Abstract
The manual annotation of brain tumor images is costly and relies heavily on physician expertise, which limits the implementation of automated and accurate brain tumor segmentation in clinical practice. Meanwhile, unlabeled images are readily available but not well-exploited. In this paper, a novel brain tumor segmentation method for improving the efficiency of labeled images is proposed, dubbed LETCP. Specifically, it presents a contrastive pre-training strategy that utilises unlabeled data for model pre-training. The segmentation model in this approach is constructed based on a self-attention transformer. Extensive evaluation of the method is performed on three public datasets. By using pre-training with unlabeled data and then fine-tuning with small amounts of labeled data, this method achieves segmentation performance surpassing other semi-supervised methods and shows competitive performance compared with supervised methods.
1. Introduction
The primary clinical treatment for brain tumors is to perform maximum surgical resection, requiring precise segmentation to provide accurate guidance for surgery [1,2,3]. However, manual labelling is a time-consuming, labour-intensive task that relies heavily on neurosurgical expertise. In recent years, tremendous developments in deep learning have opened up opportunities for automated and accurate brain tumor segmentation [4,5]; this technique relies on a large number of annotated images for model training, which is another challenge in medical image analysis [6].
To address the challenge of the shortage of annotated data, self-supervised learning has emerged as an option, which strives to enable models to learn the representations’ information from unannotated data [7,8]. Contrastive learning is an important branch of self-supervised learning; it is based on the intuition that different transformed versions of the same image have similar representations, and transformations from different images have different representations. By bringing similar representations closer together in the representation space and dissimilar representations further apart, the model exploits a contrastive loss function for effective pre-training [9,10,11,12]. Therefore, in this paper, a novel brain tumor segmentation method aiming to achieve label efficiency is proposed (see Figure 1 for an overview). Specifically, for the scenario with limited labeled data, a pre-training strategy based on contrastive learning is presented. This strategy includes a positive and negative pairs construction method applied to brain tumor images and a representation learning system for feature learning on unlabeled data.
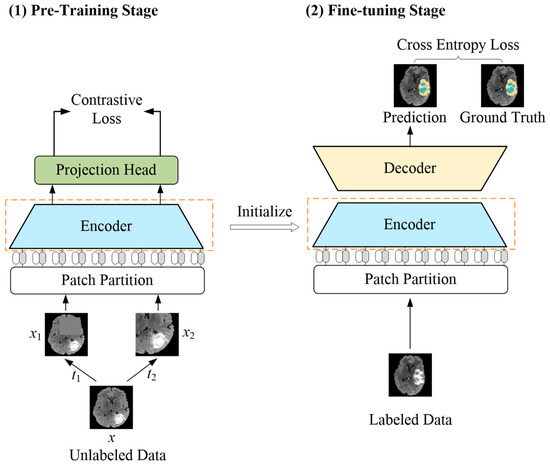
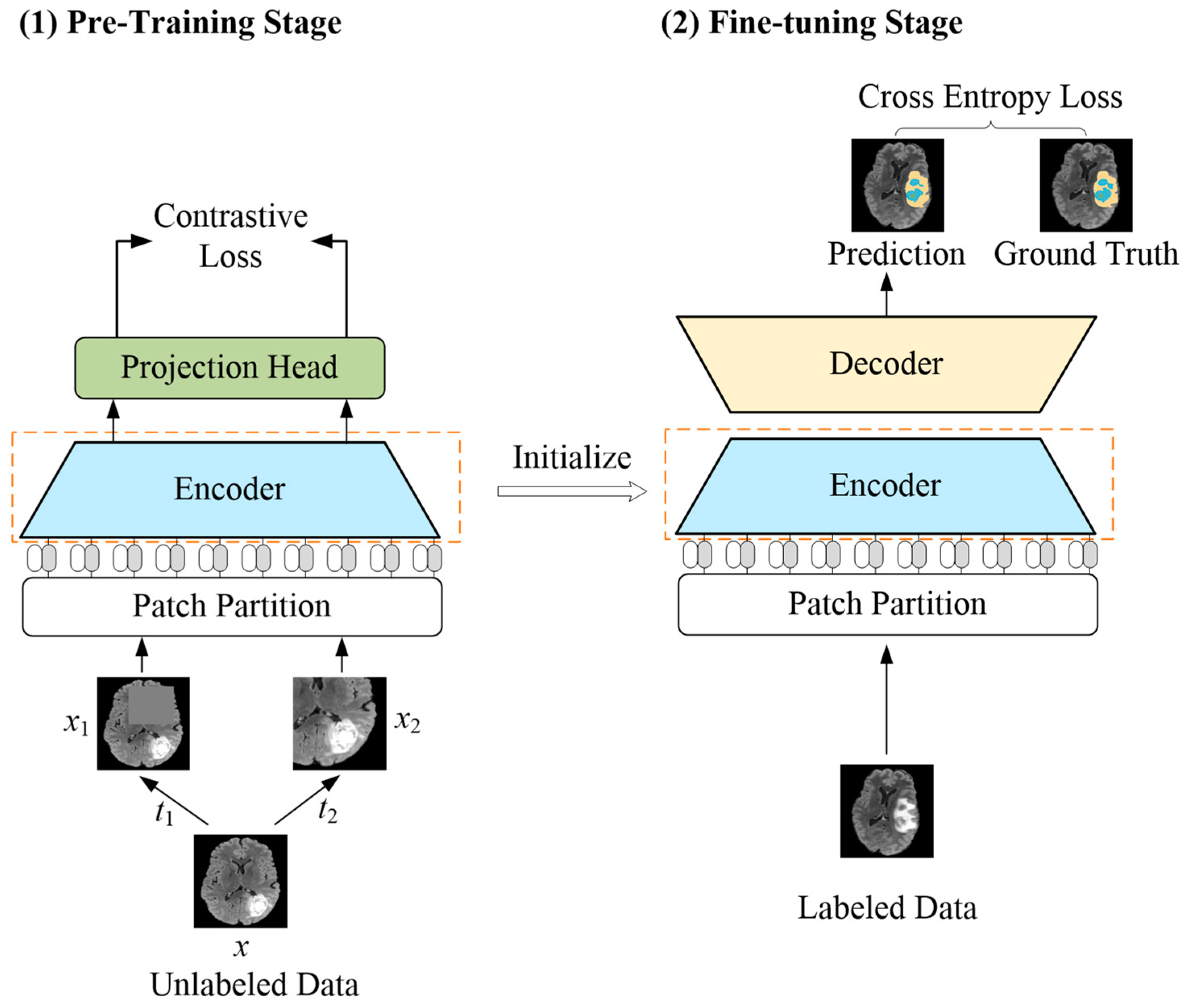
Figure 1.
Overview of the proposed LETCP method. In pre-training stage, input image x with random augmentation t1 and t2 to obtain enhanced images x1 and x2. The pre-trained encoder serves as initialization of fine-tuning stage for segmentation prediction.
A self-attention mechanism [13], which is an alternative feature extraction unit to convolution, has caused a revolution in the field of computer vision. Vision Transformer [14] was the first to introduce a standard transformer in NLP to vision, achieving excellent results in image classification. Based on this, Swin Transformer [15] proposed a shifted-window mechanism and hierarchical architecture, which introduced inductive bias for the transformer and outperforms convolution in several vision tasks, such as classification and segmentation [16,17,18,19]. Therefore, a transformer has also been introduced to the medical image analysis community [20,21,22,23,24]. Inspired by this, in the presented method, a brain tumor segmentation network based on a self-attention mechanism is proposed, which has an encoder–decoder structure design and employs skip connection.
2. Related Works
In this section, an overview of existing convolutional neural network-based, transformer-based, and hybrid architecture-based brain tumor segmentation models is presented. Then, the characteristics of convolutional and self-attention computation models are outlined, and the common structural designs of medical image segmentation models are described. Finally, pre-training methods in medical image analysis are presented.
2.1. Brain Tumor Segmentation with Convolutional Neural Network
With the revolutionary breakthrough in deep learning technology, researchers have started to concentrate on solving various practical issues with deep neural networks. Pioneering works from Zikic et al. [4] and Pereira et al. [25] aimed to design a customized deep convolutional neural network to achieve an accurate brain tumor segmentation. With the breakthrough brought by Fully Convolutional Network [26] and U-Net [27], later innovations [28] on brain tumor segmentation focus on building fully convolutional encoder–decoder networks without fully connected layers to enable end-to-end tumor segmentation. The convolutional neural network is designed with prior knowledge related to image processing; it can obtain identical features from the same images at different locations, which is referred to as inductive bias. Meanwhile, the feature extraction of the convolutional neural network relies on local receptive fields, which leads to its lack of learning ability for long-term dependencies.
2.2. Brain Tumor Segmentation with Transformer
A transformer was first used in natural language processing; the feature extraction of the transformer is based on a self-attention mechanism which facilitates the learning of long-range dependencies in sequences. Dosovitskiy et al. [14] propose to divide the image into patches, then convert patches into a token sequence using a transformer-based model for feature extraction, and it achieved good performance. After that, the performance of transformer-based models has outperformed convolutional models in natural image processing [15,16,29]. A number of studies [30,31,32] have applied transformer models to medical image analysis, including brain tumor segmentation [33,34,35]. Wang et al. [34] propose employing a 3D transformer block for encoding bottleneck layer features with the encoder and decoder still using convolutional modules. Hatamizadeh et al. [35] propose building their encoder with a 3D transformer to encode images directly, then using a CNN decoder to catch global information. Jia et al. [36] propose using bi-level transformer blocks for encoding two levels of bottleneck features with the rest of network remaining a convolutional neural network. To summarize the studies mentioned above, some apply the transformer blocks to the bottleneck layer for feature extraction [34,37] while others use the transformer blocks as additional modules after the convolutional layers [38,39]. However, these efforts do not leverage the spatial context advantages of transformers. Inspired by this, a pure transformer segmentation network is proposed in this paper to fully exploit the feature extraction ability of a self-attention mechanism.
Medical image analysis, like segmentation, requires models to extract multi-scale features for dense prediction. Skip connection-based architectures such as the U-Net [27] and pyramid network [40] are widely adopted to leverage hierarchical features; this type of structure allows the final recovered feature map to incorporate more low-level features, and it also enables the fusion of features from different scales, thus providing multi-scale prediction. The self-attention mechanism in a vision transformer [14] is computed based on a single patch size, which results in its feature extraction lacking the inductive bias inherent in CNNs; therefore, it is not appropriate for a brain tumor image with rich structural information and a high variation of regions of interest. Soon after, Swin Transformer [15] introduced a hierarchical architecture for the transformer, which is more suitable for dense prediction tasks. Therefore, the segmentation model proposed in this paper takes the Swin Transformer as a basic unit, incorporating skip connection with a U-shape encoder–decoder structure.
2.3. Pre-Training in Medical Image Analysis
Some previous research [41,42] has trained models on natural images to improve performance on medical image analysis through transfer learning. However, there is a large gap in contextual knowledge between natural images and medical images [43]. The rise of self-supervised learning, represented by contrastive learning, indicates an opportunity to exploit unlabeled data for model training. Wu et al. [7] propose an instance discrimination pretext task that treats each image as an individual class. Ye et al. [8] propose to select a small random batch of images from a dataset and obtain the positive and negative pairs of image features by an encoder. SimCLR [9] is a typical contrastive learning method that treats other images within a batch as negative pairs. As medical images are different from natural images, a special design for specific medical analysis tasks is required. Therefore, the contrastive pre-training strategy proposed in this paper includes a positive and negative pairs construction method and a representation learning system for unlabeled data.
In summary, the brain tumor segmentation method proposed in this paper aims to improve the utilization efficiency of limited annotation data and presents a new perspective that distinguishes it from other brain tumor methods. Existing approaches for brain tumor segmentation have focused either on the specific design of internal modules of the network or on the employment of multiple networks based on a cascade or integration strategy. The main contributions of this paper are summarised as follows:
- A brain tumor segmentation method is proposed to improve the efficiency of utilising limited labeled data;
- A contrastive pre-training strategy is introduced which allows model training with unlabeled data;
- A segmentation model based on a self-attention transformer is presented towards good brain tumor segmentation performance;
- It is the first attempt to incorporate contrastive learning and a transformer for brain tumor segmentation.
3. Method
A label-efficient brain tumor segmentation method was proposed in this paper, which combines the advantages of a transformer and contrastive learning, dubbed LETCP (Label-Efficient Transformer-based Contrastive Pre-training); see Figure 1 for details. It contained two stages: pre-training and fine-tuning. The first stage worked with unlabeled data to pre-train the encoder; it picked up the intrinsic representations of brain tumor images by contrastive loss. The second stage took small amounts of labeled data to fine-tune the segmentation model with cross-entropy loss. Please note that the projection head was only used in pre-training, and the encoder after pre-training served as the initialization for the fine-tuning stage.
3.1. Transformer-Based Segmentation Model
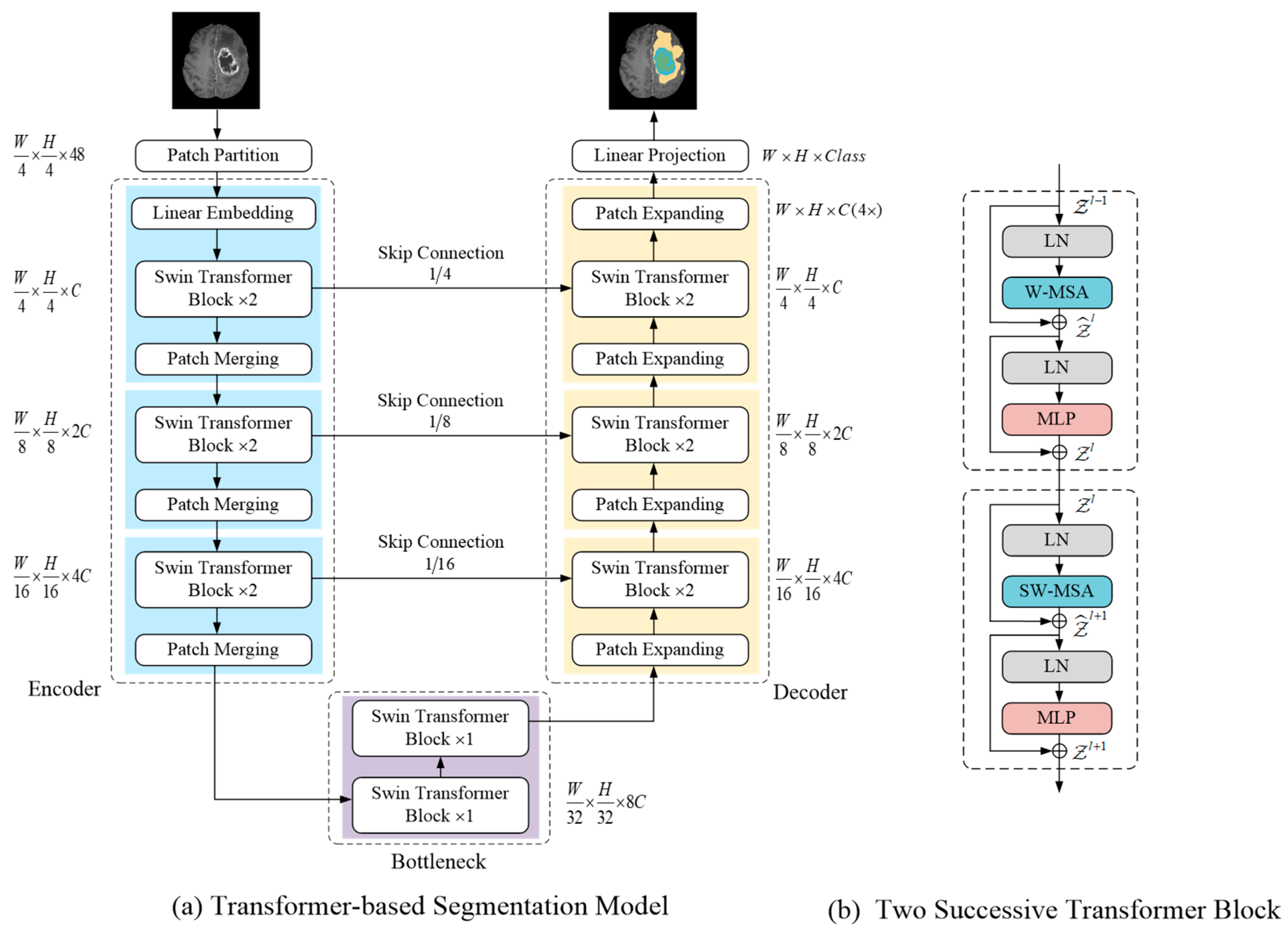
A transformer-based brain tumor segmentation model is presented in this section (see Figure 2a for an overview). To fully exploit the feature extraction capability of the self-attention mechanism inspired by previous studies [15], the segmentation network adopts Swin Transformer as a basic unit and U-shape encoder–decoder structure. The details of each component are described in the following sections.
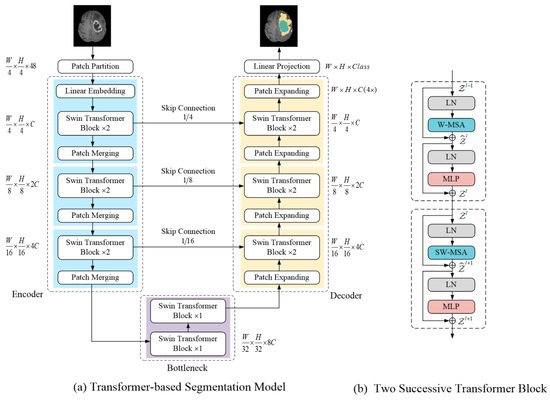
Figure 2.
(a) Illustrates the proposed segmentation model architecture; (b) Shows two consecutive Swin Transformer block structure.
3.1.1. Swin Transformer Block
Each Swin Transformer block consisted of a LayerNorm (LN) layer, a multi-head self-attention module, a residual connection, and a 2-layer MLP with GELU non-linearity. In Figure 2b, two consecutive Swin Transformer blocks are presented; the difference between both is in the multi-head self-attention module. The window-based multi-head self-attention (W-MSA) module and the shifted window-based multi-head self-attention (SW-MSA) module were applied in them, respectively. The Swin transformer block was formulated as follows:
The variables and denote two consecutive Swin Transformer blocks, respectively, while and represent the output of W-MSA and MLP in block , respectively. and represent the output of SW-MSA and MLP in block , respectively. Self-attention was calculated similarly to earlier research [44,45] as follows:
where denote the query, key, and value matrices. The variables and represent the number of patches in a window and the dimension of the query or key, respectively, and represents the relative position bias of the patch token.
3.1.2. Encoder
The encoder was designed to encode images and obtain feature representation. First, the patch partitioning layer divided the input image into non-overlapping patches, each in size, and the feature dimension of each patch was . The first stage of the encoder consisted of a linear embedding layer, two consecutive Swin Transformer blocks, and a patch merging layer. The linear embedding layer was used to a project the feature dimension into C (the Swin Transformer block processing dimension). Then, input images were converted into a patch token form so that they were ready to input the Swin Transformer block.
The Swin Transformer blocks performed representation learning on the features without changing the resolution and dimension. Patch merging layers were responsible for down-sampling by a factor of 2 and increasing the channel dimension. Specifically, it concatenated the feature of each group of 2 × 2 neighboring patch tokens, and the resulting vector was projected via a linear mapping to a space where the channel dimensionality of the patch tokens was doubled. The structure of the encoder stages 1, 2, and 3 is shown in the left part of Figure 2a. Each stage contained two consecutive Swin Transformer blocks and a patch merging layer.
3.1.3. Bottleneck and Skip Connection
Two consecutive Swin Transformer blocks were used to build the bottleneck for learning the deep feature representation. At the bottleneck, the feature dimension and resolution remain unchanged. Similar to U-Net [27], the skip connection was used to fuse multi-scale features from the encoder during up-sampling, which can reduce the loss of spatial information caused by down-sampling.
3.1.4. Decoder
Corresponding to each stage of encoder, the symmetric decoder was built based on a patch expanding layer and the Swin Transformer block. The patch expanding layer was designed to up-sample the deep feature representation. Specifically, with the patch expanding layer, the resolution of the feature map was increased by a factor of 2, and the dimension of the feature token was reduced by half. Finally, the decoder restored the feature map to the same resolution as the input image, and the segmentation results were obtained through a linear projection layer.
3.2. Contrastive Pre-Training Strategy
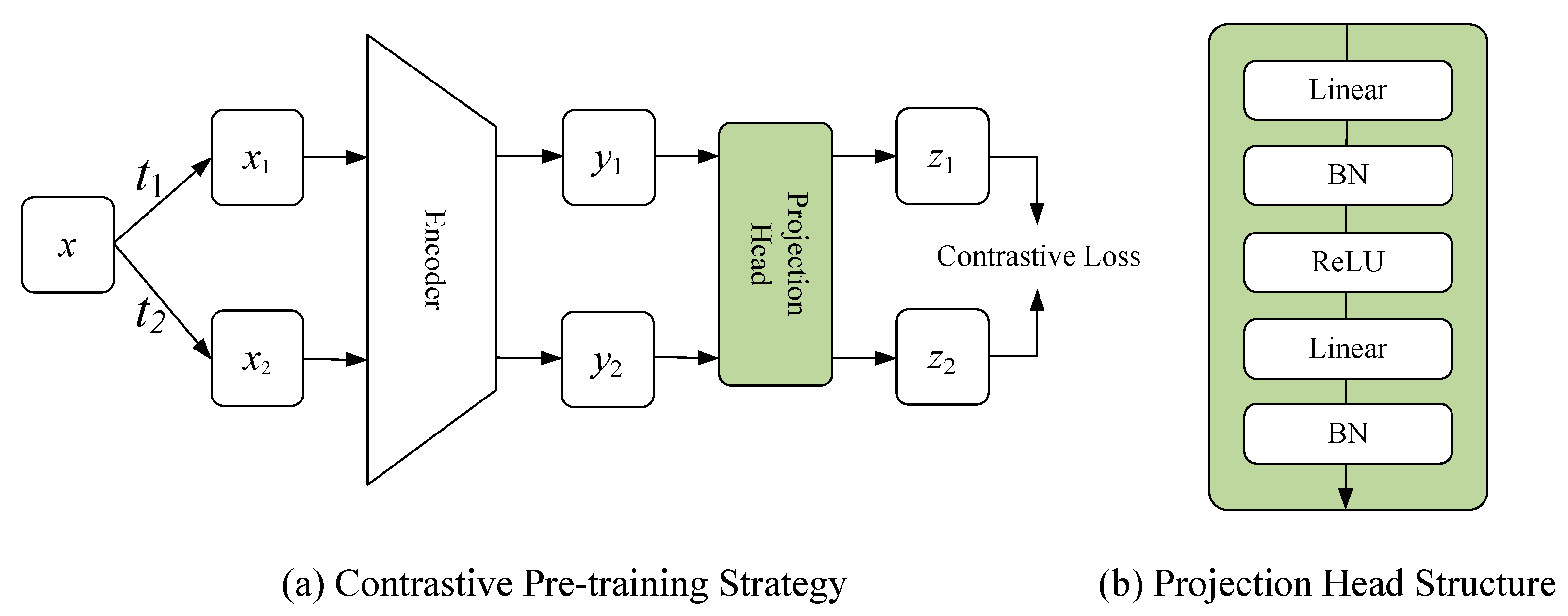
In the pre-training stage, a contrastive pre-training strategy was proposed. Specifically, this strategy contained a positive and negative pairs constructing method and a contrastive representation learning system, as illustrated in Figure 3a.
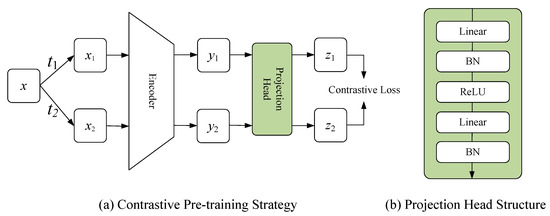
Figure 3.
(a) Shows the proposed contrastive pre-training strategy; (b) Shows the structure of projection head.
3.2.1. Positive and Negative Pairs Constructing Method
N images from the unlabeled dataset were randomly selected as a batch. The set T represented all image enhancement operations, and two operations, and , were randomly selected from it. and were applied to image in the batch, obtaining two enhanced images and , respectively. and formed a positive pair; any other images in the same batch could be treated as negative pairs of or . The enhancement operations set T included a random crop, random color, and random cutout.
3.2.2. Contrastive Representation Learning System
The proposed contrastive representation learning system contained the encoder of the segmentation network and projection head, which was used for the representation learning of unlabeled data. and represented a sample pair, which were fed into the encoder to obtain the corresponding feature representation, denoted by and , respectively. Then, and were input into the projection head to obtain the representation vectors and in the representation space, respectively. The presence of the projection head allowed the encoder to learn a more general feature representation, which facilitated the improvement of the performance of the fine-tuned segmentation (see Figure 3b for the structure of the projection head). Please note that the projection head was dropped in the fine-tuning stage.
The proposed contrastive representation learning system employed InfoNCE as a contrastive loss; by minimizing this loss, it brought positive pairs closer and pushed negative pairs further apart in the representation space. By this contrastive learning format, it enabled the encoder to learn a better feature representation of the brain tumor images. The InfoNCE loss function is formulated as follow:
where and denoted the representation vectors of positive pairs. encouraged a higher degree of similarity between positive pairs, indicating that positive pairs were closer in representation space. encouraged a lower degree of similarity between negative pairs, indicating that negative pairs were pushed away. The temperature parameter τ was experimentally best at 0.1, which represented the degree of perception of the negative pairs.
4. Experiments and Results
The proposed LETCP aims to achieve good pre-training effects with unlabeled data by using small amounts of labeled data to reach good segmentation accuracy. Experiments were performed in terms of the pre-training effect and segmentation accuracy.
4.1. Pre-Training Effect Evaluation
In the experiments of the pre-training evaluation, LETCP and other baselines were pre-trained with unlabeled data of the same size and fine-tuned using the same amount of labeled data to compare the segmentation accuracy.
4.1.1. Datasets and Image Preprocessing
The first dataset was provided by the Brain Tumor Segmentation (BraTS) Challenge 2021 [1,2,3]; it contained MRI scans from 1251 cases of patients. The dataset was divided into 1000, 200, and 51 for pre-training, fine-tuning, and testing, respectively. A 5-fold cross-validation strategy was used to evaluate the segmentation performance after pre-training. M instances were randomly selected for fine-tuning from the fine-tuning dataset of 200 instances, assuming that only these M instances were labeled. Multiple M values (e.g., 10, 20, and 40) were tested in the experiments to observe how the size of the training set during the fine-tuning stage affected the segmentation performance. The segmentation accuracy was evaluated by the average Dice Score for three regions (enhancing tumor, tumor core, and whole tumor region). The second dataset was provided by the Brain Tumor Segmentation Challenge (BraTS) 2020 [1,2,3]. It contained images of 369 patients split into 300, 50, and 19 for pre-training, fine-tuning, and testing, respectively. Other experiment settings were the same as the BraTS Challenge 2021. Image enhancement included translation, rotation, and scale operations.
4.1.2. Implementation Details
Pre-training was performed on a single Nvidia RTX 3090 GPU with 200 epochs. It used an SGD optimiser. The learning rate was 0.1, and a cosine learning rate scheduler was used. The batch size was set to 32, and the temperature parameter τ was set to 0.1. In the fine-tuning stage, the model was trained with a cross-entropy loss and 100 epochs, the batch size was set to 5, and the learning rate was . An Adam optimizer and the cosine scheduler were used.
The baseline methods for comparison were the following:
- Random: a method without any pre-training where the fine-tuning was the same as the LETCP method;
- Rotation [46]: a pre-text based method that used image rotation prediction to pre-train the encoder;
- PIRL [47]: a contrastive learning mechanism for natural image classification;
- SimCLR [9]: a method that built positive pairs by two random augmentations for the natural image classification.
4.1.3. Results and Analysis
The results of the experiments on the BraTS Challenge 2021 and BraTS Challenge 2020 are presented in Table 1 and Table 2, respectively.

Table 1.
Comparison of the proposed LETCP method with other baselines on BraTS 2021 [1,2,3]. ↑ means higher is better.

Table 2.
Comparison of the proposed LETCP method with baseline methods on BraTS 2020 [1,2,3]. ↑ means higher is better.
The following observations and analysis can be gained:
- For the same amount of fine-tuned data M, the proposed LETCP outperforms the other baselines in segmentation accuracy, and the improvement in Dice can be up to 27%, which indicates that LETCP achieves better pre-training effects and enhances labeled data efficiency in the same data conditions.
- The performance gains of LETCP are particularly high when very small amounts of training samples are used (e.g., 10 and 20). When the number of training samples in-creases, the gains are reduced. The reason is that, as training samples increase, the information gap between the fine-tuning dataset and contrastive pre-training dataset becomes small, and the fine-tuning performance saturates.
4.2. Segmentation Accuracy Evaluation
A comparison of the proposed segmentation model with other brain tumor segmentation methods is presented in order to obtain an assessment of segmentation accuracy. The pre-trained encoder serves as the initialization for the model, as it selects CNN-based methods and transformer-based methods as baselines.
4.2.1. Datasets and Implementation Details
The BraTS Challenge 2019 [1,3,48] dataset contains 335 instances of data that is split into 200 training samples, 50 validation samples, and 85 testing samples. Experiments were conducted on a single Nvidia RTX 3090 GPU; the model was trained with cross-entropy loss using an Adam optimizer with a learning rate of 1 × e−5 for 300 epochs and employing a cosin decay learning rate scheduler and a batch size of 5.
The manual annotation contains 4 classes: background (label 0), necrotic and non-enhancing tumor (label 1), peritumoral edema (label 2), and GD-enhancing tumor (label 4). The segmentation accuracy is assessed by a Dice score and Hausdorff distance (95%) metrics for enhancing the tumor region (ET, label 1), regions of the tumor core (TC, labels 1 and 4), and the whole tumor region (WT, labels 1,2 and 4).
4.2.2. Results and Analysis
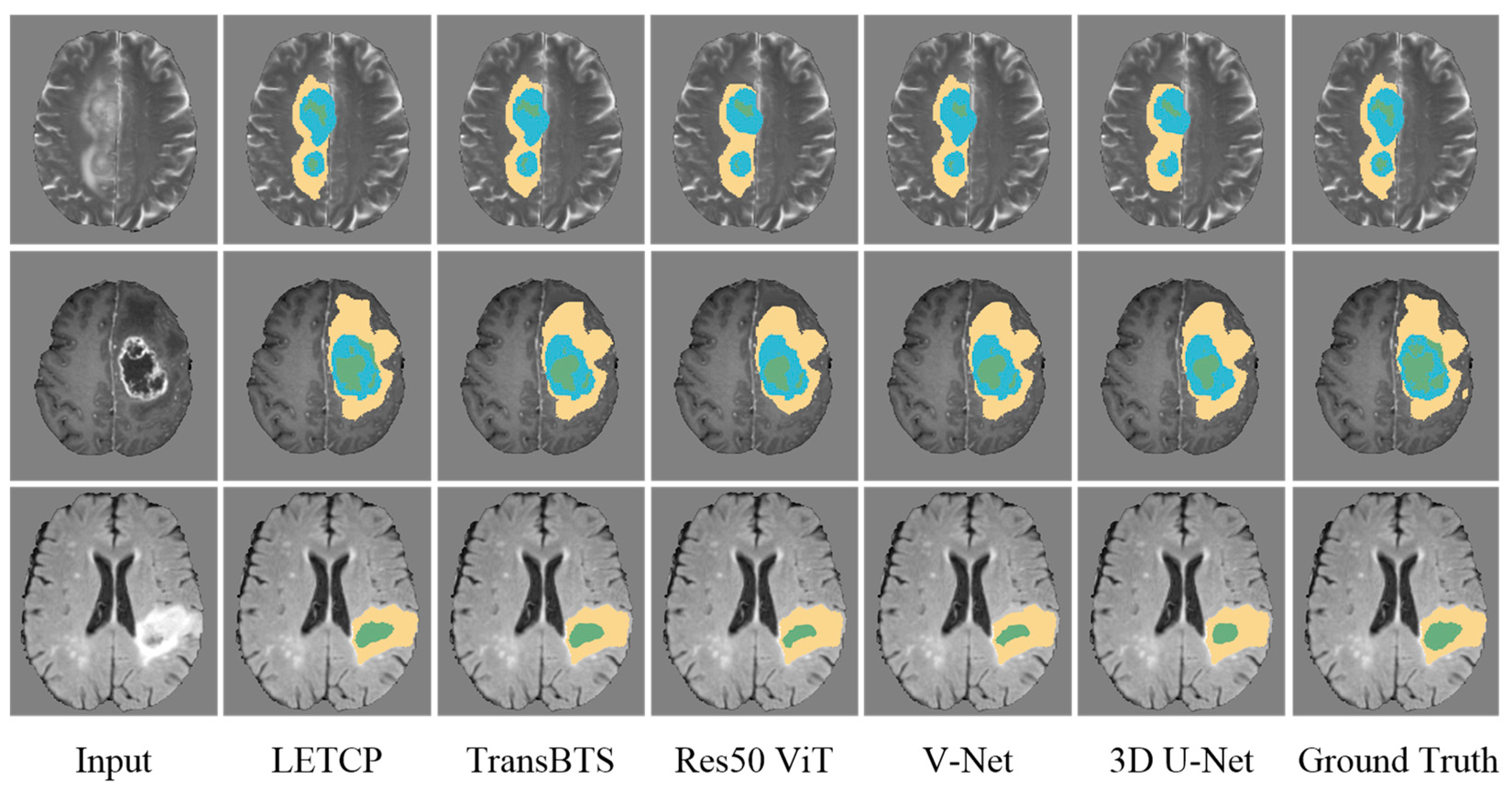
Table 3 and Table 4 presents the segmentation results of LETCP and other baselines on the BraTS Challenge 2019. Figure 4 shows qualitative segmentation results on the test samples of unseen patient data.

Table 3.
Dice Score results comparison of LETCP and baselines on BraTS 2019 [1,3,48]. ↑ means higher is better.

Table 4.
Hausdorff Distance results comparison of LETCP and baselines on BraTS 2019 [1,3,48]. ↓ means lower is better.
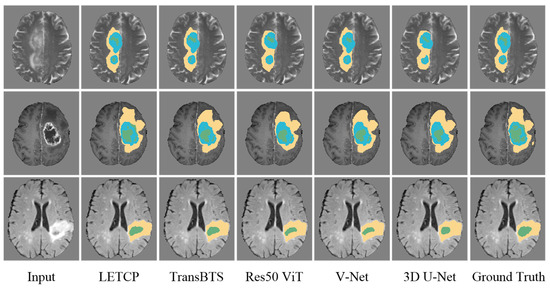
Figure 4.
Qualitative comparison of segmentation performance between LETCP and other baselines. Yellow represents whole tumor region, blue represents enhancing tumor region and green represents region of tumor core.
LETCP achieves Dice scores of 87.89%, 85.11%, and 91.89% and an HD of 6.13 mm, 7.32 mm, and 12.74 mm on ET, TC, and WT, which are comparable to or higher results than previous SOTA methods. Moreover, LETCP outperforms other methods in terms of the average value of both metrics. It is evident from Figure 4 that LETCP can describe brain tumors more accurately and generate much better segmentation masks. This clearly reveals the benefit of leveraging a self-attention transformer for modeling the global relationships.
5. Discussion
In clinical applications, the available images of brain tumors are usually fewer than natural images. The contrastive pre-training strategy proposed in this paper is based on contrastive learning and refined for brain tumor segmentation. The experimental results show that our method is more appropriate for scenarios with small-scale training data than PIRL [47] and SimCLR [9], which are used for nature image processing. Compared to the CNN based 3D U-Net [49] and hybrid structure based TransBTS [34], the pure transformer segmentation model proposed in this paper demonstrates excellent local detail processing ability. This is because the feature learning power of the self-attention mechanism needs to be demonstrated with more training data, and the contrastive pre-training strategy provides the opportunity for the model to learn from unlabeled data. Future works could try to verify the improvement of the pre-training effect with more data enhancement operations.
6. Conclusions
In this paper, a novel brain tumor segmentation method aiming to achieve labeled efficiency is presented. In order to leverage the power of unlabeled data, a pre-training strategy based on contrastive learning is proposed. To exploit the strength of the transformer, a self-attention-based segmentation model is introduced using Swin Transformer blocks as the basic unit for feature representation and long-range information interactive learning. Extensive experiments on three public brain tumor datasets demonstrate that the proposed LETCP improves the exploitation efficiency of labeled data and has excellent performance.
Author Contributions
Conceptualization, S.C. and J.Z.; methodology, S.C.; validation, S.C.; formal analysis, S.C.; writing—original draft preparation, S.C.; writing—review and editing, J.Z. and T.Z.; supervision, J.Z. and T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (Youth) under Grant No. 52001039, National Natural Science Foundation of China under Grant No. 52171310, Funding of Shandong Natural Science Foundation in China under Grant No. ZR2019LZH005, Research fund from Science and Technology on Underwater Vehicle Technology Laboratory under Grant No. 2021JCJQ-SYSJJ-LB06903.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank Editor-in-Chief, Editor and anonymous Reviewers for their valuable reviews.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the Cancer Genome Atlas Glioma MRI Collections with Expert Segmentation Labels and Radiomic Features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Zikic, D.; Ioannou, Y.; Brown, M.; Criminisi, A. Segmentation of Brain Tumor Tissues with Convolutional Neural Networks. Proc. MICCAI-BRATS 2014, 36, 36–39. [Google Scholar]
- Magadza, T.; Viriri, S. Deep Learning for Brain Tumor Segmentation: A Survey of State-of-the-Art. J. Imaging 2021, 7, 19. [Google Scholar] [CrossRef]
- Liu, Z.; Tong, L.; Chen, L.; Jiang, Z.; Zhou, F.; Zhang, Q.; Zhang, X.; Jin, Y.; Zhou, H. Deep Learning Based Brain Tumor Segmentation: A Survey. Complex Intell. Syst. 2022, 1–26. [Google Scholar] [CrossRef]
- Wu, Z.; Xiong, Y.; Yu, S.X.; Lin, D. Unsupervised Feature Learning via Non-Parametric Instance Discrimination. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 3733–3742. [Google Scholar]
- Ye, M.; Zhang, X.; Yuen, P.C.; Chang, S.-F. Unsupervised Embedding Learning via Invariant and Spreading Instance Feature. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 6203–6212. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–15 July 2020; pp. 1597–1607. [Google Scholar]
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R. Momentum Contrast for Unsupervised Visual Representation Learning. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 9726–9735. [Google Scholar]
- Chen, T.; Kornblith, S.; Swersky, K.; Norouzi, M.; Hinton, G.E. Big Self-Supervised Models Are Strong Semi-Supervised Learners. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 22243–22255. [Google Scholar]
- Chen, X.; Fan, H.; Girshick, R.; He, K. Improved Baselines with Momentum Contrastive Learning. arXiv 2020, arXiv:2003.04297. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 2021, Montreal, QC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L.; et al. Swin Transformer V2: Scaling up Capacity and Resolution. arXiv 2021, arXiv:2111.09883. [Google Scholar]
- Xu, X.; Feng, Z.; Cao, C.; Li, M.; Wu, J.; Wu, Z.; Shang, Y.; Ye, S. An Improved Swin Transformer-Based Model for Remote Sensing Object Detection and Instance Segmentation. Remote Sens. 2021, 13, 4779. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jegou, H. Training Data-Efficient Image Transformers & Distillation through Attention. In Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Ye, L.; Rochan, M.; Liu, Z.; Wang, Y. Cross-Modal Self-Attention Network for Referring Image Segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 10502–10511. [Google Scholar]
- Prangemeier, T.; Reich, C.; Koeppl, H. Attention-Based Transformers for Instance Segmentation of Cells in Microstructures. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 700–707. [Google Scholar]
- Shen, Z.; Fu, R.; Lin, C.; Zheng, S. COTR: Convolution in Transformer Network for End to End Polyp Detection. In Proceedings of the 2021 7th International Conference on Computer and Communications (ICCC), Chengdu, China, 10–13 December 2021; pp. 1757–1761. [Google Scholar]
- Zhang, Z.; Yu, L.; Liang, X.; Zhao, W.; Xing, L. TransCT: Dual-Path Transformer for Low Dose Computed Tomography. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 55–64. [Google Scholar]
- Dai, Y.; Gao, Y.; Liu, F. TransMed: Transformers Advance Multi-Modal Medical Image Classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, Y.; Frey, E.C.; Li, Y.; Du, Y. ViT-V-Net: Vision Transformer for Unsupervised Volumetric Medical Image Registration. arXiv 2021, arXiv:2104.06468. [Google Scholar]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Isensee, F.; Jäger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. Automated Design of Deep Learning Methods for Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. In Computer Vision—ECCV 2020; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 213–229. [Google Scholar]
- Xie, Y.; Zhang, J.; Xia, Y.; Wu, Q. UniMiSS: Universal Medical Self-Supervised Learning via Breaking Dimensionality Barrier. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Mondal, A.K.; Bhattacharjee, A.; Singla, P.; Prathosh, A.P. XViTCOS: Explainable Vision Transformer Based COVID-19 Screening Using Radiography. IEEE J. Transl. Eng. Health Med. 2022, 10, 1–10. [Google Scholar] [CrossRef]
- Yu, S.; Ma, K.; Bi, Q.; Bian, C.; Ning, M.; He, N.; Li, Y.; Liu, H.; Zheng, Y. MIL-VT: Multiple Instance Learning Enhanced Vision Transformer for Fundus Image Classification. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 12908, pp. 45–54. ISBN 978-3-030-87236-6. [Google Scholar]
- Zhou, H.-Y.; Guo, J.; Zhang, Y.; Yu, L.; Wang, L.; Yu, Y. NnFormer: Interleaved Transformer for Volumetric Segmentation. arXiv 2022, arXiv:2109.03201. [Google Scholar]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 12901, pp. 109–119. ISBN 978-3-030-87192-5. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2021. [Google Scholar]
- Jia, Q.; Shu, H. BiTr-Unet: A CNN-Transformer Combined Network for MRI Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 3–14. [Google Scholar]
- Li, J.; Wang, W.; Chen, C.; Zhang, T.; Zha, S.; Wang, J.; Yu, H. TransBTSV2: Towards Better and More Efficient Volumetric Segmentation of Medical Images. arXiv 2022, arXiv:2201.12785. [Google Scholar]
- Meng, X.; Zhang, X.; Wang, G.; Zhang, Y.; Shi, X.; Dai, H.; Wang, Z.; Wang, X. Exploiting Full Resolution Feature Context for Liver Tumor and Vessel Segmentation via Integrate Framework: Application to Liver Tumor and Vessel 3D Reconstruction under Embedded Microprocessor. arXiv 2022, arXiv:2111.13299. [Google Scholar]
- Zhang, Y.; Liu, H.; Hu, Q. TransFuse: Fusing Transformers and CNNs for Medical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Seferbekov, S.; Iglovikov, V.; Buslaev, A.; Shvets, A. Feature Pyramid Network for Multi-Class Land Segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018; pp. 272–275. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-Ray8: Hospital-Scale Chest X-Ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar]
- Abràmoff, M.D.; Lou, Y.; Erginay, A.; Clarida, W.; Amelon, R.; Folk, J.C.; Niemeijer, M. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset through Integration of Deep Learning. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5200–5206. [Google Scholar] [CrossRef]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding Transfer Learning for Medical Imaging. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2019; Volume 32. [Google Scholar]
- Hu, H.; Zhang, Z.; Xie, Z.; Lin, S. Local Relation Networks for Image Recognition. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Korea, 27 October–2 November 2019; pp. 3463–3472. [Google Scholar]
- Hu, H.; Gu, J.; Zhang, Z.; Dai, J.; Wei, Y. Relation Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 July 2018; pp. 3588–3597. [Google Scholar]
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised Representation Learning by Predicting Image Rotations. arXiv 2018, arXiv:1803.07728. [Google Scholar]
- Misra, I.; van der Maaten, L. Self-Supervised Learning of Pretext-Invariant Representations. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 6706–6716. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2019, arXiv:1811.02629. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).