Abstract
In clinical diagnosis, radiological reports are essential to guide the patient’s treatment. However, writing radiology reports is a critical and time-consuming task for radiologists. Existing deep learning methods often ignore the interplay between medical findings, which may be a bottleneck limiting the quality of generated radiology reports. Our paper focuses on the automatic generation of medical reports from input chest X-ray images. In this work, we mine the associations between medical discoveries in the given texts and construct a knowledge graph based on the associations between medical discoveries. The patient’s chest X-ray image and clinical history file were used as input to extract the image–text hybrid features. Then, this feature is used as the input of the adjacency matrix of the knowledge graph, and the graph neural network is used to aggregate and transfer the information between each node to generate the situational representation of the disease with prior knowledge. These disease situational representations with prior knowledge are fed into the generator for self-supervised learning to generate radiology reports. We evaluate the performance of the proposed method using metrics from natural language generation and clinical efficacy on two public datasets. Our experiments show that our method outperforms state-of-the-art methods with the help of a knowledge graph constituted by prior knowledge of the patient.
1. Introduction
Medical images are important to diagnose and detect underlying diseases, and radiological reports are essential to aid clinical decision making [1]. They describe some observations of the image such as the extent, size, and location of the disease. The physician communicates findings and diagnoses from the patient’s medical scan through the medical report. This process is often laborious, taking an average of 5 to 10 min to write a medical report [2]. The daily task of a radiologist involves analyzing a large number of medical images, which helps the physician to locate the lesion more accurately. Due to the increasing demand for medical images, radiologists still have a large workload. However, the process of writing radiology reports can be time-consuming and tedious for radiologists [3], and it can also be error-prone when writing a report. In addition, the ability to automatically generate accurate reports helps radiologists and physicians to make quick and meaningful diagnoses. Its potential efficiency and benefits can be substantial, especially in critical situations such as outbreaks of COVID or similar pandemics. In order to reduce the burden on radiologists, it is important to be able to generate reports accurately and automatically. These reasons provide a good motivation for our research into the automatic generation of medical reports.
With the development of image captioning and the availability of large-scale datasets, the application of deep learning in the automatic generation of medical reports has been continuously deepened. However, how to generate accurate radiology reports is still a challenging task, because the radiology report generation task is quite different from the image captioning task. First, a radiology report is generated to output a paragraph, which is usually composed of several sentences, while image captioning generally only needs to generate one sentence. Secondly, the generation of radiology reports requires extensive domain knowledge to generate clinically coherent text and the use of medical terms to describe normal and abnormal medical observations [4]. In addition, in the image caption, the model needs to cover all the details of the image as much as possible to generate rich captions. However, in radiology report generation, the model only needs to focus on abnormal areas and infer potential diseases to generate radiology reports. The relationship between potential diseases can determine the accuracy of the generated radiology reports.
Most existing methods focus on the image-to-fluent text aspect of the medical report generation problem. These methods generally perform reasonably well in addressing verbal fluency, but their results are significantly unsatisfactory in terms of clinical accuracy. The possible reason is that their results are far from proficient in revealing the topics related to the expected diseases and symptoms in the generated texts, and they ignore the associations between these underlying diseases. Adding a knowledge graph can alleviate this problem. A knowledge graph describes the relationships between concepts, entities, and their keys in the objective world in a structured way. Knowledge graphs are also often used as prior knowledge, which can provide complementary information for accurate reporting. Medical reports typically consist of many long sentences describing various disease-related symptoms and related topics in precise and domain-specific terms, which may potentially influence each other. Deep learning methods often suffer from a lack of knowledge when not explicitly taught compared to experienced radiologists, which limits the accuracy of the generation. Modeling the associations between medical observations in the form of a knowledge graph allows us to further leverage prior knowledge to generate high-quality reports. To this end, Zhang et al. [5] and Li et al. [6] combined knowledge reasoning based on the knowledge graph with encoders and decoders for radiology report generation. However, their prior knowledge is manually predefined and, therefore, requires domain experts to be closely involved in the design and implementation of the system, which is a waste of time and effort. Due to the nature of the graph, their method can usually achieve high accuracy, but may miss some important findings. While it is feasible to manually identify and implement a high-quality knowledge graph to obtain good accuracy, it is often impractical to exhaustively encode all nodes and relationships in this way. Our work uses prior knowledge from text mining to build a generic knowledge graph to alleviate some of these concerns.
We propose an innovative framework for automatic report generation based on prior knowledge, which seamlessly integrates prior and linguistic knowledge at different levels. First, we investigate a data-driven approach to automatically obtain associations between disease labels in radiology reports. This prior knowledge is a natural extension of human-designed knowledge. Disease labels are defined as nodes in the knowledge graph, which are related and influence each other during the propagation of the graph. Secondly, we establish a graph convolutional neural network to aggregate and transmit information between each node to obtain prior knowledge [7]. Specifically, a set of multi-view chest X-ray images are sent to the convolutional neural network for image feature extraction, and then the content of the clinical document instructed by the doctor is used for text feature extraction using Transformer [8]. The two extracted features are summed and normalized to wound together to obtain a hybrid image–text feature, called a contextualized embedding. The image–text hybrid features and the adjacency matrix constructed according to the knowledge graph are transferred to the three-layer graph convolutional network, and the special features of each knowledge graph node are learned to obtain the episodic representation with prior knowledge. Then, these node features are transferred to two branches, a linear classifier for disease classification, and a generator made of a transformer to generate reports. After generating the report, the generated report is passed into the text classifier again to fine-tune the generated report. Unlike previous studies, additional text mining concepts are added to the model as labels for classification as well as nodes in the knowledge graph, and the expressive power of the model is enriched by training on the chest X-ray image dataset with structured labels.
We evaluate our proposed method on the publicly accessible Open-I [9] and MIMIC-CXR [10] datasets, where we employ Natural Language Generation (NLG) and clinical efficacy (CE) metrics to analyze the quality of clinically generated reports. The results show that the proposed method achieves good performance in both natural language generation and clinical efficacy indicators. It is also shown that the addition of prior knowledge helps to improve the quality and accuracy of the automatic generation of radiology reports.
Our contributions are outlined below:
- We mine and model the text, and according to the mined information, we use prior knowledge to build the knowledge graph and construct the corresponding adjacency matrix (Section 3.1.1).
- We combine text–image hybrid features with knowledge reasoning based on the knowledge graph to improve the quality and accuracy of radiology report generation (Section 3.1.4).
- Our experiments show that our proposed model outperforms state-of-the-art methods. The knowledge graph composed of prior knowledge of patients plays a crucial role in improving the quality of generated reports.
2. Related Work
2.1. Image-Based Captioning and Medical Report Generation
Most of the work on image-based captioning is based on the classical structure of CNN + LSTM, which aims to generate real sentences or relevant paragraphs of a topic to summarize the visual content in an image or video [11,12,13,14]. With the development of computer vision and natural language processing technology, many works combine radiology images and free text to automatically generate radiology reports to help clinical radiologists make a quick and meaningful diagnosis [15]. Radiology report generation takes X-ray images as input and generates descriptive reports to support inference of better diagnostic conclusions beyond disease labels. Many radiology report generation methods follow the practice of image captioning models [16,17,18]. For example, ref. [19] adopted an encoder–decoder architecture and proposed a hierarchical generator and attention mechanism to generate long reports. Xue et al. [20] fused the visual features and semantic features of the previous sentence through the attention mechanism, used the fused features to generate the next sentence, and then generated the whole report in a loop. Wang et al. [21] proposed an embedding network with text and image as input to jointly learn text and image information and train the CNN-LSTM architecture end-to-end, which was then combined with a multi-level attention model to generate a chest X-ray report. Chen et al. [22] recorded important information during the generation process and then further assisted the generation of radiology reports by providing memory-driven transformers. Jing et al. [23] used reinforcement learning to exploit structural information between and within reports to generate high-quality radiology reports. Liu et al. [24] combined self-key sequence training and reinforcement learning to optimize the emergence of disease keywords in radiology reports. Shin et al. [25] adopted the CNN-RNN framework to generate radiological reports describing detected diseases based on visual features on chest X-ray image datasets.
Our work is mainly similar to that of Hoang et al. [26], who proposed a fully distinguishable end-to-end structural model, which mainly consists of three complementary modules for classifier, generator, and interpreter, which increase the linguistic fluency and clinical accuracy of generated reports. However, it does not add the association between disease labels, and lacks the association between disease labels in the classification. We use the association between labels learned from the text knowledge base to promote the semantic alignment between disease labels and images, which can better show the correlation between disease labels in classification, improve the accuracy of label classification, and further improve the accuracy of report generation.
2.2. Knowledge Graph
The concept of the knowledge graph was formally proposed by Google in 2012 to achieve a more intelligent search engine. Since 2013, it has gained popularity in academia and industry and plays an important role in intelligent question answering, intelligence analysis, anti-fraud, and other applications. A knowledge graph is essentially a knowledge base known as a semantic network, which is a knowledge base with a directed graph structure where the nodes of the graph represent entities or concepts. The edges of the graph represent semantic relations between entities/concepts, such as similarity relations between two entities, or syntactical correspondences.
In our knowledge graph, we use the tool SentencePiece [27] to obtain entities and then determine the relationship between entities based on the number of co-occurrences to build a knowledge graph, which is a structured way to represent knowledge graphically. In a knowledge graph, information is represented as a set of nodes, which are connected by a set of labeled directed lines to represent the relationship between nodes. A knowledge graph can well represent the relationship between nodes.
2.3. Transformer
Transformer was first introduced in the context of machine translation with the aim of speeding up training and improving remote dependency modeling. It is implemented by parallel processing of sequential data and an attention mechanism, which consists of a multi-head self-attention module and a feed-forward layer. By considering multi-head self-attention mechanisms and graph attention networks [28], recent transformer-based models have shown considerable progress in many difficult tasks, such as image generation [29], question answering, and linguistic reasoning [30]. Radiology reports are usually composed of several long sentences. As the traditional RNN is not suitable for generating long sentences and paragraphs, Chang et al. [31] designed a hierarchical RNN architecture as the decoder to generate long sentences, but the effect was not satisfactory. The recently emerged Transformer architecture can alleviate this problem. Therefore, we mainly use Transformers to compose our text codec in our work.
3. Our Approach
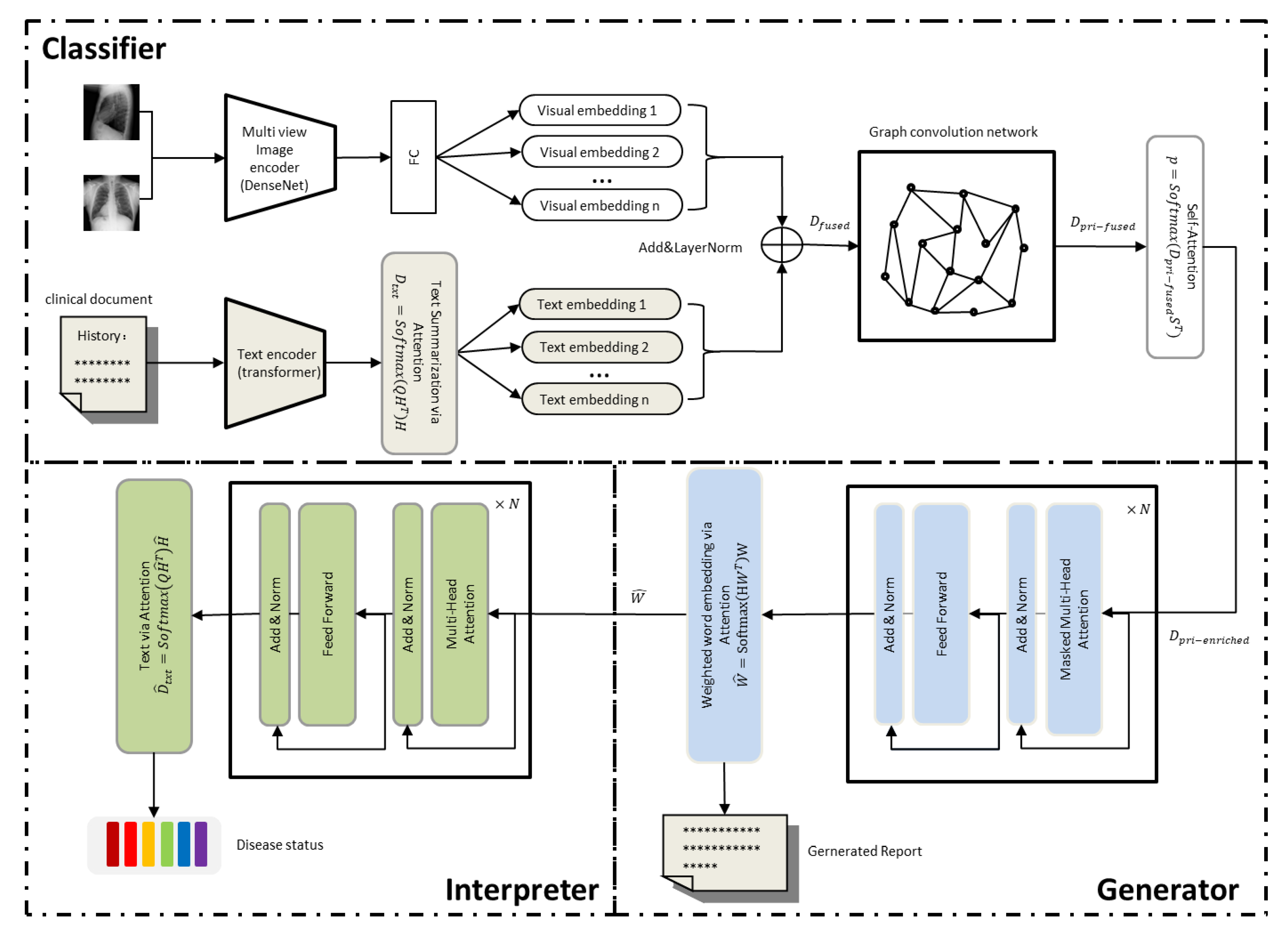
Our framework consists of a classification module, a generation module, and an interpretation module, as shown in Figure 1. The classification module consists of a multi-view image encoder, a text encoder, and a graph convolutional network based on a knowledge graph. We first build a knowledge graph in a data-driven manner (Section 3.1.1), then use a multi-view image encoder to read multiple chest X-ray images and extract the global visual feature representation, which is passed to a fully connected layer to decouple the global visual feature representation into multiple low-dimensional visual embedding (Section 3.1.2). At the same time, the text encoder reads the clinical documents and summarizes the content into text summary embedding, and then uses the “Add &LayerNorm” operation to wrap the visual embedding and text summary embedding together to obtain the image–text hybrid features, which are referred to as context-related embedding of the disease topic (Section 3.1.3). The episodic embedding is passed to a graph convolutional network (GCN) based on a knowledge graph that propagates semantic correlations between disease topics based on the knowledge graph to inject prior knowledge into concept representation learning (Section 3.1.4). The generation module takes as initial input the rich disease embedding that passes through the graph convolutional network to generate the text (Section 3.2). Finally, the generated text is sent to the interpreter for fine-tuning to align with the disease-related topics of the classification module (Section 3.3). In what follows, we elaborate on these three modules.

Figure 1.
Our model mainly includes three parts. The classifier is used to read chest X-ray images and clinical history to extract visual features and text features, and then the image–text hybrid features and the adjacency matrix of the knowledge graph are passed into the graph convolutional network to obtain the features containing prior knowledge. The obtained features are combined with topic features and state features and fed into the generator based on the visual self-attention model to generate the radiology report. The generated report is then passed into the interpreter to adjust the generated radiology report.
3.1. Classifier
3.1.1. Construction of Knowledge Graph
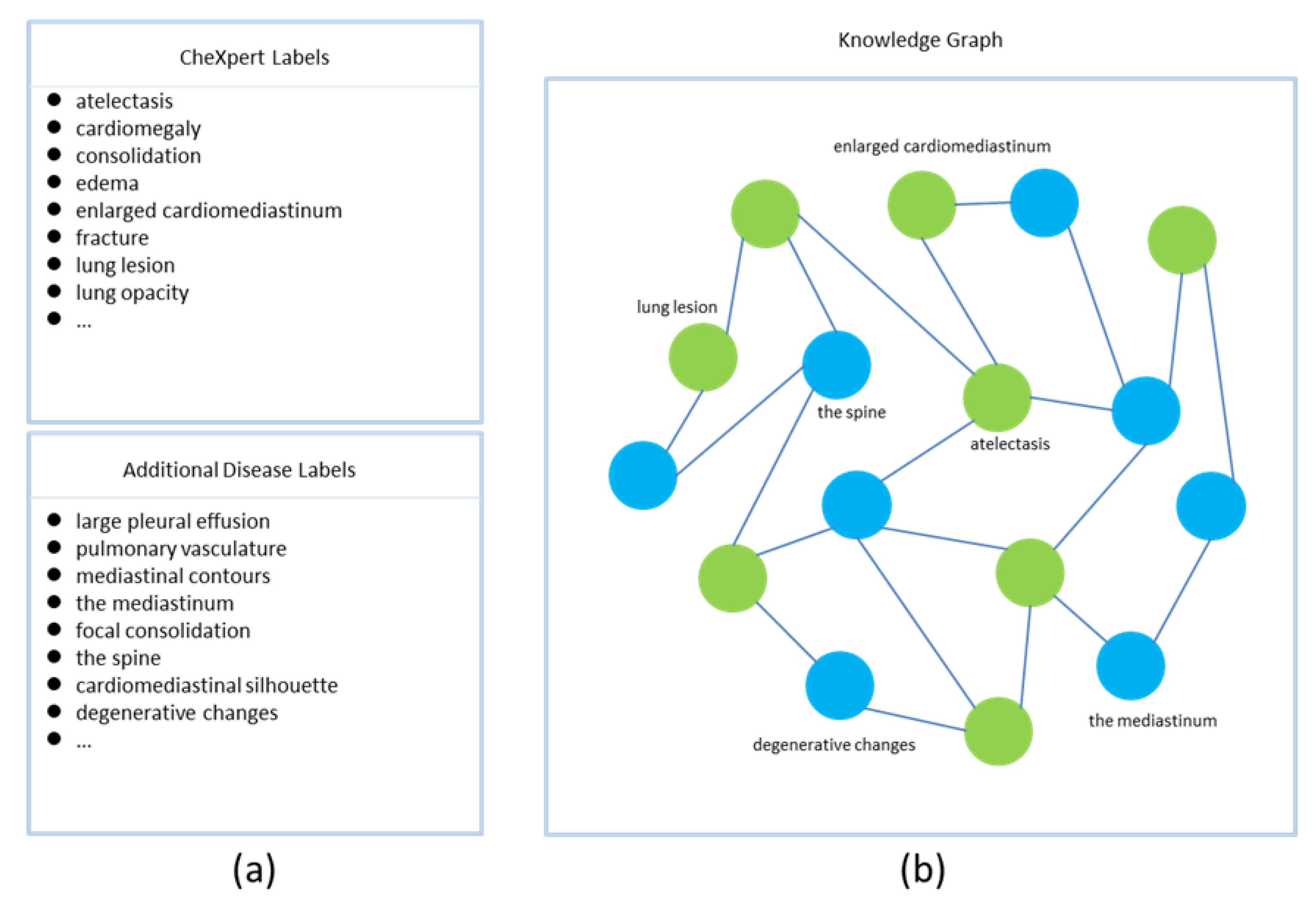
In our study, the nodes of the knowledge graph are disease-related topics. Edges are semantic associations between concepts. Our knowledge graph consists of two parts. The first part is defined by the CheXpert [32] tagger, a rule-based system that extracts and classifies medical reports into 14 common diseases. The label of each disease has four states, namely positive, negative, uncertain, or unmentioned. The MIMIC-CXR dataset has been annotated by the CheXpert tagger. The second part consists of supplementary concepts and their interrelationships, mined from radiology reports in a data-driven manner. Specifically, we count nouns in radiology reports using the SentencePiece tool, which is an unsupervised text tagger and de-tagger, and then we select nouns with top-k occurrences as additional disease labels, if they are not included in the fourteen disease labels defined by the CheXpert tagger. We establish the knowledge graph according to the co-occurrence of labels in radiology reports (Figure 2), and then construct the incidence matrix and binarize the matrix to form the adjacency matrix of the knowledge graph [33]. Specifically, we build a matrix, where each row or column represents a label, and then calculate the values in the matrix based on the number of co-occurrences of the labels in the radiology report. If the number of co-occurrences between two labels is greater than the average number of co-occurrences, the two labels are considered to be associated, and the corresponding value of the matrix is assigned a value of 1. On the contrary, if the number of co-occurrences between two labels is less than the average number of co-occurrences, the two labels are regarded as not associated, and the corresponding value of the matrix is assigned a value of 0. This matrix is then regarded as the adjacency matrix of the knowledge graph.

Figure 2.
Construction of the knowledge graph. The knowledge graph is built based on the co-occurrence of the tags in the radiology report, and only the tag names of some nodes are shown. In the figure, part (a) represents all node names in the knowledge graph; part (b) represents the knowledge graph, where the green nodes represent the 14 common disease labels defined by CheXpert, and the blue nodes represent the additional disease labels defined by us.
3.1.2. Multi-View Image Encoder
Each medical study consists of chest X-ray images , as DenseNet can achieve better performance than ResNet [34] with fewer parameters and computational cost. We extract its corresponding latent features via a Densenet-121 [35] image encoder with shared weights, where is the number of features. Then, we obtain multi-view latent features by combining latent feature sets , referring to the method proposed by Su et al. [36]. When , the multi-view encoder reduces to a single-image encoder.
3.1.3. Text Encoder and Disease Contextualized Representation
Let be a text document of length with word embedding , where denotes the -th word in the text and is the embedding dimension. We use the Transformer encoder as our text feature extractor to extract a set of hidden states , where denotes the attention feature of the -th word to other words in the text.
disease-related topics are queried from the whole document , which is summarized as . We refer to this retrieval process as text summarization embedding :
is randomly initialized and updated by the attention mechanism. represents the word-attention heat-map of query diseases in the document.
As shown in Figure 1, we decouple the multi-view latent features extracted by the image encoder into a low-dimensional disease representation , where each row is a vector . is defined as follows:
where and are trainable parameters for the -th class of disease representation. denotes the number of disease representations and is the dimension of the embedding. Then, the visual embedding and the text summary embedding are twisted together to form the disease situational representation :
The fusion of visual and textual information allows our model to simulate the workflow of a hospital to screen the visual manifestations of a disease based on a patient’s clinical history.
3.1.4. Graph Convolutional Networks and Contextualized Representations of Diseases with Prior Knowledge
We use the GCN to model the intrinsic association between diseases or topics, and the adjacency matrix is built based on the knowledge graph detailed above. The GCN updates its node representation via message passing, and graph convolution is represented as [37]:
where is the state in layer and is initialized using the disease contextualized representation. is the adjacency matrix with self-connection, is the adjacency matrix of the knowledge graph, is the identity matrix of order , is the degree matrix of the graph, is the batch normalization, and is a trainable layer-specific weight matrix. We extract to the semantic information between nodes by the GCN, and obtain the disease situational representation with prior knowledge.
3.1.5. Rich Disease Representation with Prior Knowledge
To further improve the accuracy of generated reports, we introduce rich disease representations with prior knowledge. The main idea behind rich disease representations with prior knowledge is to further encode informative attributes about the disease state, such as positive, negative, uncertain, or unmentioned. Formally, let k be the number of states and the state embedding be , then the confidence of each disease classification as one of disease states is:
is a trainable parameter initialized randomly. is used as the feature of multi-label classification, and the classification loss function is as follows:
where and represent the true and predicted values of the -th disease label, respectively. The state embedding can be calculated as follows:
Finally, is the disease representation enriched with prior knowledge, and is composed of disease embedding states, disease name topics, and episodic disease representations with prior knowledge.
where is randomly initialized to represent the disease or topic to be generated and is a trainable parameter. The rich disease representation with prior knowledge provides a clear and accurate disease description, which provides strong data support for the subsequent generation module.
3.2. Generator
Our report generator is derived from Transformer. As shown in Figure 1, the network consists of the masked multi-head self-attention module and the feed-forward layer superimposed on each other times. The previous disease embedding and word embedding are then used to calculate the hidden state for each word of the medical reporter species, and the disease embedding is denoted as :
Then, we predict the possible words based on the hidden state .
is this vocabulary embedding, is the vocabulary size, is the length of the document, and represents the confidence that the -th position in the generated medical report selects the -th word in the vocabulary. The loss function of the generator is the cross entropy of the real word and the predicted word .
The final generated report is:
3.3. Interpreter
To make the generated report more consistent with the original output of the classifier, we refer to the idea of CycleGAN [38]. We build a text classifier based on the text encoder above, input the generated report into the text classifier, output the state of the disease-related topic, compare it with the original output of the classifier module, and then fine-tune the generated report by adjusting the word representation output .
First, the text encoder summarizes the current medical report and outputs the report summary embedding of the queried disease .
where is calculated from the medical report using Equation (1). Each report summary embedding is classified into one of k disease-related states, and is the -th line of .
The loss function of the interpreter is similar to the loss function of the classifier.
Here, denotes the confidence of the true disease label, and denotes the confidence of the predicted label. Overall, the training loss function of the whole model can be summarized as follows.
4. Experiments
The experimental section evaluates the medical report generation task from two aspects: verbal performance and clinical accuracy performance. The evaluation of the experiments is performed on two widely used chest X-ray datasets, the Open-I and MIMIC-CXR datasets.
4.1. Datasets
4.1.1. MIMIC-CXR Dataset
The MIMIC-CXR dataset is a large publicly available dataset of chest X-ray images in JPG format, containing 377,110 images and 227,835 medical reports of 65,379 patients from multiple viewpoints. Chest X-ray images from three main perspectives are reported: anterior-posterior (AP), posterior-anterior (PA), and lateral (LA). Each study included a comparison, clinical history, indication, reason for examination, impression, and findings section. In our approach, we use multi-view images and concatenate the clinical history, reason for examination, and indication sections as contextual information. For consistency, we follow the experimental setup of [39] and focus on generating the “findings” section as the corresponding radiology report.
4.1.2. Open-I Dataset
The Open-I dataset, collected at Indiana University Hospital, is a public radiology dataset containing 3955 radiology studies corresponding to 7470 frontal and lateral chest X-ray images. These radiological studies are related to one or more chest X-ray images. Each study reported impression, findings, comparison, and indication sections. Similar to the MIMIC-CXR dataset, we use multi-view chest X-ray images (frontal and lateral) and the indicator part as context input. In our approach, we follow the approach of the existing literature [2] and concatenate the impression part and the survey results part as the correct generated report.
4.2. Implementation Detail
We use Densenet-121 as the core of our CNN model, and all images are resized to 256 × 256. We use Transformer as the core of the text encoder. Both generators and interpreters are implemented based on Transformers and trained from scratch, all hyper-parameters are selected based on the performance on the validation set, and the number of reporting encoder layers is set to 12. We train the classification and generation reports on the Open-I and MIMIC-CXR datasets using the Adam optimizer with an initial learning rate of 3 × 10−4 and weight decay of 1 × 10−2. For the interpreter, the Open-I dataset is trained using a learning rate of 3 × 10−5, and the MIMIC-CXR dataset is trained using an Adam optimizer with a learning rate of 3 × 10−6 and a weight decay of 1 × 10−2. We train the model with epochs of 50 for both Open-I and MIMIC-CXR datasets. We evaluate the proposed model on the validation set. Our experiments are trained in parallel on two RTX3090 sheets, and the experiment for each dataset is mainly divided into two parts. The first part is the training of the classifier and generator, and then the trained model is added to the interpreter for the second part of training. Each part is trained for 50 epochs in our experiments. The training time of the first and second parts is about 54 s and 59 s per round for the Open-I dataset, and about 34 min and 35 min per epoch for the MIMIC-CXR dataset, respectively.
4.3. Experimental Results
4.3.1. Language Generation Performance
We employ the widely used NLG metric to evaluate the proposed model, which includes scores from BLEU-1 to BLEU-4 [40], ROUGE-L [41], and METEOR [42]. We use the nlg-eval library [43] to calculate the BLEU-1 to BLEU-4 scores, ROUGE-L scores, and METEOR scores. In Table 1, our experimental results are compared with other state-of-the-art methods, and all metrics have a certain improvement. The scores of BLEU-1 to BLEU-4 are obtained by analyzing the sequence of consecutive words appearing in the prediction report. In our results, BLEU-1 to BLEU-4 are significantly improved, indicating that our method ignores some meaningless words and focuses more on describing diseases with long sentences. ROUGE-L and METEOR are also much better than previous excellent methods, which mean that our method can generate accurate reports and the framework is effective.

Table 1.
A comparison of our method and many existing methods, using different linguistic metrics: BLEU-1 to BLEU-4, METEOR, and ROUGE-L, with the best results highlighted in bold.
4.3.2. Clinical Accuracy Performance
We use the CheXpert [10] label as a measure to evaluate the clinical accuracy of generated reports. We compare 14 common diseases proposed in CheXpert and MIMIC-CXR based on precision, precision, recall, and F-1 metrics. We show the macro and micro scores, respectively. A high macro score indicates an improvement in the detection of all 14 diseases, while a higher micro score indicates an improvement in the impact caused by the imbalance of the dataset, such as the higher frequency of some diseases than others. The results of our comparison are shown in Table 2. Compared with other experiments, our clinical performance has improved in most of the indicators in the macro and micro scores.

Table 2.
The clinical accuracy of the generated reports was quantitatively compared by evaluating 14 common diseases defined together in the CheXpert and MIMIC-CXR datasets, with the best results highlighted using bold font.
4.4. Ablation Studies
The quantitative results of our method in the Open-I dataset are shown in Table 3. Because the MIMIC-CXR dataset is too large and the effects of our method in both datasets are improved, we mainly focus on the smaller dataset Open-I when analyzing the quantitative results. By observing Table 3, it can be seen that after adding rich disease embedding containing prior knowledge to the classifier, all evaluation indicators are improved, and after adding the interpreter on this basis, the indicators are again improved to a certain extent. Compared with the model that only uses rich disease embedding and adds prior knowledge, the BLEU-1 value of the highest index is increased by 6.7% from 0.445 to 0.475. After adding rich disease embedding with prior knowledge to the classifier, our model adds an interpreter to obtain the best performance. Compared with the basic model with the interpreter, our final model also has a great improvement in various indicators. The BLEU-4 value is improved compared with the basic model without the interpreter and the model with the interpreter, and it is 7% higher than the basic model with the interpreter. It can be seen that the prior knowledge we incorporate is aided by the automatic generation of accurate radiology reports.

Table 3.
Ablation studies. Base with refers to the model composed of the classifier and generator mentioned in this paper, and the classifier uses enriched disease embedding. Base with refers to the model composed of the classifier and generator mentioned in this paper. Rich disease embedding with prior knowledge is used in the classifier. Interpreter is the Interpreter mentioned above. The best results are highlighted in bold.
4.5. Qualitative Results
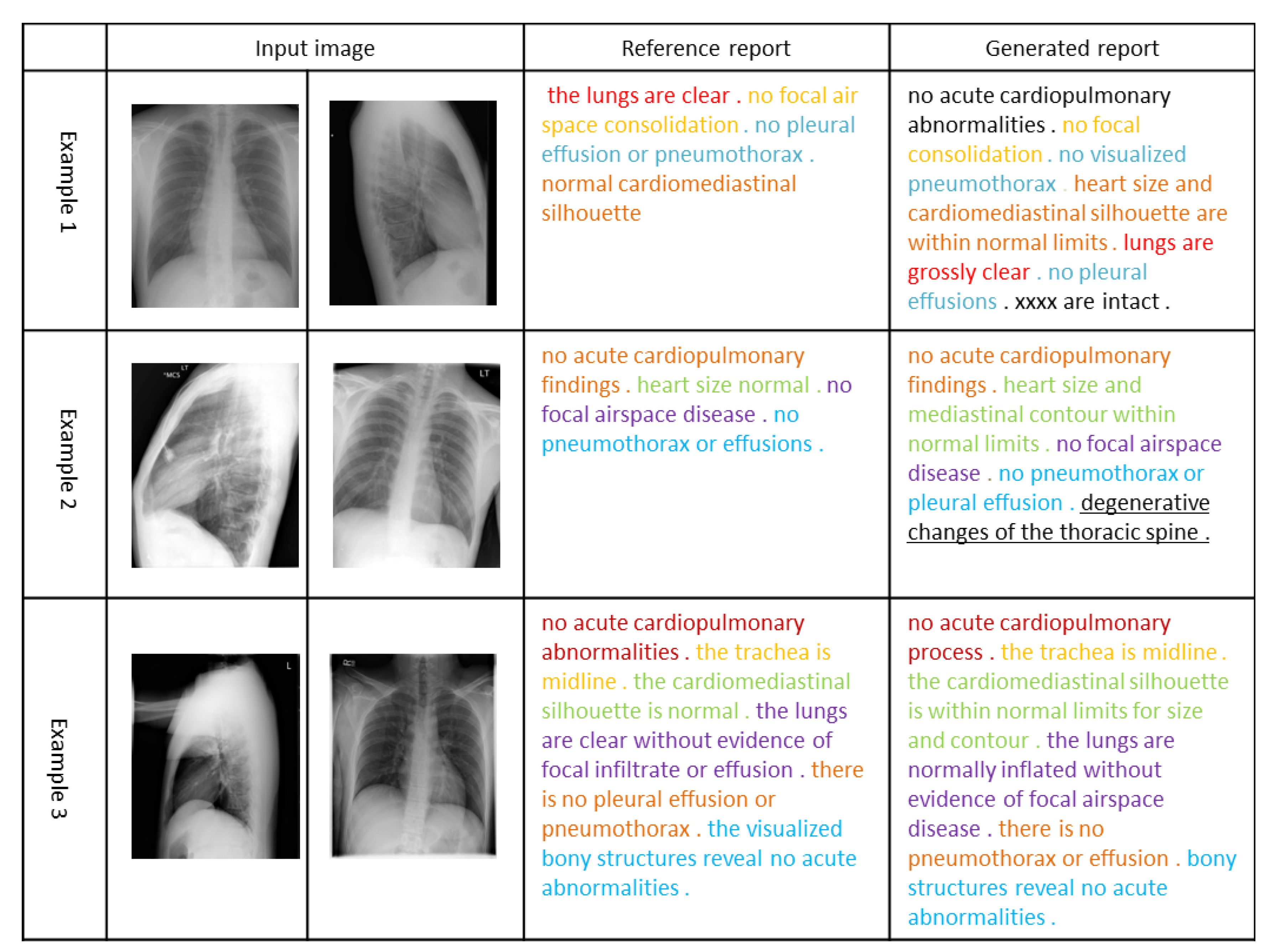
In Figure 3, we visualize the radiology reports of the three sets of images. The same color in the reference report and the generated report indicates the same description of the chest X-ray image, and the underscore indicates additional findings of our model for the chest X-ray image. In the first example, the reports generated by our model are almost identical to the actual situation. In the second example, the report generated by our model completely covers the actual situation and finds an additional disease based on the input image. Although this disease may indeed not exist, it can be used as a reminder to assist the doctor to make a more comprehensive diagnosis. In the third example, our model generates a longer report based on the obtained features, and it is almost the same as the actual situation, indicating that our model also has a relatively good performance in generating complex sentences. It can be seen that our method can extract features from images and clinical documents combined with prior knowledge to generate readable and accurate reports.

Figure 3.
Examples of three visual reports selected from the Open-I dataset. The same color emphasizes the same description of the chest X-ray image. Additional findings of our model for images are highlighted by underlines.
5. Conclusions and Outlook
In this work, we propose a model to enhance the accuracy of generated medical reports based on prior knowledge. We validate the proposed model experimentally, and we validate the effectiveness of our added prior knowledge on the Open-I and MIMIC-CXR datasets. The experimental results show that our model achieves relatively excellent performance in the indicators of natural language generation and clinical efficacy. Ablation experiments show that our model can learn visual features and text features better after adding prior knowledge, so it can generate medical reports more accurately. In addition, the establishment of our knowledge graph is built according to the dataset, which does not need additional experts to build, so it can be more convenient to apply to other datasets.
In our work, we have not considered the influence of location information on the generation of radiology reports, which is important. In the future, we will explore the impact of including location information in disease classification on improving the accuracy of generated reports. Next, we will explore how to improve the accuracy of our classifier, which is related to the accuracy of our automated reports. Specifically, we will pre-train our image encoder and text encoder using a public dataset and then try to incorporate location information into the classifier.
Author Contributions
Conceptualization, D.Z., A.R., and Q.L.; methodology, A.R., Q.L., J.L., Y.M., and D.Z.; investigation, A.R. and Y.M.; visualization, H.W., A.R., and J.L.; project administration, D.Z. and Q.L.; writing—original draft preparation, A.R., D.Z., J.L., and H.W.; writing—review and editing, Q.L., D.Z., A.R., J.L., Y.M., and H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by (i) Natural Science Foundation China (NSFC) under Grant No. 61402397, 61263043, 61562093 and 61663046; (ii) Open Foundation of Key Laboratory in Media Convergence of Yunnan Province under Grant No. 220225201. (iii) Open Foundation of Key Laboratory in Software Engineering of Yunnan Province: 2020SE304. (iv) Practical innovation project of Yunnan University, Project No. 2021z34, No. 2021y128 and 2021y129.
Acknowledgments
This research was supported by the Yunnan Provincial Key Laboratory of Software Engineering, the Kunming Provincial Key Laboratory of Data Science and Intelligent Computing and the Key Laboratory in Media Convergence of Yunnan Province.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Handbook of Medical Image Computing and Computer Assisted Intervention; Academic Press: Cambridge, MA, USA, 2019.
- Jing, B.; Xie, P.; Xing, E. On the automatic generation of medical imaging reports. arXiv 2017, arXiv:1711.08195. [Google Scholar]
- Bruno, M.A.; Walker, E.A.; Abujudeh, H.H. Understanding and confronting our mistakes: The epidemiology of error in radiology and strategies for error reduction. Radiographics 2015, 35, 1668–1676. [Google Scholar] [CrossRef]
- Shin, H.C.; Lu, L.; Kim, L.; Seff, A.; Yao, J.; Summers, R.M. Interleaved text/image deep mining on a very large-scale radiology database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1090–1099. [Google Scholar]
- Zhang, Y.; Wang, X.; Xu, Z.; Yu, Q.; Yuille, A.; Xu, D. When radiology report generation meets knowledge graph. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 12910–12917. [Google Scholar]
- Li, C.Y.; Liang, X.; Hu, Z.; Xing, E.P. Knowledge-driven encode, retrieve, paraphrase for medical image report generation. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 6666–6673. [Google Scholar]
- Yao, T.; Pan, Y.; Li, Y.; Mei, T. Exploring visual relationship for image captioning. In Proceedings of the European conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 684–699. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 2016, 23, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.-y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vinyals, O.; Toshev, A.; Bengio, S.; Erhan, D. Show and tell: A neural image caption generator. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3156–3164. [Google Scholar]
- Goyal, Y.; Khot, T.; Summers-Stay, D.; Batra, D.; Parikh, D. Making the v in vqa matter: Elevating the role of image understanding in visual question answering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6904–6913. [Google Scholar]
- Rennie, S.J.; Marcheret, E.; Mroueh, Y.; Ross, J.; Goel, V. Self-critical sequence training for image captioning. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7008–7024. [Google Scholar]
- Tran, A.; Mathews, A.; Xie, L. Transform and tell: Entity-aware news image captioning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 13035–13045. [Google Scholar]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. Proc. IEEE 2021, 109, 820–838. [Google Scholar] [CrossRef]
- Xu, K.; Ba, J.; Kiros, R.; Cho, K.; Courville, A.; Salakhudinov, R.; Zemel, R.; Bengio, Y. Show, attend and tell: Neural image caption generation with visual attention. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 7–9 July 2015; pp. 2048–2057. [Google Scholar]
- Lu, J.; Xiong, C.; Parikh, D.; Socher, R. Knowing when to look: Adaptive attention via a visual sentinel for image captioning. In Proceedings of the IEEE conference on computer vision and pattern recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 375–383. [Google Scholar]
- Anderson, P.; He, X.; Buehler, C.; Teney, D.; Johnson, M.; Gould, S.; Zhang, L. Bottom-up and top-down attention for image captioning and visual question answering. In Proceedings of the IEEE conference on computer vision and pattern recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6077–6086. [Google Scholar]
- Yuan, J.; Liao, H.; Luo, R.; Luo, J. Automatic radiology report generation based on multi-view image fusion and medical concept enrichment. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Cham, Switzerland, 2019; pp. 721–729. [Google Scholar]
- Xue, Y.; Xu, T.; Rodney Long, L.; Xue, Z.; Antani, S.; Thoma, G.R.; Huang, X. Multimodal recurrent model with attention for automated radiology report generation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Cham, Switzerland, 2018; pp. 457–466. [Google Scholar]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Summers, R.M. Tienet: Text-image embedding network for common thorax disease classification and reporting in chest X-rays. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 9049–9058. [Google Scholar]
- Chen, Z.; Song, Y.; Chang, T.-H.; Wan, X. Generating radiology reports via memory-driven transformer. arXiv 2020, arXiv:2010.16056. [Google Scholar]
- Jing, B.; Wang, Z.; Xing, E. Show, describe and conclude: On exploiting the structure information of chest X-ray reports. arXiv 2020, arXiv:2004.12274. [Google Scholar]
- Liu, G.; Hsu TM, H.; McDermott, M.; Boag, W.; Weng, W.-H.; Szolovits, P.; Ghassemi, M. Clinically accurate chest X-ray report generation. In Proceedings of the Machine Learning for Healthcare Conference, PMLR, Ann Arbor, MI, USA, 9–10 August 2019; pp. 249–269. [Google Scholar]
- Shin, H.C.; Roberts, K.; Lu, L.; Demner-Fushman, D.; Yao, J.; Summers, R.M. Learning to read chest X-rays: Recurrent neural cascade model for automated image annotation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2497–2506. [Google Scholar]
- Nguyen, H.; Nie, D.; Badamdorj, T.; Liu, Y.; Zhu, Y.; Truong, J.; Cheng, L. Automated generation of accurate\& fluent medical X-ray reports. arXiv 2021, arXiv:2108.12126. [Google Scholar]
- Kudo, T.; Richardson, J. Sentencepiece: A simple and language independent subword tokenizer and detokenizer for neural text processing. arXiv 2018, arXiv:1808.06226. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph attention networks. arXiv 2017, arXiv:1710.10903. [Google Scholar]
- Chen, M.; Radford, A.; Child, R.; Wu, J.; Jun, H.; Luan, D.; Sutskever, I. Generative pretraining from pixels. In Proceedings of the International Conference on Machine Learning, Virtual Event, 13–18 July 2020; pp. 1691–1703. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Yin, C.; Qian, B.; Wei, J.; Li, X.; Zhang, X.; Li, Y.; Zheng, Q. Automatic generation of medical imaging diagnostic report with hierarchical recurrent neural network. In Proceedings of the 2019 IEEE International Conference on Data Mining (ICDM), Beijing, China, 8–11 November 2019; pp. 728–737. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 590–597. [Google Scholar]
- Chen, Z.M.; Wei, X.S.; Wang, P.; Guo, P. Multi-label image recognition with graph convolutional networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 5177–5186. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Su, H.; Maji, S.; Kalogerakis, E.; Learned-Miller, E. Multi-view convolutional neural networks for 3d shape recognition. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 945–953. [Google Scholar]
- Jia, N.; Tian, X.; Zhang, Y.; Wang, F. Semi-supervised node classification with discriminable squeeze excitation graph convolutional networks. IEEE Access 2020, 8, 148226–148236. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Lovelace, J.; Mortazavi, B. Learning to generate clinically coherent chest X-ray reports. In Proceedings of the Findings of the Association for Computational Linguistics: EMNLP 2020, Punta Cana, Dominican Republic, 8–12 November 2020; Volume 2020, pp. 1235–1243. [Google Scholar]
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.-J. Bleu: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics, Philadelphia, PA, USA, 7–12 July 2002; pp. 311–318. [Google Scholar]
- Lin, C.Y. Rouge: A Package for Automatic Evaluation of Summaries; Text Summarization Branches Out; Association for Computational Linguistics: Barcelona, Spain, 2004; pp. 74–81. [Google Scholar]
- Banerjee, S.; Lavie, A. METEOR: An automatic metric for MT evaluation with improved correlation with human judgments. In Proceedings of the Acl Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization, Ann Arbor, MI, USA, 29 June 2005; pp. 65–72. [Google Scholar]
- Sharma, S.; Asri, L.E.; Schulz, H.; Zumer, J. Relevance of unsupervised metrics in task-oriented dialogue for evaluating natural language generation. arXiv 2017, arXiv:1706.09799. [Google Scholar]
- Li, Y.; Liang, X.; Hu, Z.; Xing, E.P. Hybrid retrieval-generation reinforced agent for medical image report generation. In Proceedings of the Neural Information Processing Systems 2018, held at Palais des Congres de Montreal, Montreal CANADA, 2–8 December 2018 Advances in Neural Information Processing Systems 2018, Montreal, QC, Canada, 2–8 December 2018; Volume 31. [Google Scholar]
- Liu, F.; Wu, X.; Ge, S.; Fan, W.; Zou, Y. Exploring and distilling posterior and prior knowledge for radiology report generation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 13753–13762. [Google Scholar]
- Li, J.; Li, S.; Hu, Y.; Tao, H. A Self-Guided Framework for Radiology Report Generation. arXiv 2022, arXiv:2206.09378. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).