Age Estimation from fMRI Data Using Recurrent Neural Network
Abstract
:1. Introduction
2. Methods
2.1. Data Acquisition
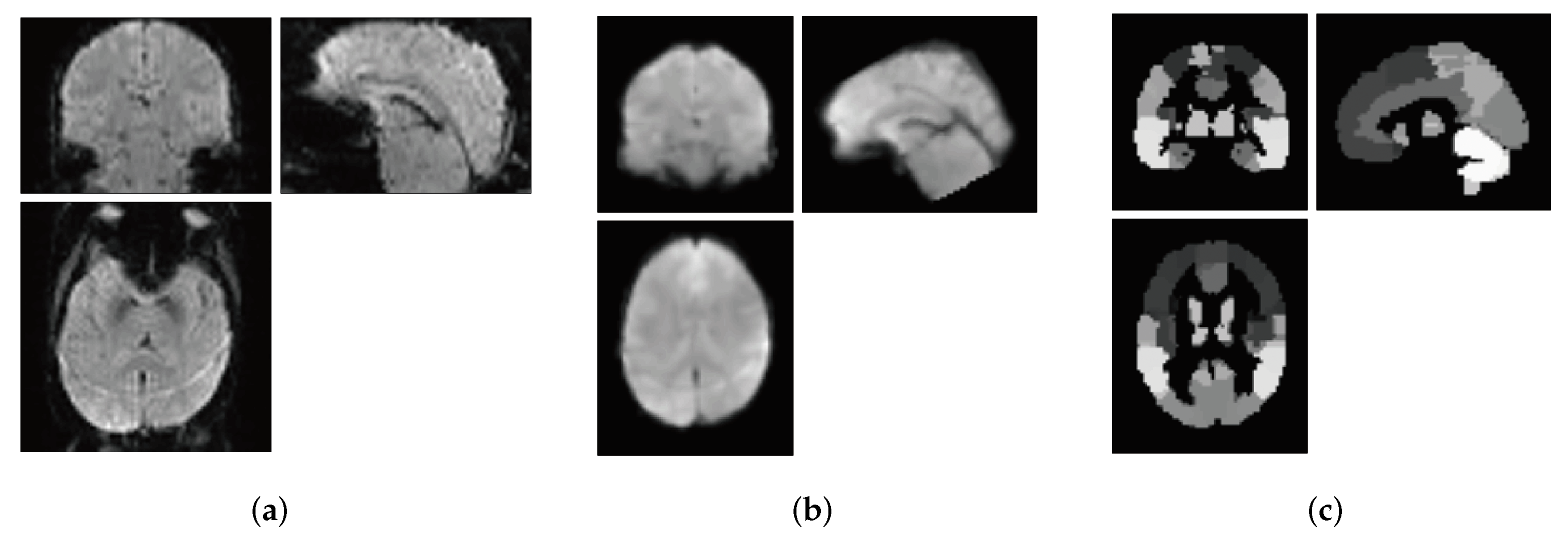
2.2. Data Preprocessing
3. Age Estimation
3.1. Preliminaries
3.2. Proposed Model
Other Baseline Models
4. Experiments
4.1. Model Parameters
4.2. Training
4.3. Implementations
4.4. Results
4.5. Discussion
4.5.1. Age Bias
4.5.2. Gender Difference
5. Ablation Study
5.1. Effect of Brain Regions
5.2. Effect of Dropout
5.3. Choice of Loss Function
5.4. Effect of Scan Time
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Havighurst, R.J. Successful aging. Process. Aging Soc. Psychol. Perspect. 1963, 1, 299–320. [Google Scholar]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [Green Version]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Rowe, J.W.; Kahn, R.L. Successful aging. Gerontologist 1997, 37, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Blagosklonny, M.V. Aging-suppressants: Cellular senescence (hyperactivation) and its pharmacologic deceleration. Cell Cycle 2009, 8, 1883–1887. [Google Scholar] [CrossRef] [Green Version]
- Ellingham, S.; Adserias-Garriga, J. Chapter 1—Complexities and considerations of human age estimation. In Age Estimation, 1st ed.; Adserias-Garriga, J., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–15. [Google Scholar] [CrossRef]
- Seok, J.; Kasa-Vubu, J.; DiPietro, M.; Girard, A. Expert system for automated bone age determination. Expert Syst. Appl. 2016, 50, 75–88. [Google Scholar] [CrossRef]
- Zaghbani, S.; Boujneh, N.; Bouhlel, M.S. Age estimation using deep learning. Comput. Electr. Eng. 2018, 68, 337–347. [Google Scholar] [CrossRef]
- Dimri, G.P.; Lee, X.; Basile, G.; Acosta, M.; Scott, G.; Roskelley, C.; Medrano, E.E.; Linskens, M.; Rubelj, I.; Pereira-Smith, O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [Google Scholar] [CrossRef] [Green Version]
- Krishnamurthy, J.; Torrice, C.; Ramsey, M.R.; Kovalev, G.I.; Al-Regaiey, K.; Su, L.; Sharpless, N.E. Ink4a/Arf expression is a biomarker of aging. J. Clin. Investig. 2004, 114, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Greider, C.W.; Szostak, J.W. Telomeres and telomerase: The path from maize, Tetrahymena and yeast to human cancer and aging. Nat. Med. 2006, 12, 1133. [Google Scholar] [CrossRef] [PubMed]
- Raz, N.; Gunning, F.M.; Head, D.; Dupuis, J.H.; McQuain, J.; Briggs, S.D.; Loken, W.J.; Thornton, A.E.; Acker, J.D. Selective aging of the human cerebral cortex observed in vivo: Differential vulnerability of the prefrontal gray matter. Cereb. Cortex 1997, 7, 268–282. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.; Backman, L.; Nilsson, L. Part 3, Chapter 6—The aging brain: Structural changes and their implications for cognitive aging. In New Frontiers in Cognitive Aging, 1st ed.; Raz, N., Ed.; Oxford University Press: Oxford, UK, 2004; pp. 115–133. [Google Scholar] [CrossRef]
- Salat, D.H.; Buckner, R.L.; Snyder, A.Z.; Greve, D.N.; Desikan, R.S.; Busa, E.; Morris, J.C.; Dale, A.M.; Fischl, B. Thinning of the cerebral cortex in aging. Cereb. Cortex 2004, 14, 721–730. [Google Scholar] [CrossRef] [Green Version]
- Park, D.C.; Reuter-Lorenz, P. The adaptive brain: Aging and neurocognitive scaffolding. Annu. Rev. Psychol. 2009, 60, 173–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fjell, A.M.; Walhovd, K.B. Structural brain changes in aging: Courses, causes and cognitive consequences. Rev. Neurosci. 2010, 21, 187–222. [Google Scholar] [CrossRef] [PubMed]
- Montagne, A.; Barnes, S.R.; Sweeney, M.D.; Halliday, M.R.; Sagare, A.P.; Zhao, Z.; Toga, A.W.; Jacobs, R.E.; Liu, C.Y.; Amezcua, L.; et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 2015, 85, 296–302. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.H.; Kim, H.W.; Shin, S.; Kim, J.; Jeong, Y.H.; Moon, J. Decipher reliable biomarkers of brain aging by integrating literature-based evidence with interactome data. Exp. Mol. Med. 2018, 50, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koenigsberg, R.A.; Bianco, B.A.; Faro, S.H.; Stickles, S.; Hershey, B.L.; Siegal, T.L.; Mohamed, F.B.; Dastur, C.K.; Tsai, F.Y. Chapter 23—Neuroimaging. In Textbook of Clinical Neurology, 3rd ed.; Goetz, C.G., Ed.; W.B. Saunders: Philadelphia, PA, USA, 2007; pp. 437–476. [Google Scholar] [CrossRef]
- Goldstein, J.M.; Seidman, L.J.; Horton, N.J.; Makris, N.; Kennedy, D.N.; Caviness, V.S., Jr.; Faraone, S.V.; Tsuang, M.T. Normal sexual dimorphism of the adult human brain assessed by in vivo magnetic resonance imaging. Cereb. Cortex 2001, 11, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Giedd, J.N.; Snell, J.W.; Lange, N.; Rajapakse, J.C.; Casey, B.; Kozuch, P.L.; Vaituzis, A.C.; Vauss, Y.C.; Hamburger, S.D.; Kaysen, D.; et al. Quantitative magnetic resonance imaging of human brain development: Ages 4–18. Cereb. Cortex 1996, 6, 551–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfefferbaum, A.; Mathalon, D.H.; Sullivan, E.V.; Rawles, J.M.; Zipursky, R.B.; Lim, K.O. A quantitative magnetic resonance imaging study of changes in brain morphology from infancy to late adulthood. Arch. Neurol. 1994, 51, 874–887. [Google Scholar] [CrossRef]
- Giedd, J.N.; Blumenthal, J.; Jeffries, N.O.; Castellanos, F.X.; Liu, H.; Zijdenbos, A.; Paus, T.; Evans, A.C.; Rapoport, J.L. Brain development during childhood and adolescence: A longitudinal MRI study. Nat. Neurosci. 1999, 2, 861. [Google Scholar] [CrossRef]
- Sowell, E.R.; Peterson, B.S.; Thompson, P.M.; Welcome, S.E.; Henkenius, A.L.; Toga, A.W. Mapping cortical change across the human life span. Nat. Neurosci. 2003, 6, 309. [Google Scholar] [CrossRef]
- Giedd, J.N. The teen brain: Insights from neuroimaging. J. Adolesc. Health 2008, 42, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.D.; Savoy, R.L. Functional magnetic resonance imaging (fMRI)”brain reading”: Detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage 2003, 19, 261–270. [Google Scholar] [CrossRef]
- Logothetis, N.K.; Pauls, J.; Augath, M.; Trinath, T.; Oeltermann, A. Neurophysiological investigation of the basis of the fMRI signal. Nature 2001, 412, 150. [Google Scholar] [CrossRef]
- Madden, D.J.; Spaniol, J.; Whiting, W.L.; Bucur, B.; Provenzale, J.M.; Cabeza, R.; White, L.E.; Huettel, S.A. Adult age differences in the functional neuroanatomy of visual attention: A combined fMRI and DTI study. Neurobiol. Aging 2007, 28, 459–476. [Google Scholar] [CrossRef] [Green Version]
- Dennis, E.L.; Thompson, P.M. Functional brain connectivity using fMRI in aging and Alzheimer’s disease. Neuropsychol. Rev. 2014, 24, 49–62. [Google Scholar] [CrossRef]
- Brown, R.W.; Cheng, Y.C.N.; Haacke, E.M.; Thompson, M.R.; Venkatesan, R. Magnetic Resonance Imaging: Physical Principles and Sequence Design; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Huettel, S.A.; Song, A.W.; McCarthy, G. Functional Magnetic Resonance Imaging; Sinauer Associates: Sunderland, MA, USA, 2004; Volume 1. [Google Scholar]
- Giorgio, A.; Santelli, L.; Tomassini, V.; Bosnell, R.; Smith, S.; De Stefano, N.; Johansen-Berg, H. Age-related changes in grey and white matter structure throughout adulthood. Neuroimage 2010, 51, 943–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonov, V.; Evans, A.C.; Botteron, K.; Almli, C.R.; McKinstry, R.C.; Collins, D.L.; The Brain Development Cooperative Group. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 2011, 54, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Meunier, D.; Achard, S.; Morcom, A.; Bullmore, E. Age-related changes in modular organization of human brain functional networks. Neuroimage 2009, 44, 715–723. [Google Scholar] [CrossRef]
- Ota, M.; Obata, T.; Akine, Y.; Ito, H.; Ikehira, H.; Asada, T.; Suhara, T. Age-related degeneration of corpus callosum measured with diffusion tensor imaging. Neuroimage 2006, 31, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.E.; Sanchez, C.; Phillips-Meek, M.; Xie, W. A database of age-appropriate average MRI templates. Neuroimage 2016, 124, 1254–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franke, K.; Ziegler, G.; Klöppel, S.; Gaser, C. The Alzheimer’s Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. Neuroimage 2010, 50, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Huang, T.W.; Chen, H.T.; Fujimoto, R.; Ito, K.; Wu, K.; Sato, K.; Taki, Y.; Fukuda, H.; Aoki, T. Age estimation from brain MRI images using deep learning. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 849–852. [Google Scholar]
- Qi, Q.; Du, B.; Zhuang, M.; Huang, Y.; Ding, X. Age estimation from MR images via 3D convolutional neural network and densely connect. In Proceedings of the International Conference on Neural Information Processing (ICONIP 2018), Bangkok, Thailand, 23–27 November 2020; Springer: Cham, Switzerland, 2018; pp. 410–419. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Jiang, H.; Guo, J.; Du, H.; Xu, J.; Qiu, B. Transfer learning on T1-weighted images for brain age estimation. Math. Biosci. Eng. MBE 2019, 16, 4382–4398. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.D.; Snyder, A.Z.; Vincent, J.L.; Corbetta, M.; Van Essen, D.C.; Raichle, M.E. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. USA 2005, 102, 9673–9678. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Criaud, M.; Cho, S.S.; Díez-Cirarda, M.; Mihaescu, A.; Coakeley, S.; Ghadery, C.; Valli, M.; Jacobs, M.F.; Houle, S.; et al. Abnormal intrinsic brain functional network dynamics in Parkinson’s disease. Brain 2017, 140, 2955–2967. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.J. Functional MRI of brain physiology in aging and neurodegenerative diseases. Neuroimage 2019, 187, 209–225. [Google Scholar] [CrossRef]
- Yao, Y.; Lu, W.; Xu, B.; Li, C.; Lin, C.; Waxman, D.; Feng, J. The increase of the functional entropy of the human brain with age. Sci. Rep. 2013, 3, 2853. [Google Scholar] [CrossRef]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Cho, K.; van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations using RNN Encoder–Decoder for Statistical Machine Translation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1724–1734. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Gated feedback recurrent neural networks. In Proceedings of the International Conference on Machine Learning (ICML 2015), Lille, France, 6–11 July 2015; pp. 2067–2075. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:1409.0473. [Google Scholar]
- Kim, Y.; Denton, C.; Hoang, L.; Rush, A.M. Structured attention networks. arXiv 2017, arXiv:1702.00887. [Google Scholar]
- Wei, D.; Zhuang, K.; Ai, L.; Chen, Q.; Yang, W.; Liu, W.; Wang, K.; Sun, J.; Qiu, J. Structural and functional brain scans from the cross-sectional Southwest University adult lifespan dataset. Sci. Data 2018, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Woolrich, M.W.; Jbabdi, S.; Patenaude, B.; Chappell, M.; Makni, S.; Behrens, T.; Beckmann, C.; Jenkinson, M.; Smith, S.M. Bayesian analysis of neuroimaging data in FSL. Neuroimage 2009, 45, S173–S186. [Google Scholar] [CrossRef]
- Smith, S.M.; Jenkinson, M.; Woolrich, M.W.; Beckmann, C.F.; Behrens, T.E.; Johansen-Berg, H.; Bannister, P.R.; De Luca, M.; Drobnjak, I.; Flitney, D.E.; et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004, 23, S208–S219. [Google Scholar] [CrossRef] [Green Version]
- Rolls, E.T.; Joliot, M.; Tzourio-Mazoyer, N. Implementation of a new parcellation of the orbitofrontal cortex in the automated anatomical labeling atlas. Neuroimage 2015, 122, 1–5. [Google Scholar] [CrossRef]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to forget: Continual prediction with LSTM. Neural Comput. 2000, 12, 2451–2471. [Google Scholar] [CrossRef]
- Jozefowicz, R.; Zaremba, W.; Sutskever, I. An empirical exploration of recurrent network architectures. In Proceedings of the International Conference on Machine Learning (ICML 2015), Lille, France, 6–11 July 2015; pp. 2342–2350. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Power, J.D.; Cohen, A.L.; Nelson, S.M.; Wig, G.S.; Barnes, K.A.; Church, J.A.; Vogel, A.C.; Laumann, T.O.; Miezin, F.M.; Schlaggar, B.L.; et al. Functional network organization of the human brain. Neuron 2011, 72, 665–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, E.M.; Laumann, T.O.; Adeyemo, B.; Huckins, J.F.; Kelley, W.M.; Petersen, S.E. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 2016, 26, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Seitzman, B.A.; Gratton, C.; Marek, S.; Raut, R.V.; Dosenbach, N.U.; Schlaggar, B.L.; Petersen, S.E.; Greene, D.J. A set of functionally-defined brain regions with improved representation of the subcortex and cerebellum. Neuroimage 2020, 206, 116290. [Google Scholar] [CrossRef] [PubMed]




| Project | Subjects | Age | M | F | |
|---|---|---|---|---|---|
| AnnArbor_a | 18 | 13–40 | 17 | 1 | 283 |
| Baltimore | 12 | 30–40 | 6 | 6 | 111 |
| Bangor | 3 | 19 | 3 | 0 | 253 |
| Beijing_Zang | 39 | 18–24 | 21 | 18 | 213 |
| Berlin_Margulies | 14 | 23–44 | 7 | 7 | 183 |
| Cambridge_Buckner | 81 | 18-30 | 30 | 51 | 107 |
| Dallas | 14 | 20–71 | 8 | 6 | 103 |
| ICBM | 35 | 19–79 | 15 | 20 | 116 |
| Leiden_2180 | 3 | 20–27 | 3 | 0 | 203 |
| Leiden_2200 | 4 | 18–25 | 4 | 0 | 203 |
| Leipzig | 15 | 22–42 | 7 | 8 | 183 |
| Milwaukee_b | 46 | 44–65 | 15 | 31 | 163 |
| Newark | 7 | 21–39 | 4 | 3 | 123 |
| NewHaven_a | 2 | 18–38 | 2 | 0 | 237 |
| NewHaven_b | 6 | 18–42 | 4 | 2 | 169 |
| NewYork_a | 37 | 10–49 | 20 | 17 | 180 |
| NewYork_a_ADHD | 16 | 24–50 | 15 | 1 | 180 |
| NewYork_b | 11 | 18–46 | 7 | 4 | 163 |
| Orangeburg | 14 | 31–55 | 11 | 3 | 153 |
| Oulu | 17 | 21–22 | 4 | 13 | 233 |
| Oxford | 8 | 30–35 | 6 | 2 | 163 |
| PaloAlto | 12 | 26–46 | 1 | 11 | 223 |
| Pittsburgh | 1 | 27 | 1 | 0 | 263 |
| Queensland | 6 | 21–34 | 4 | 2 | 178 |
| SaintLouis | 5 | 21–28 | 3 | 2 | 115 |
| SALD | 369 | 19–80 | 140 | 229 | 230 |
| Training/Validation | Test |
|---|---|
| 640 | 155 |
| Model Name | Regions | Correlation | MSE | MAE | ICC |
|---|---|---|---|---|---|
| MLP | All | 0.780 | 140.09 | 8.645 | 0.754 |
| RNN | All | 0.797 | 110.28 | 7.935 | 0.774 |
| LSTM | All | 0.858 | 116.48 | 8.627 | 0.812 |
| GRU | All | 0.905 | 70.76 | 6.507 | 0.883 |
| GRU | Right | 0.769 | 132.49 | 8.630 | 0.754 |
| GRU | Left | 0.835 | 110.69 | 8.850 | 0.807 |
| Transformer | All | 0.883 | 74.44 | 6.566 | 0.884 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; No, A. Age Estimation from fMRI Data Using Recurrent Neural Network. Appl. Sci. 2022, 12, 749. https://doi.org/10.3390/app12020749
Gao Y, No A. Age Estimation from fMRI Data Using Recurrent Neural Network. Applied Sciences. 2022; 12(2):749. https://doi.org/10.3390/app12020749
Chicago/Turabian StyleGao, Yunfei, and Albert No. 2022. "Age Estimation from fMRI Data Using Recurrent Neural Network" Applied Sciences 12, no. 2: 749. https://doi.org/10.3390/app12020749
APA StyleGao, Y., & No, A. (2022). Age Estimation from fMRI Data Using Recurrent Neural Network. Applied Sciences, 12(2), 749. https://doi.org/10.3390/app12020749






