRef]
- Han, S.; Zhang, Y.; Zhang, X.; Zhang, H.; Meng, S.; Kong, M.; Liu, X.; Ma, X. Single-Cell RNA Sequencing of the Nucleus Pulposus Reveals Chondrocyte Differentiation and Regulation in Intervertebral Disc Degeneration. Front. Cell Dev. Biol. 2022, 10, 824771. [Google Scholar] [CrossRef] [PubMed]
- Cherif, H.; Mannarino, M.; Pacis, A.S.; Ragoussis, J.; Rabau, O.; Ouellet, J.A.; Haglund, L. Single-Cell RNA-Seq Analysis of Cells from Degenerating and Non-Degenerating Intervertebral Discs from the Same Individual Reveals New Biomarkers for Intervertebral Disc Degeneration. Int. J. Mol. Sci. 2022, 23, 3993. [Google Scholar] [CrossRef] [PubMed]
- Nerlich, A.G.; Weiler, C.; Zipperer, J.; Narozny, M.; Boos, N. Immunolocalization of Phagocytic Cells in Normal and Degenerated Intervertebral Discs. Spine 2002, 27, 2484–2490. [Google Scholar] [CrossRef] [PubMed]
- Sobajima, S.; Shimer, A.L.; Chadderdon, R.C.; Kompel, J.F.; Kim, J.S.; Gilbertson, L.G.; Kang, J.D. Quantitative analysis of gene expression in a rabbit model of intervertebral disc degeneration by real-time polymerase chain reaction. Spine J. 2005, 5, 14–23. [Google Scholar] [CrossRef]
- Kim, J.-S.; Kroin, J.S.; Li, X.; An, H.S.; Buvanendran, A.; Yan, D.; Tuman, K.J.; van Wijnen, A.J.; Chen, D.; Im, H.-J. The rat intervertebral disk degeneration pain model: Relationships between biological and structural alterations and pain. Arthritis Res. Ther. 2011, 13, R165. [Google Scholar] [CrossRef]
- Lai, A.; Moon, A.; Purmessur, D.; Skovrlj, B.; Winkelstein, B.A.; Cho, S.K.; Hecht, A.C.; Iatridis, J.C. Assessment of functional and behavioral changes sensitive to painful disc degeneration. J. Orthop. Res. 2015, 33, 755–764. [Google Scholar] [CrossRef]
- Miyagi, M.; Ishikawa, T.; Orita, S.; Eguchi, Y.; Kamoda, H.; Arai, G.; Suzuki, M.; Inoue, G.; Aoki, Y.; Toyone, T.; et al. Disk Injury in Rats Produces Persistent Increases in Pain-Related Neuropeptides in Dorsal Root Ganglia and Spinal Cord Glia but Only Transient Increases in Inflammatory Mediators: Pathomechanism of Chronic Diskogenic Low Back Pain. Spine 2011, 6, 26. [Google Scholar]
- Elliott, D.M.; Yerramalli, C.S.; Beckstein, J.C.; Boxberger, J.I.; Johannessen, W.; Vresilovic, E.J. The Effect of Relative Needle Diameter in Puncture and Sham Injection Animal Models of Degeneration. Spine 2008, 33, 588–596. [Google Scholar] [CrossRef]
- Risbud, M.V.; Schoepflin, Z.R.; Mwale, F.; Kandel, R.A.; Grad, S.; Iatridis, J.C.; Sakai, D.; Hoyland, J.A. Defining the phenotype of young healthy nucleus pulposus cells: Recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J. Orthop. Res. 2014, 33, 283–293. [Google Scholar] [CrossRef]
- Minogue, B.M.; Richardson, S.M.; Zeef, L.A.; Freemont, A.J.; Hoyland, J.A. Transcriptional profiling of bovine inter-vertebral disc cells: Implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 2010, 12, R22. [Google Scholar] [CrossRef]
- Rutges, J.; Creemers, L.; Dhert, W.; Milz, S.; Sakai, D.; Mochida, J.; Alini, M.; Grad, S. Variations in gene and protein expression in human nucleus pulposus in comparison with annulus fibrosus and cartilage cells: Potential associations with aging and degeneration. Osteoarthr. Cartil. 2010, 18, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Sakai, D.; Nakai, T.; Toyama, K.; Mochida, J.; Alini, M.; Grad, S. A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur. Spine J. 2007, 16, 2174–2185. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Miyamoto, T.; Imai, J.-I.; Hosogane, N.; Suzuki, T.; Yagi, M.; Morita, K.; Ninomiya, K.; Miyamoto, K.; Takaishi, H.; et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem. Biophys. Res. Commun. 2005, 338, 1890–1896. [Google Scholar] [CrossRef]
- Tang, X.; Jing, L.; Richardson, W.J.; Isaacs, R.E.; Fitch, R.D.; Brown, C.R.; Erickson, M.M.; Setton, L.A.; Chen, J. Identifying molecular phenotype of nucleus pulposus cells in human intervertebral disc with aging and degeneration. J. Orthop. Res. 2016, 34, 1316–1326. [Google Scholar] [CrossRef]
- Yang, S.-H.; Hu, M.-H.; Wu, C.-C.; Chen, C.-W.; Sun, Y.-H.; Yang, K.-C. CD24 expression indicates healthier phenotype and less tendency of cellular senescence in human nucleus pulposus cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3021–3028. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, B.; Li, Y.; Duan, H.Q.; Sun, C.; Xu, Y.Q.; Feng, S.Q. Gene expression profile identifies potential biomarkers for human intervertebral disc degeneration. Mol. Med. Rep. 2017, 16, 8665–8672. [Google Scholar] [CrossRef] [PubMed]
- Brisby, H.; Olmarker, K.; Rosengren, L.; Cederlund, C.; Rydevik, B. Markers of Nerve Tissue Injury in the Cerebrospinal Fluid in Patients with Lumbar Disc Herniation and Sciatica. Spine 1999, 24, 742–746. [Google Scholar] [CrossRef]
- Freemont, A.J.; Watkins, A.; Le Maitre, C.; Baird, P.; Jeziorska, M.; Knight, M.T.N.; Ross, E.R.S.; O’Brien, J.P.; Hoyland, J.A. Nerve growth factor expression and innervation of the painful intervertebral disc. J. Pathol. 2002, 197, 286–292. [Google Scholar] [CrossRef]
- Porta, C.; Riboldi, E.; Ippolito, A.; Sica, A. Molecular and epigenetic basis of macrophage polarized activation. Semin. Immunol. 2015, 27, 237–248. [Google Scholar] [CrossRef]
- Abdelaziz, M.H.; Abdelwahab, S.F.; Wan, J.; Cai, W.; Huixuan, W.; Jianjun, C.; Kumar, K.D.; Vasudevan, A.; Sadek, A.; Su, Z.; et al. Alternatively activated macrophages; a double-edged sword in allergic asthma. J. Transl. Med. 2020, 18, 1–12. [Google Scholar] [CrossRef]
- Rousseau, M.-A.A.; Ulrich, J.A.; Bass, E.C.; Rodriguez, A.G.; Liu, J.J.; Lotz, J.C. Stab Incision for Inducing Intervertebral Disc Degeneration in the Rat. Spine 2007, 32, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Kanerva, A.; Kommonen, B.; Grönblad, M.; Tolonen, J.; Habtemariam, A.; Virri, J.; Karaharju, E. Inflammatory Cells in Experimental Intervertebral Disc Injury. Spine 1997, 22, 2711–2715. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Shapiro, I.M. Role of cytokines in intervertebral disc degeneration: Pain and disc content. Nat. Rev. Rheumatol. 2013, 10, 44–56. [Google Scholar] [CrossRef]
- Leimer, E.M.; Gayoso, M.G.; Jing, L.; Tang, S.; Gupta, M.C.; Setton, L.A. Behavioral Compensations and Neuronal Remodeling in a Rodent Model of Chronic Intervertebral Disc Degeneration. Sci. Rep. 2019, 9, 3759. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y. Osteoarthritis year in review 2021: Biology. Osteoarthr. Cartil. 2021, 30, 207–215. [Google Scholar] [CrossRef]
- Kawaguchi, S.; Yamashita, T.; Yokogushi, K.; Murakami, T.; Ohwada, O.; Sato, N. Immunophenotypic Analysis of the Inflammatory Infiltrates in Herniated Intervertebral Discs. Spine 2001, 26, 1209–1214. [Google Scholar] [CrossRef]
- Doita, M.; Kanatani, T.; Harada, T.; Mizuno, K. Immunohistologic Study of the Ruptured Intervertebral Disc of the Lumbar Spine. Spine 1996, 21, 235–241. [Google Scholar] [CrossRef]
- Doita, M.; Kanatani, T.; Ozaki, T.; Matsui, N.; Kurosaka, M.; Yoshiya, S. Influence of Macrophage Infiltration of Herniated Disc Tissue on the Production of Matrix Metalloproteinases Leading to Disc Resorption. Spine 2001, 26, 1522–1527. [Google Scholar] [CrossRef]
- Grönblad, M.; Virri, J.; Tolonen, J.; Seitsalo, S.; Kääpä, E.; Kankare, J.; Myllynen, P.; Karaharju, E.O. A Controlled Immunohistochemical Study of Inflammatory Cells in Disc Herniation Tissue. Spine 1994, 19, 2744–2751. [Google Scholar] [CrossRef]
- Shamji, M.F.; Setton, L.A.; Jarvis, W.; So, S.; Chen, J.; Jing, L.; Bullock, R.; Isaacs, R.E.; Brown, C.; Richardson, W.J. Proinflammatory cytokine expression profile in degenerated and herniated human intervertebral disc tissues. Arthritis Rheum. 2010, 62, 1974–1982. [Google Scholar]
- Jefferies, W.A.; Green, J.R.; Williams, A.F. Authentic T helper CD4 (W3/25) antigen on rat peritoneal macrophages. J. Exp. Med. 1985, 162, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/macrosialin: Not just a histochemical marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Lee, C.; Schindler, C. Deletion of the murine scavenger receptor CD68. J. Lipid Res. 2011, 52, 1542–1550. [Google Scholar] [CrossRef]
- Tesfaigzi, Y.; Daheshia, M. CD14. Encyclopedia of Respiratory Medicine; Laurent, G.J., Shapiro, S.D., Eds.; Academic Press: Oxford, UK, 2006; pp. 343–347. [Google Scholar]
- Markart, P.; Faust, N.; Graf, T.; Na, C.-L.; Weaver, T.E.; Akinbi, H.T. Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem. J. 2004, 380, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T.; Natoli, G. Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 2011, 11, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Shubinsky, G.; Schlesinger, M. The CD38 Lymphocyte Differentiation Marker: New Insight into Its Ectoenzymatic Activity and Its Role as a Signal Transducer. Immunity 1997, 7, 315–324. [Google Scholar] [CrossRef]
- Oliver, A.M.; Martin, F.; Kearney, J.F. Mouse CD38 is down-regulated on germinal center B cells and mature plasma cells. J. Immunol. 1997, 158, 1108–1115. [Google Scholar]
- Becker, M.; Hobeika, E.; Jumaa, H.; Reth, M.; Maity, P.C. CXCR4 signaling and function require the expression of the IgD-class B-cell antigen receptor. Proc. Natl. Acad. Sci. USA 2017, 114, 5231–5236. [Google Scholar] [CrossRef]
- Könnecke, I.; Serra, A.; El Khassawna, T.; Schlundt, C.; Schell, H.; Hauser, A.; Ellinghaus, A.; Volk, H.D.; Radbruch, A.; Duda, G.N.; et al. T and B cells participate in bone repair by infiltrating the fracture callus in a two-wave fashion. Bone 2014, 64, 155–165. [Google Scholar] [CrossRef]
- Muire, P.J.; Mangum, L.H.; Wenke, J.C. Time Course of Immune Response and Immunomodulation During Normal and Delayed Healing of Musculoskeletal Wounds. Front. Immunol. 2020, 11, 1056. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, F.R.; Kurzhagen, J.T.; Rabb, H. Reparative T lymphocytes in organ injury. J. Clin. Investig. 2019, 129, 2608–2618. [Google Scholar] [CrossRef] [PubMed]
- Skaper, S.D. Nerve growth factor: A neuroimmune crosstalk mediator for all seasons. Immunology 2017, 151, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Safieh-Garabedian, B.; Poole, S.; Allchorne, A.; Winter, J.; Woolf, C.J. Contribution of interleukin-1β to the inflamma-tion-induced increase in nerve growth factor levels and inflammatory hyperalgesia. Br. J. Pharmacol. 1995, 115, 1265–1275. [Google Scholar] [CrossRef]
- Aoki, Y.; Takahashi, Y.; Ohtori, S.; Moriya, H.; Takahashi, K. Distribution and immunocytochemical characterization of dorsal root ganglion neurons innervating the lumbar intervertebral disc in rats: A review. Life Sci. 2004, 74, 2627–2642. [Google Scholar] [CrossRef]
- Shu, X.; Mendell, L.M. Nerve growth factor acutely sensitizes the response of adult rat sensory neurons to capsaicin. Neurosci. Lett. 1999, 274, 159–162. [Google Scholar] [CrossRef]
- Mukai, M.; Sakuma, Y.; Suzuki, M.; Orita, S.; Yamauchi, K.; Inoue, G.; Aoki, Y.; Ishikawa, T.; Miyagi, M.; Kamoda, H.; et al. Evaluation of behavior and expression of NaV1.7 in dorsal root ganglia after sciatic nerve compression and application of nucleus pulposus in rats. Eur. Spine J. 2013, 23, 463–468. [Google Scholar] [CrossRef]
- Otten, U.; Ehrhard, P.; Peck, R. Nerve growth factor induces growth and differentiation of human B lymphocytes. Proc. Natl. Acad. Sci. USA 1989, 86, 10059–10063. [Google Scholar] [CrossRef]
- Thorpe, L.W.; Perez-Polo, J.R. The influence of nerve growth factor on the in vitro proliferative response of rat spleen lymphocytes. J. Neurosci. Res. 1987, 18, 134–139. [Google Scholar] [CrossRef]
- Lotz, J.C.; Ulrich, J.A. Innervation, Inflammation, and Hypermobility May Characterize Pathologic Disc Degeneration: Review of Animal Model Data. J. Bone Jt. Surg. 2006, 88, 76–82. [Google Scholar] [CrossRef]
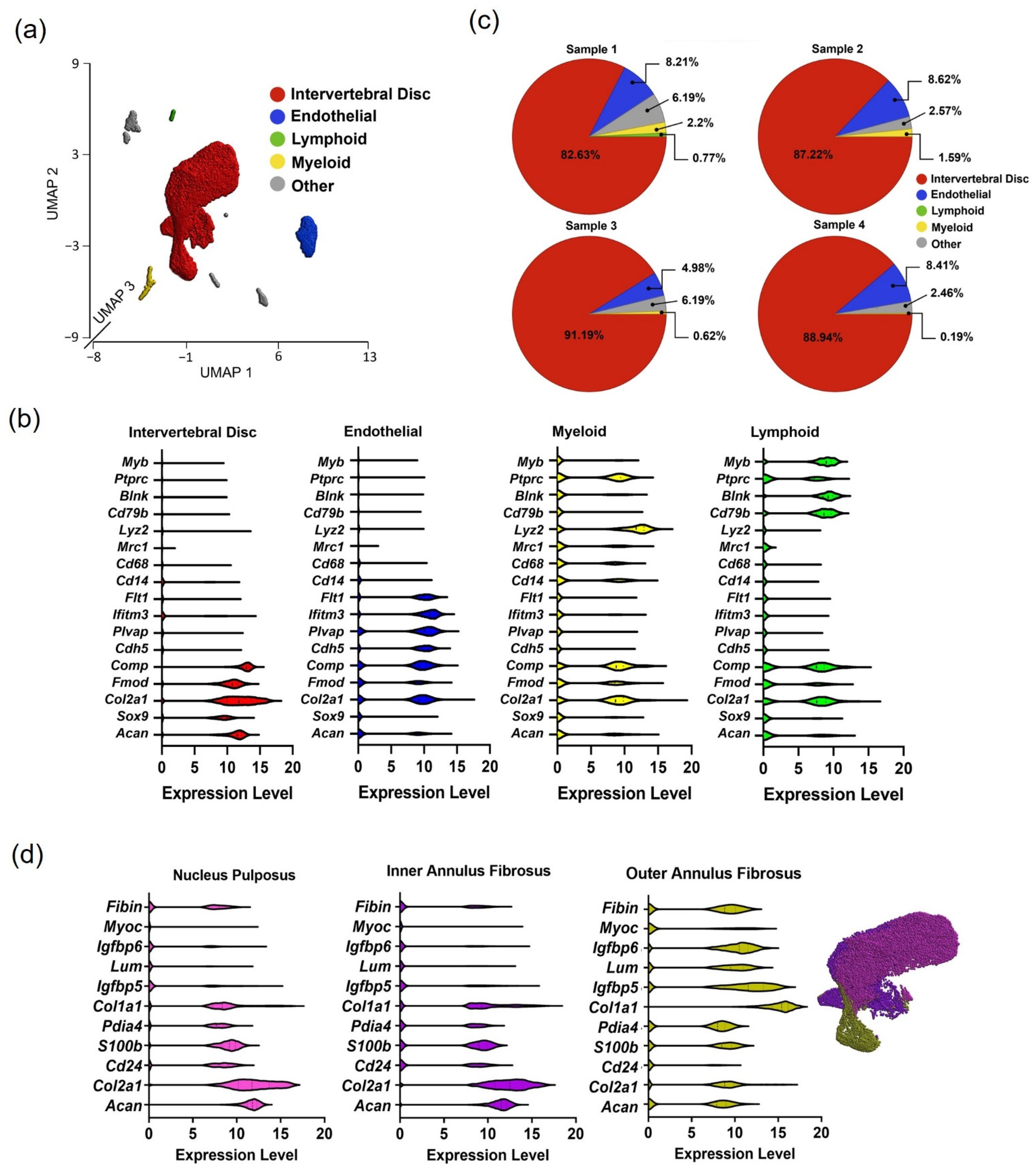
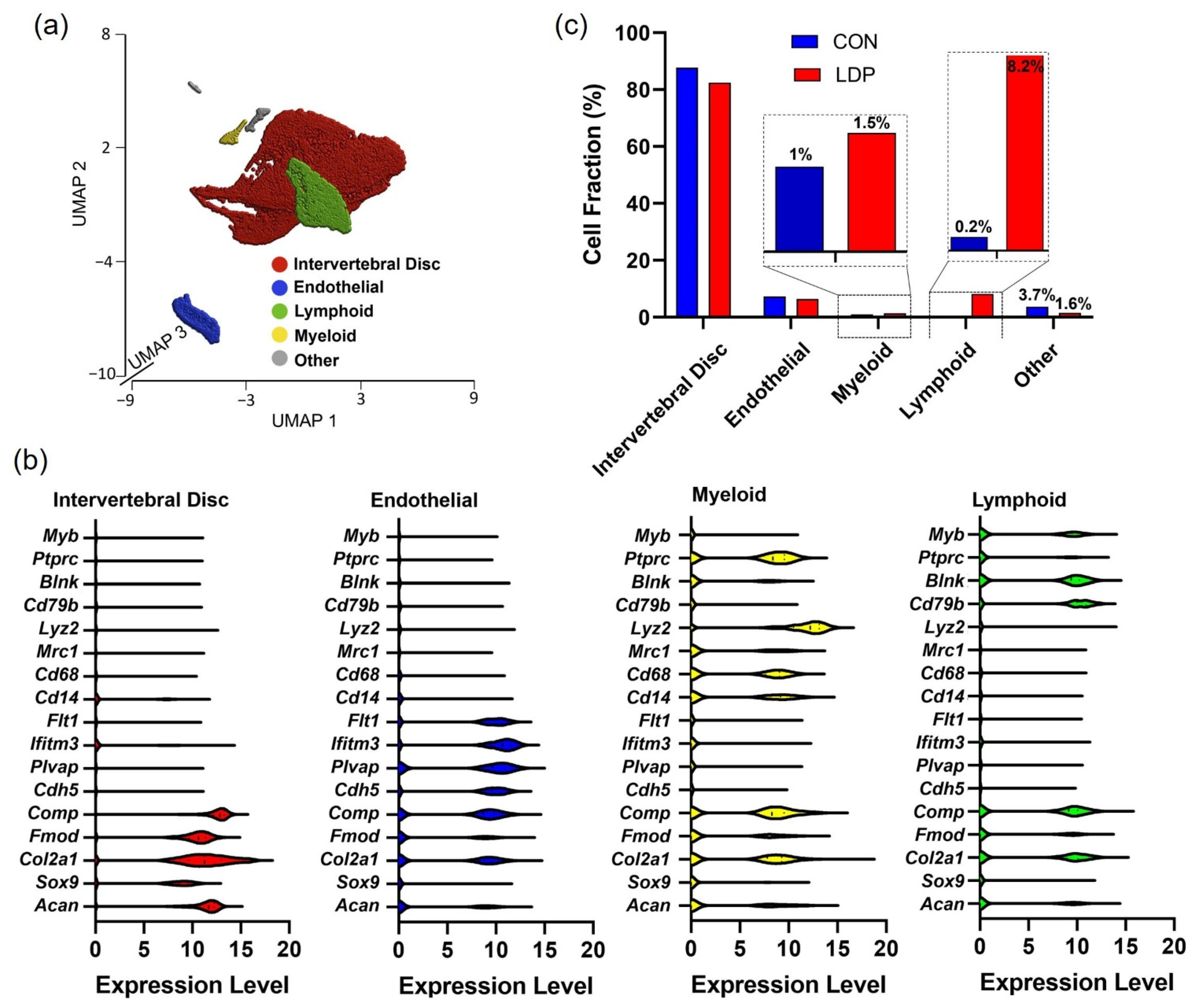
| Cell Type | CON | LDP | Gene Marker |
|---|---|---|---|
| Intervertebral disc cells | 2 week (87.7%) 8 week (83.7%) | 2 week (82.4%) 8 week (87.4%) | Can—aggrecan Col2a1—collagen type II alpha 1 chain Sox9—SRY-box 9 Fmod—fibromodulin Comp—cartilage oligomeric matrix protein |
| Endothelial cells | 2 week (7.3%) 8 week (0%) | 2 week (6.41%) 8 week (2.3%) | Cdh5—cadherin 5 Plvap—plasmalemma vesicle-associated protein Ifitm3—interferon-induced transmembrane protein 3 Flt1—tyrosine–protein kinase receptor |
| Myeloid cells | 2 week (1.0%) 8 week (1.2%) | 2 week (1.4%) 8 week (0.8%) | Cd14—monocyte differentiation antigen CD14 Mrc1—mannose receptor C-Type 1 Cd68—CD68 antigen Lyz2—lysozyme 2 |
| Lymphoid cells | 2 week (0.2%) 8 week (8.4%) | 2 week (8.2%) 8 week (5.1%) | Cd79b—B-cell antigen receptor complex-associated protein beta chain Blnk—B cell linker Ptprc—protein tyrosine phosphatase receptor type C Myb—MYB proto-oncogene, transcription factor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rohanifar, M.; Clayton, S.W.; Easson, G.W.D.; Patil, D.S.; Lee, F.; Jing, L.; Barcellona, M.N.; Speer, J.E.; Stivers, J.J.; Tang, S.Y.; et al. Single Cell RNA-Sequence Analyses Reveal Uniquely Expressed Genes and Heterogeneous Immune Cell Involvement in the Rat Model of Intervertebral Disc Degeneration. Appl. Sci. 2022, 12, 8244. https://doi.org/10.3390/app12168244
Rohanifar M, Clayton SW, Easson GWD, Patil DS, Lee F, Jing L, Barcellona MN, Speer JE, Stivers JJ, Tang SY, et al. Single Cell RNA-Sequence Analyses Reveal Uniquely Expressed Genes and Heterogeneous Immune Cell Involvement in the Rat Model of Intervertebral Disc Degeneration. Applied Sciences. 2022; 12(16):8244. https://doi.org/10.3390/app12168244
Chicago/Turabian StyleRohanifar, Milad, Sade W. Clayton, Garrett W.D. Easson, Deepanjali S. Patil, Frank Lee, Liufang Jing, Marcos N. Barcellona, Julie E. Speer, Jordan J. Stivers, Simon Y. Tang, and et al. 2022. "Single Cell RNA-Sequence Analyses Reveal Uniquely Expressed Genes and Heterogeneous Immune Cell Involvement in the Rat Model of Intervertebral Disc Degeneration" Applied Sciences 12, no. 16: 8244. https://doi.org/10.3390/app12168244






