Deep Learning for Orthopedic Disease Based on Medical Image Analysis: Present and Future
Abstract
:1. Introduction
2. Deep Learning for Fractures
3. Deep Learning for Osteoarthritis and Prediction of Arthroplasty Implants
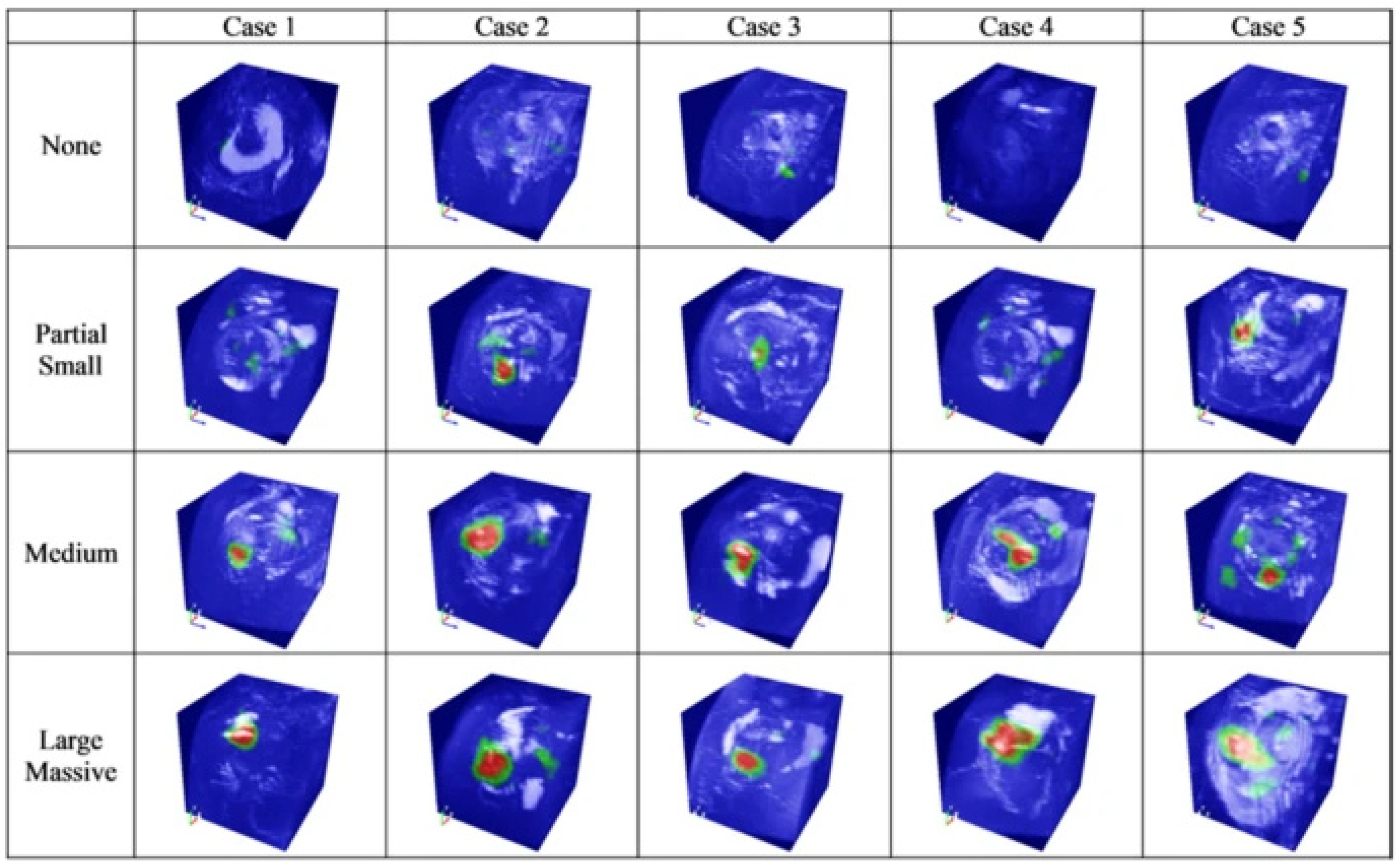
4. Deep Learning for Joint-Specific Soft Tissue Disease
5. Miscellaneous
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hubel, D.H.; Wiesel, T.N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 1962, 160, 106–154. [Google Scholar] [CrossRef] [PubMed]
- Hornik, K.; Stinchcombe, M.; White, H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989, 2, 359–366. [Google Scholar] [CrossRef]
- Zhao, Z.-Q.; Zheng, P.; Xu, S.-T.; Wu, X. Object detection with deep learning: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Casalino, L.P.; Khullar, D. Deep learning in medicine—Promise, progress, and challenges. JAMA Intern. Med. 2019, 179, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Budd, S.; Robinson, E.C.; Kainz, B. A survey on active learning and human-in-the-loop deep learning for medical image analysis. Med. Image Anal. 2021, 71, 102062. [Google Scholar] [CrossRef]
- Fujita, H. AI-based computer-aided diagnosis (AI-CAD): The latest review to read first. Radiol. Phys. Technol. 2020, 13, 6–19. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, I.; Rundo, L.; Codari, M.; Leo, G.D.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Med. 2021, 83, 9–24. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4793–4813. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 30 June 2016; pp. 2921–2929. [Google Scholar]
- Shim, E.; Kim, J.Y.; Yoon, J.P.; Ki, S.-Y.; Lho, T.; Kim, Y.; Chung, S.W. Automated rotator cuff tear classification using 3D convolutional neural network. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef]
- Kim, B.; Wattenberg, M.; Gilmer, J.; Cai, C.; Wexler, J.; Viegas, F. Interpretability beyond feature attribution: Quantitative testing with concept activation vectors (tcav). In Proceedings of the 35th International Conference on Machine Learning, PMLR, Stockholm, Sweden, 10–15 July 2018; pp. 2668–2677. [Google Scholar]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; The CAMELYON16 Consortium. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Chung, S.W.; Han, S.S.; Lee, J.W.; Oh, K.-S.; Kim, N.R.; Yoon, J.P.; Kim, J.Y.; Moon, S.H.; Kwon, J.; Lee, H.-J.; et al. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop. 2018, 89, 468–473. [Google Scholar] [CrossRef] [Green Version]
- Demir, S.; Key, S.; Tuncer, T.; Dogan, S. An exemplar pyramid feature extraction based humerus fracture classification method. Med. Hypotheses 2020, 140, 109663. [Google Scholar] [CrossRef]
- Urakawa, T.; Tanaka, Y.; Goto, S.; Matsuzawa, H.; Watanabe, K.; Endo, N. Detecting intertrochanteric hip fractures with orthopedist-level accuracy using a deep convolutional neural network. Skelet. Radiol. 2019, 48, 239–244. [Google Scholar] [CrossRef]
- Yamada, Y.; Maki, S.; Kishida, S.; Nagai, H.; Arima, J.; Yamakawa, N.; Iijima, Y.; Shiko, Y.; Kawasaki, Y.; Kotani, T.; et al. Automated classification of hip fractures using deep convolutional neural networks with orthopedic surgeon-level accuracy: Ensemble decision-making with antero-posterior and lateral radiographs. Acta Orthop. 2020, 91, 699–704. [Google Scholar] [CrossRef]
- Lee, C.; Jang, J.; Lee, S.; Kim, Y.S.; Jo, H.J.; Kim, Y. Classification of femur fracture in pelvic X-ray images using meta-learned deep neural network. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.; Akbarian, E.; Olsson, S.; Nåsell, H.; Sköldenberg, O.; Razavian, A.S.; Gordon, M. Artificial intelligence for the classification of fractures around the knee in adults according to the 2018 AO/OTA classification system. PLoS ONE 2021, 16, e0248809. [Google Scholar] [CrossRef] [PubMed]
- Farda, N.A.; Lai, J.-Y.; Wang, J.-C.; Lee, P.-Y.; Liu, J.-W.; Hsieh, I.-H. Sanders classification of calcaneal fractures in CT images with deep learning and differential data augmentation techniques. Injury 2020, 52, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Ozkaya, E.; Topal, F.E.; Bulut, T.; Gursoy, M.; Ozuysal, M.; Karakaya, Z. Evaluation of an artificial intelligence system for diagnosing scaphoid fracture on direct radiography. Eur. J. Trauma Emerg. Surg. 2020, 1–8. [Google Scholar] [CrossRef]
- Langerhuizen, D.W.G.; Bulstra, A.E.J.; Janssen, S.J.; Ring, D.; Kerkhoffs, G.M.M.J.; Jaarsma, R.L.; Doornberg, J.N. Is deep learning on par with human observers for detection of radiographically visible and occult fractures of the scaphoid? Clin. Orthop. Relat. Res. 2020, 478, 2653–2659. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Hsu, B.W.-Y.; Yin, Y.-K.; Lin, F.-H.; Yang, T.-H.; Yang, R.-S.; Lee, C.-K.; Tseng, V.S. Application of deep learning algorithm to detect and visualize vertebral fractures on plain frontal radiographs. PLoS ONE 2021, 16, e0245992. [Google Scholar] [CrossRef] [PubMed]
- Yabu, A.; Hoshino, M.; Tabuchi, H.; Takahashi, S.; Masumoto, H.; Akada, M.; Morita, S.; Maeno, T.; Iwamae, M.; Inose, H.; et al. Using artificial intelligence to diagnose fresh osteoporotic vertebral fractures on magnetic resonance images. Spine J. 2021, 21, 1652–1658. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.L.; Jung, S.H.; Choi, G.Y. Ecaluation of focal bone mineral density using three-dimensional of Hounsfield units in the proximal humerus. CiSE. 2015, 18, 86–90. [Google Scholar]
- Xue, Y.; Zhang, R.; Deng, Y.; Chen, K.; Jiang, T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS ONE 2017, 12, e0178992. [Google Scholar] [CrossRef] [Green Version]
- Üreten, K.; Arslan, T.; Gültekin, K.E.; Demir, A.N.D.; Özer, H.F.; Bilgili, Y. Detection of hip osteoarthritis by using plain pelvic radiographs with deep learning methods. Skelet. Radiol. 2020, 49, 1369–1374. [Google Scholar] [CrossRef]
- Tiulpin, A.; Thevenot, J.; Rahtu, E.; Lehenkari, P.; Saarakkala, S. Automatic Knee Osteoarthritis Diagnosis from Plain Radiographs: A Deep Learning-Based Approach. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Swiecicki, A.; Li, N.; O’Donnell, J.; Said, N.; Yang, J.; Mather, R.C.; Jiranek, D.A.; Mazurowski, M.A. Deep learning-based algorithm for assessment of knee osteoarthritis severity in radiographs matches performance of radiologists. Comput. Biol. Med. 2021, 133, 104334. [Google Scholar] [CrossRef] [PubMed]
- Pedoia, V.; Lee, J.; Norman, B.; Link, T.M.; Majumdar, S. Diagnosing osteoarthritis from T2 maps using deep learning: An analysis of the entire Osteoarthritis Initiative baseline cohort. Osteoarthr. Cartil. 2019, 27, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lee, K.J.; Choi, D.; Lee, J.I.; Choi, H.G.; Lee, Y.S. Can Additional Patient Information Improve the Diagnostic Performance of Deep Learning for the Interpretation of Knee Osteoarthritis Severity. J. Clin. Med. 2020, 9, 3341. [Google Scholar] [CrossRef]
- Karnuta, J.M.; Luu, B.C.; Roth, A.L.; Haeberle, H.S.; Chen, A.F.; Iorio, R.; Schaffer, J.L.; Mont, M.A.; Patterson, B.M.; Krebs, V.E.; et al. Artificial Intelligence to Identify Arthroplasty Implants From Radiographs of the Knee. J. Arthroplast. 2021, 36, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Borjali, A.; Chen, A.F.; Muratoglu, O.K.; Morid, M.A.; Varadarajan, K.M. Detecting total hip replacement prosthesis design on plain radiographs using deep convolutional neural network. J. Orthop. Res. 2020, 38, 1465–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.-J.; Yoo, J.-I.; Cha, Y.-H.; Park, C.H.; Kim, J.-T. Machine learning–based identification of hip arthroplasty designs. J. Orthop. Transl. 2020, 21, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Urban, G.; Porhemmat, S.; Stark, M.; Feeley, B.; Okada, K.; Baldi, P. Classifying shoulder implants in X-ray images using deep learning. Comput. Struct. Biotechnol. J. 2020, 18, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Sultan, H.; Owais, M.; Park, C.; Mahmood, T.; Haider, A.; Park, K.R. Artificial Intelligence-Based Recognition of Different Types of Shoulder Implants in X-ray Scans Based on Dense Residual Ensemble-Network for Personalized Medicine. J. Pers. Med. 2021, 11, 482. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, Y.; Georgiou, T.; Lew, M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 2018, 7, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Wang, G. Skeletal bone age prediction based on a deep residual network with spatial transformer. Comput. Methods Programs Biomed. 2020, 197, 105754. [Google Scholar] [CrossRef]
- Kim, J.Y.; Ro, K.; You, S.; Nam, B.R.; Yook, S.; Park, H.S.; Yoo, J.C.; Park, E.; Cho, K.; Cho, B.H.; et al. Development of an automatic muscle atrophy measuring algorithm to calculate the ratio of supraspinatus in supraspinous fossa using deep learning. Comput. Methods Programs Biomed. 2019, 182, 105063. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, E.; Truffer, O.; Becce, F.; Eminian, S.; Gidoin, S.; Terrier, A.; Farron, A.; Büchler, P. Deep learning for the rapid automatic quantification and characterization of rotator cuff muscle degeneration from shoulder CT datasets. Eur. Radiol. 2021, 31, 181–190. [Google Scholar] [CrossRef]
- Medina, G.; Buckless, C.G.; Thomasson, E.; Oh, L.S.; Torriani, M. Deep learning method for segmentation of rotator cuff muscles on MR images. Skelet. Radiol. 2021, 50, 683–692. [Google Scholar] [CrossRef]
- Lee, K.; Kim, J.Y.; Lee, M.H.; Choi, C.-H.; Hwang, J.Y. Imbalanced Loss-Integrated Deep-Learning-Based Ultrasound Image Analysis for Diagnosis of Rotator-Cuff Tear. Sensors 2021, 21, 2214. [Google Scholar] [CrossRef]
- Couteaux, V.; Si-Mohamed, S.; Nempont, O.; Lefevre, T.; Popoff, A.; Pizaine, G.; Villain, N.; Bloch, I.; Cotten, A.; Boussel, L. Automatic knee meniscus tear detection and orientation classification with Mask-RCNN. Diagn. Interv. Imaging 2019, 100, 235–242. [Google Scholar] [CrossRef]
- Roblot, V.; Giret, Y.; Antoun, M.B.; Morillot, C.; Chassin, X.; Cotten, A.; Zerbib, J.; Fournier, L. Artificial intelligence to diagnose meniscus tears on MRI. Diagn. Interv. Imaging 2019, 100, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.D.; Wong, T.T.; Rasiej, M.J. Deep Learning for Detection of Complete Anterior Cruciate Ligament Tear. J. Digit. Imaging 2019, 32, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Flannery, S.W.; Kiapour, A.M.; Edgar, D.J.; Murray, M.M.; Fleming, B.C. Automated magnetic resonance image segmentation of the anterior cruciate ligament. J. Orthop. Res. 2021, 39, 831–840. [Google Scholar] [CrossRef]
- Mahmoodi, S.; Sharif, B.S.; Chester, E.G.; Owen, J.P.; Lee, R. Skeletal growth estimation using radiographic image processing and analysis. IEEE Trans. Inf. Technol. Biomed. 2000, 4, 292–297. [Google Scholar] [CrossRef] [Green Version]
- Kyung, B.S.; Lee, S.H.; Jeong, W.K.; Park, S.Y. Disparity between clinical and ultrasound examinations in neonatal hip screening. CiOS 2016, 8, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.-C.; Sun, J.; Liu, C.-B.; Fang, J.-H.; Xie, H.-T.; Ning, B. Clinical application of artificial intelligence-assisted diagnosis using anteroposterior pelvic radiographs in children with developmental dysplasia of the hip. Bone Jt. J. 2020, 102, 1574–1581. [Google Scholar] [CrossRef]
- Rhyou, I.H.; Lee, J.H.; Park, K.J.; Kang, H.S.; Kim, K.W. The ulnar collateral ligament is always torn in the posterolateral elbow dislocation: A suggestion on the new mechanism of dislocation using MRI findings. CiSE 2011, 14, 193–198. [Google Scholar] [CrossRef]
- England, J.R.; Gross, J.S.; White, E.A.; Patel, D.B.; England, J.T.; Cheng, P.M. Detection of Traumatic Pediatric Elbow Joint Effusion Using a Deep Convolutional Neural Network. Am. J. Roentgenol. 2018, 211, 1361–1368. [Google Scholar] [CrossRef]
- Zhang, B.; Yu, K.; Ning, Z.; Wang, K.; Dong, Y.; Liu, X.; Liu, S.; Wang, J.; Zhu, C.; Yu, Q.; et al. Deep learning of lumbar spine X-ray for osteopenia and osteoporosis screening: A multicenter retrospective cohort study. Bone 2020, 140, 115561. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Sukegawa, S.; Kitamura, A.; Goto, R.; Noda, T.; Nakano, K.; Takabatake, K.; Kawai, H.; Nagatsuka, H.; Kawasaki, K.; et al. Deep Learning for Osteoporosis Classification Using Hip Radiographs and Patient Clinical Covariates. Biomolecules 2020, 10, 1534. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Yang, W.; Wei, S.; Cai, R.; Li, J.; Guo, S.; Li, Q.; Wang, J.; Li, X. Automated measurement of hip–knee–ankle angle on the unilateral lower limb X-rays using deep learning. Phys. Eng. Sci. Med. 2021, 44, 53–62. [Google Scholar] [CrossRef]
- Rouzrokh, P.; Wyles, C.C.; Philbrick, K.A.; Ramazanian, T.; Weston, A.D.; Cai, J.C.; Taunton, M.J.; Lewallen, D.G.; Berry, D.J.; Erickson, B.J.; et al. A Deep Learning Tool for Automated Radiographic Measurement of Acetabular Component Inclination and Version After Total Hip Arthroplasty. J. Arthroplast. 2021, 36, 2510–2517.e6. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.J.; Kim, S.T.; Yoon, M.G.; Kim, S.S.; Moon, M.S. Chronic osteomyelitis of the lumbar transverse process. CiOS 2011, 3, 254–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Fang, Z.; Lang, N.; Yuan, H.; Su, M.-Y.; Baldi, P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput. Biol. Med. 2017, 84, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Chmelik, J.; Jakubicek, R.; Walek, P.; Jan, J.; Ourednicek, P.; Lambert, L.; Amadori, E.; Gavelli, G. Deep convolutional neural network-based segmentation and classification of difficult to define metastatic spinal lesions in 3D CT data. Med. Image Anal. 2018, 49, 76–88. [Google Scholar] [CrossRef]
- Kim, K.; Kim, S.; Lee, Y.H.; Lee, S.H.; Lee, H.S.; Kim, S. Performance of the deep convolutional neural network based magnetic resonance image scoring algorithm for differentiating between tuberculous and pyogenic spondylitis. Sci. Rep. 2018, 8, 13124. [Google Scholar] [CrossRef]
- Won, D.; Lee, H.-J.; Lee, S.-J.; Park, S.H. Spinal Stenosis Grading in Magnetic Resonance Imaging Using Deep Convolutional Neural Networks. Spine 2020, 45, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Rouzrokh, P.; Ramazanian, T.; Wyles, C.C.; Philbrick, K.A.; Cai, J.C.; Taunton, M.J.; Kremers, H.M.; Lewallen, D.G.; Erickson, B.J. Deep Learning Artificial Intelligence Model for Assessment of Hip Dislocation Risk Following Primary Total Hip Arthroplasty From Postoperative Radiographs. J. Arthroplast. 2021, 36, 2197–2203.e3. [Google Scholar] [CrossRef]
- Huang, W.; Zhou, F. DA-CapsNet: Dual attention mechanism capsule network. Sci. Rep. 2020, 10, 11383. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, F.; Kakimoto, A.; Ota, N.; Ito, S.; Nishizawa, S. Automated segmentation of 2D low-dose CT images of the psoas-major muscle using deep convolutional neural networks. Radiol. Phys. Technol. 2019, 12, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, N.; Li, J.; Kume, M.; Fujita, H.; Shen, D.; Zheng, G. Fully automatic segmentation of paraspinal muscles from 3D torso CT images via multi-scale iterative random forest classifications. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Cao, X.; Liang, J.; Chen, X.; Zhan, Y. Medical Image Segmentation based on U-Net: A Review. J. Imaging Sci. Technol. 2020, 64, 20508. [Google Scholar] [CrossRef]
- Hiasa, Y.; Otake, Y.; Takao, M.; Ogawa, T.; Sugano, N.; Sato, Y. Automated Muscle Segmentation from Clinical CT Using Bayesian U-Net for Personalized Musculoskeletal Modeling. IEEE Trans. Med. Imaging 2020, 39, 1030–1040. [Google Scholar] [CrossRef] [Green Version]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Rundo, L.; Han, C.; Nagano, Y.; Zhang, J.; Hataya, R.; Militello, C.; Tangherloni, A.; Nobile, M.S.; Ferretti, C.; Besozzi, D.; et al. USE-Net: Incorporating Squeeze-and-Excitation blocks into U-Net for prostate zonal segmentation of multi-institutional MRI datasets. Neurocomputing 2019, 365, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Focus U-Net: A novel dual attention-gated CNN for polyp segmentation during colonoscopy. Comput. Biol. Med. 2021, 137, 104815. [Google Scholar] [CrossRef] [PubMed]
- Kolachalama, V.B.; Garg, P.S. Machine learning and medical education. NPJ Digit. Med. 2018, 1, 54. [Google Scholar] [CrossRef]
- Nie, D.; Trullo, R.; Lian, J.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical image synthesis with context-aware generative adversarial networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Quebec City, QC, Canada, 11–13 September 2017; Springer: Cham, Switzerland; pp. 417–425. [Google Scholar]
- Ker, J.; Wang, L.; Rao, J.; Lim, C.T. Deep Learning Applications in Medical Image Analysis. IEEE Access 2017, 6, 9375–9389. [Google Scholar] [CrossRef]
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. Welcoming new guidelines for AI clinical research. Nat. Med. 2020, 26, 1318–1320. [Google Scholar] [CrossRef]
- Liu, X.; Rivera, S.C.; Moher, D.; Calvert, M.J.; Denniston, A.K. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: The CONSORT-AI Extension. BMJ 2020, 370, m3164. [Google Scholar] [CrossRef] [PubMed]
- Grauhan, N.F.; Niehues, S.M.; Gaudin, R.A.; Keller, S.; Vahldiek, J.L.; Adams, L.C.; Bressem, K.K. Deep learning for accurately recognizing common causes of shoulder pain on radiographs. Skelet. Radiol. 2021, 51, 355–362. [Google Scholar] [CrossRef]
- Kang, Y.; Choi, D.; Lee, K.J.; Oh, J.H.; Kim, B.R.; Ahn, J.M. Evaluating subscapularis tendon tears on axillary lateral radiographs using deep learning. Eur. Radiol. 2021, 31, 9408–9417. [Google Scholar] [CrossRef] [PubMed]


| Fracture Site | Image Used | Author. Year | CNN Used | Work | Dataset Size | Accuracy | AUC | Winner |
|---|---|---|---|---|---|---|---|---|
| Hip (femur neck) | X-ray | Matthew et al. 2019 | GooLeNet | Binary classification | 805 | 94% | 0.98 | |
| Hip | X-ray | Cheng et al. 2019 | DenseNet | Binary classification | 3605 | 91% | 0.98 | Orthopedist > CNN |
| Hip | X-ray | Takaaki et al. 2019 | VGG-16 | Binary classification | 3346 | CNN > Orthopedist | ||
| Hip | X-ray | Yamada et al. 2020 | Xception, ImageNet | Binary classification | 3123 | 98% | CNN > Orthopedist | |
| Hip | X-ray | Lee et al. 2020 | GoogLeNet-inception v3 | Classification | 686 | 86.8% | ||
| Hip | X-ray | Tanzi et al. 2020 | InceptionV3, VGG-16, ResNet50 | Classification | 2453 | 86% (3 class) 81% (5 class) | ||
| Hip (Atypical fracture) | X-ray | Zdolsek et al. 2021 | VGG19, InceptionV3, ResNet | Binary classification | 982 | 91% (ResNet50) 83% (VGG19) 89% (InceptionV3) | ||
| Shoulder (proximal humerus) | X-ray | Chung et al. 2018 | ResNet | Binary classification Classification | 1891 | 95% | 0.99 | Orthopedist > CNN (specialized in the shoulder) |
| Knee | X-ray | Lind et al. 2021 | ResNet-based CNN | Classification | 6768 | 0.87 (Proximal tibia) 0.89 (Patella) 0.89 (Distal femur) | ||
| Ankle | X-ray | Gene et al. 2019 | Xception | Binary classification | 596 | 75% | ||
| Ankle (Malleolar) | X-ray | Olczak et al. 2021 | ResNet | Classification | 5495 | 0.90 | ||
| Ankle (Calcaneal) | CT | Farda et al. 2021 | PCANet | Classification, Segmentation | 5534 | 72% | ||
| Wrist | X-ray | Kim et al. 2017 | Inception | Binary classification | 1389 | 0.95 | ||
| Wrist | X-ray | Thian et al. 2019 | ResNet | Binary classification | 7356 | 88.9% | 0.90 | |
| Wrist (Scaphoid) | X-ray | Langerhuizen et al. 2020 | VGG-16 | Binary classification | 300 | 72% | 0.77 | Orthopedist > CNN |
| Wrist (Scaphoid) | X-ray | Ozkaya et al. 2020 | ResNet50 | Binary classification | 390 | 084 | Orthopedist > CNN | |
| Vertebra | X-ray | Chen et al. 2021 | ImageNet, ResNeXt | Binary classification | 1306 | 73.6% | 0.72 | Orthopedist > CNN |
| Vertebra | MRI | Yabu et al. 2021 | VGG-16,19, Inception V3, ResNet50 | Binary classification | 1624 | 0.95 | CNN > Orthopedist |
| Location | Image Used | Author. Year | CNN Used | Work | Dataset Size | Accuracy | AUC |
|---|---|---|---|---|---|---|---|
| Knee | X-ray | Tiulpin et al. 2018 | Siamese CNN | Classification | 5960 | 66.7% | |
| Knee | X-ray | Pedoia et al. 2019 | DenseNet | Classification | 5042 | 75% | 0.83 |
| Knee | X-ray | Kim et al. 2020 | SE-ResNet | Classification | 4366 | 61.6% (with additional information) | 0.75 (with additional information) |
| Knee | X-ray | Swiecicki et al. 2021 | Faster R-CNN | Classification | 2802 | 71.9% | |
| Hip | X-ray | Xue et al. 2017 | VGG-16 | Binary classification | 420 | 92.8% | |
| Hip | X-ray | Ureten et al. 2020 | VGG-16 | Binary classification | 434 | 90.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Chung, S.W. Deep Learning for Orthopedic Disease Based on Medical Image Analysis: Present and Future. Appl. Sci. 2022, 12, 681. https://doi.org/10.3390/app12020681
Lee J, Chung SW. Deep Learning for Orthopedic Disease Based on Medical Image Analysis: Present and Future. Applied Sciences. 2022; 12(2):681. https://doi.org/10.3390/app12020681
Chicago/Turabian StyleLee, JiHwan, and Seok Won Chung. 2022. "Deep Learning for Orthopedic Disease Based on Medical Image Analysis: Present and Future" Applied Sciences 12, no. 2: 681. https://doi.org/10.3390/app12020681






