The Impact of Vairimorpha (Nosema) ceranae Natural Infection on Honey Bee (Apis mellifera) and Bee Bread Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site, Sample Collection and Spore Count
2.2. V. ceranae Molecular Identification
2.3. Bee Bread DNA Extraction
2.4. Honey Bee DNA Extraction
2.5. PCR Amplification and Next-Generation Sequencing (NGS)
2.6. Bioinformatics and Statistical Analysis
3. Results
3.1. V. ceranae Quantification and Molecular Identification
3.2. Next-Generation Sequencing Analysis
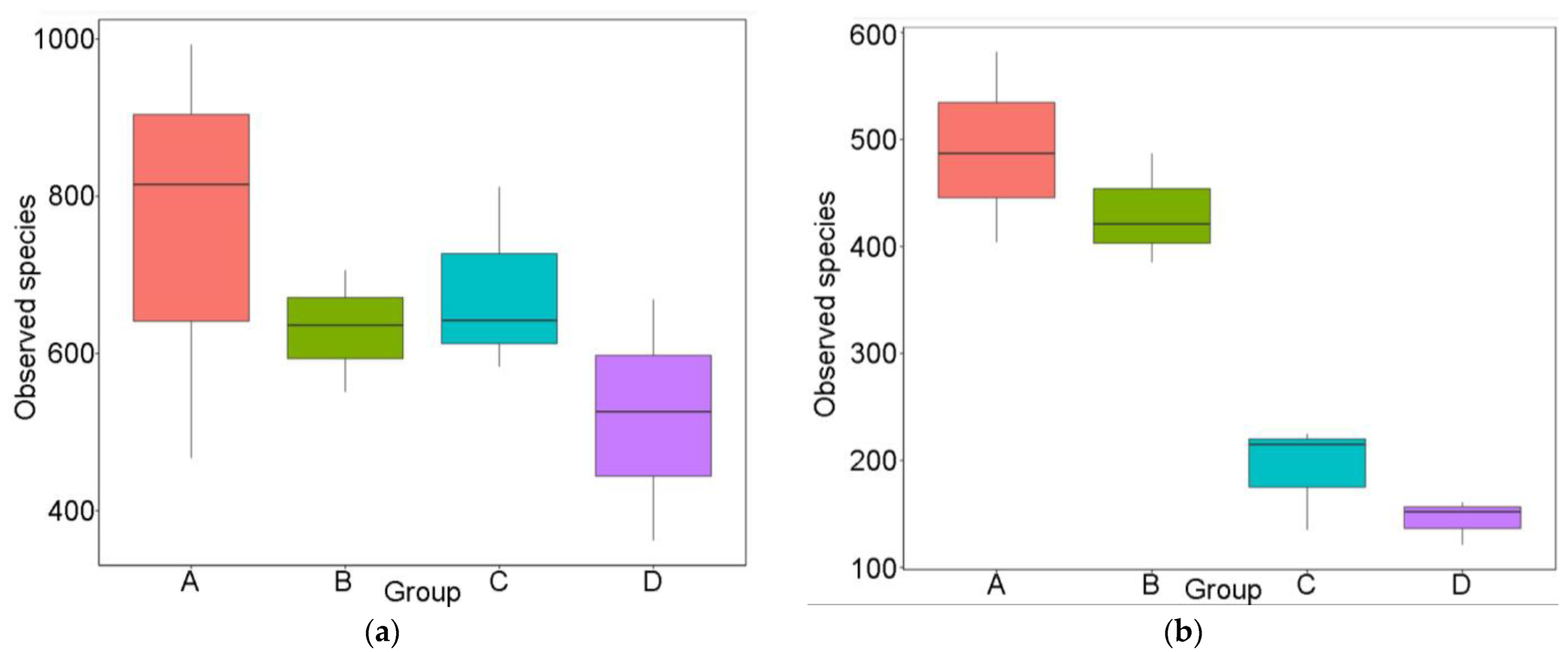
3.2.1. Observed Unique Operating Taxonomic Units (OTUs) and Alpha Diversity Indices
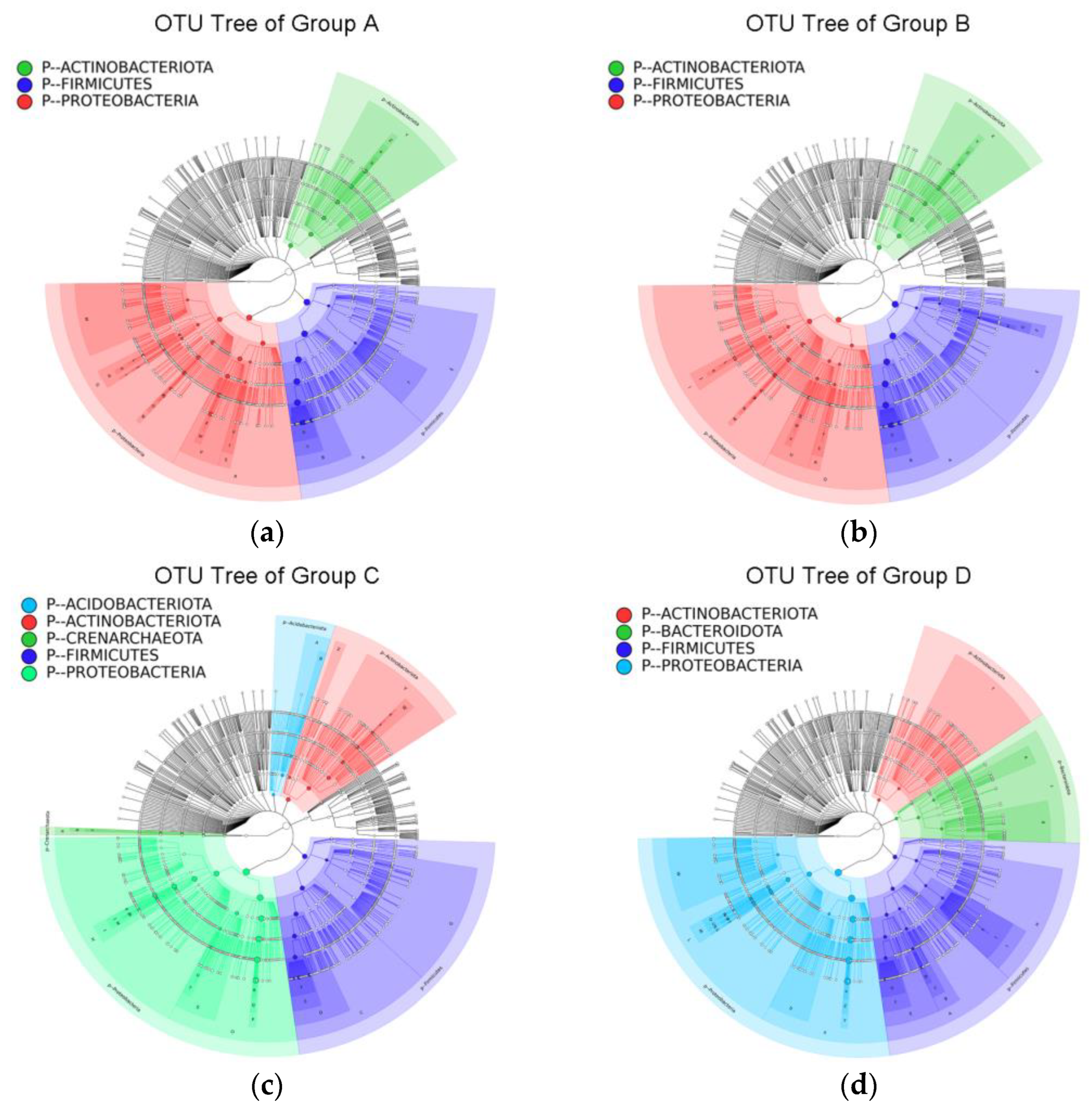
3.2.2. OTU Identification and Taxonomic Annotation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Papa, G.; Maier, R.; Durazzo, A.; Lucarini, M.; Karabagias, I.K.; Plutino, M.; Bianchetto, E.; Aromolo, R.; Pignatti, G.; Ambrogio, A.; et al. The Honey Bee Apis mellifera: An Insect at the Interface between Human and Ecosystem Health. Biology 2022, 11, 233. [Google Scholar] [CrossRef] [PubMed]
- van Engelsdorp, D.; Meixner, M.D. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 2010, 103, S80–S95. [Google Scholar] [CrossRef] [PubMed]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín-Hernández, R.; Meana, A. Nosema ceranae in Europe: An emergent type C nosemosis. Apidologie 2010, 41, 375–392. [Google Scholar] [CrossRef]
- Dussaubat, C.; Brunet, J.-L.; Higes, M.; Colbourne, J.K.; Lopez, J.; Choi, J.-H.; Martín-Hernández, R.; Botías, C.; Cousin, M.; McDonnell, C.; et al. Gut Pathology and Responses to the Microsporidium Nosema ceranae in the Honey Bee Apis mellifera. PLoS ONE 2012, 7, e37017. [Google Scholar] [CrossRef]
- Paris, L.; El Alaoui, H.; Delbac, F.; Diogon, M. Effects of the gut parasite Nosema ceranae on honey bee physiology and behavior. Curr. Opin. Insect Sci. 2018, 26, 149–154. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Dong, Z.-X.; Li, H.-Y.; Chen, Y.-F.; Wang, F.; Deng, X.-Y.; Lin, L.-B.; Zhang, Q.-L.; Li, J.-L.; Guo, J. Colonization of the gut microbiota of honey bee (Apis mellifera) workers at different developmental stages. Microbiol. Res. 2020, 231, 126370. [Google Scholar] [CrossRef]
- Yun, J.-H.; Jung, M.-J.; Kim, P.S.; Bae, J.-W. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci. Rep. 2018, 8, 2019. [Google Scholar] [CrossRef]
- Kakumanu, M.L.; Reeves, A.M.; Anderson, T.D.; Rodrigues, R.R.; Williams, M.A. Honey Bee Gut Microbiome Is Altered by In-Hive Pesticide Exposures. Front. Microbiol. 2016, 7, 1255. [Google Scholar] [CrossRef] [PubMed]
- Paris, L.; Peghaire, E.; Moné, A.; Diogon, M.; Debroas, D.; Delbac, F.; El Alaoui, H. Honeybee gut microbiota dysbiosis in pesticide/parasite co-exposures is mainly induced by Nosema ceranae. J. Invertebr. Pathol. 2020, 172, 107348. [Google Scholar] [CrossRef] [PubMed]
- Didaras, N.A.; Karatasou, K.; Dimitriou, T.G.; Amoutzias, G.D.; Mossialos, D. Antimicrobial Activity of Bee-Collected Pollen and Beebread: State of the Art and Future Perspectives. Antibiotics 2020, 9, 811. [Google Scholar] [CrossRef] [PubMed]
- Didaras, N.A.; Kafantaris, I.; Dimitriou, T.G.; Mitsagga, C.; Karatasou, K.; Giavasis, I.; Stagos, D.; Amoutzias, G.D.; Hatjina, F.; Mossialos, D. Biological Properties of Bee Bread Collected from Apiaries Located across Greece. Antibiotics 2021, 10, 555. [Google Scholar] [CrossRef] [PubMed]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol. Evol. 2018, 8, 4743–4756. [Google Scholar] [CrossRef]
- Disayathanoowat, T.; Li, H.; Supapimon, N.; Suwannarach, N.; Lumyong, S.; Chantawannakul, P.; Guo, J. Different Dynamics of Bacterial and Fungal Communities in Hive-Stored Bee Bread and Their Possible Roles: A Case Study from Two Commercial Honey Bees in China. Microorganisms 2020, 8, 264. [Google Scholar] [CrossRef]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.; Mott, B.M.; Maes, P.; Corby-Harris, V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- Mazur, E.D.; Gajda, A.M. Nosemosis in Honeybees: A Review Guide on Biology and Diagnostic Methods. Appl. Sci. 2022, 12, 5890. [Google Scholar] [CrossRef]
- Charistos, L.; Parashos, N.; Hatjina, F. Long term effects of a food supplement HiveAliveTM on honey bee colony strength and Nosema ceranae spore counts. J. Apic. Res. 2015, 54, 420–426. [Google Scholar] [CrossRef]
- Cantwell, G.E. Standard methods for counting Nosema spores. Am. Bee J. 1970, 110, 222–223. [Google Scholar]
- Fries, I.; Chauzat, M.P.; Chen, Y.P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martín-Hernández, R.; Natsopoulou, M.; et al. Standard methods for Nosema research. J. Apic. Res. 2015, 52, 1–28. [Google Scholar] [CrossRef]
- Martín-Hernández, R.; Meana, A.; Prieto, L.; Salvador, A.M.; Garrido-Bailón, E.; Higes, M. Outcome of Colonization of Apis mellifera by Nosema ceranae. Appl. Environ. Microbiol. 2007, 73, 6331–6338. [Google Scholar] [CrossRef]
- Casas, I.; Powell, L.; Klapper, P.E.; Cleator, G.M. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 1995, 53, 25–36. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Fries, I.; Feng, F.; Da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Tokarev, Y.S.; Huang, W.-F.; Solter, L.F.; Malysh, J.M.; Becnel, J.J.; Vossbrinck, C.R. A formal redefinition of the genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and reassignment of species based on molecular phylogenetics. J. Invertebr. Pathol. 2020, 169, 107279. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Martín, R.; Meana, A. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J. Invertebr. Pathol. 2006, 92, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Calderón, R.A.; Sanchez, L.A.; Yañez, O.; Fallas, N. Presence of Nosema ceranae in Africanized honey bee colonies in Costa Rica. J. Apic. Res. 2008, 47, 328–329. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Smith, I.B.; Pettis, J.S. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008, 97, 186–188. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Zhou, L.; Boncristiani, H.; Kimura, K.; Xiao, T.; Litkowski, A.M.; Pettis, J.S. Asymmetrical coexistence of Nosema ceranae and Nosema apis in honey bees. J. Invertebr. Pathol. 2009, 101, 204–209. [Google Scholar] [CrossRef]
- Alaux, C.; Brunet, J.-L.; Dussaubat, C.; Mondet, F.; Tchamitchan, S.; Cousin, M.; Brillard, J.; Baldy, A.; Belzunces, L.P.; Le Conte, Y. Interactions between Nosema microspores and a neonicotinoid weaken honeybees ( Apis mellifera ). Environ. Microbiol. 2010, 12, 774–782. [Google Scholar] [CrossRef]
- Doublet, V.; Natsopoulou, M.E.; Zschiesche, L.; Paxton, R.J. Within-host competition among the honey bees pathogens Nosema ceranae and Deformed wing virus is asymmetric and to the disadvantage of the virus. J. Invertebr. Pathol. 2015, 124, 31–34. [Google Scholar] [CrossRef]
- Vidau, C.; Diogon, M.; Aufauvre, J.; Fontbonne, R.; Viguès, B.; Brunet, J.-L.; Texier, C.; Biron, D.G.; Blot, N.; El Alaoui, H.; et al. Exposure to Sublethal Doses of Fipronil and Thiacloprid Highly Increases Mortality of Honeybees Previously Infected by Nosema ceranae. PLoS ONE 2011, 6, e21550. [Google Scholar] [CrossRef]
- Zheng, H.-Q.; Gong, H.-R.; Huang, S.-K.; Sohr, A.; Hu, F.-L.; Chen, Y.P. Evidence of the synergistic interaction of honey bee pathogens Nosema ceranae and Deformed wing virus. Vet. Microbiol. 2015, 177, 1–6. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Nosemosis of honey bees. In Terrestrial Manual; WOAH: Paris, France, 2018; pp. 744–749. ISBN 978-92-95108-18-9. [Google Scholar]
- Meana, A.; Martín-Hernández, R.; Higes, M. The reliability of spore counts to diagnose Nosema ceranae infections in honey bees. J. Apic. Res. 2010, 49, 212–214. [Google Scholar] [CrossRef]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Herb, M.; Schramm, M. Functions of ROS in Macrophages and Antimicrobial Immunity. Antioxidants 2021, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Armitage, S.A.; Genersch, E.; McMahon, D.P.; Rafaluk-Mohr, C.; Rolff, J. Tripartite interactions: How immunity, microbiota and pathogens interact and affect pathogen virulence evolution. Curr. Opin. Insect Sci. 2022, 50, 100871. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Wolbachia and the insect immune system: What reactive oxygen species can tell us about the mechanisms of Wolbachia–host interactions. Front. Microbiol. 2015, 6, 1201. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, M.; Wang, L.; Huang, S.; Su, S.; Huang, W.-F. Vairimorpha (Nosema) ceranae Infection Alters Honey Bee Microbiota Composition and Sustains the Survival of Adult Honey Bees. Biology 2021, 10, 905. [Google Scholar] [CrossRef]
- Lenaerts, M.; Goelen, T.; Paulussen, C.; Herrera-Malaver, B.; Steensels, J.; Van den Ende, W.; Verstrepen, K.J.; Wäckers, F.; Jacquemyn, H.; Lievens, B. Nectar bacteria affect life history of a generalist aphid parasitoid by altering nectar chemistry. Funct. Ecol. 2017, 31, 2061–2069. [Google Scholar] [CrossRef]
- Linde, M.; Shishkoff, N. Powdery Mildew. In Encyclopedia of Rose Science; Elsevier: Amsterdam, The Netherlands, 2003; pp. 158–165. [Google Scholar]

| Hive Coding Number | V. ceranae Average Spore Count Per Bee | Sample Designation for Metataxonomics | Group Name | |
|---|---|---|---|---|
| Bee Samples | 11 Bee | 6000 | A1 | A |
| 14 Bee | 24,000 | A2 | ||
| 15 Bee | 39,000 | A3 | ||
| 12 Bee | 4,500,000 | B1 | B | |
| 27 Bee | 2,800,000 | B2 | ||
| 112 Bee | 4,200,000 | B3 | ||
| Bee Bread samples | 11 BB | - | C1 | C |
| 14 BB | C2 | |||
| 15 BB | C3 | |||
| 12 BB | D1 | D | ||
| 27 BB | D2 | |||
| 112 BB | D3 |
| Group | 16S rRNA (Bacteria) | ITS (Fungi) |
|---|---|---|
| A | 13.926 | 43.700 |
| B | 11.244 | 36.593 |
| C | 20.711 | 25.942 |
| D | 6.329 | 39.886 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgi, I.; Asoutis Didaras, N.; Nikolaidis, M.; Dimitriou, T.G.; Charistos, L.; Hatjina, F.; Amoutzias, G.D.; Mossialos, D. The Impact of Vairimorpha (Nosema) ceranae Natural Infection on Honey Bee (Apis mellifera) and Bee Bread Microbiota. Appl. Sci. 2022, 12, 11476. https://doi.org/10.3390/app122211476
Georgi I, Asoutis Didaras N, Nikolaidis M, Dimitriou TG, Charistos L, Hatjina F, Amoutzias GD, Mossialos D. The Impact of Vairimorpha (Nosema) ceranae Natural Infection on Honey Bee (Apis mellifera) and Bee Bread Microbiota. Applied Sciences. 2022; 12(22):11476. https://doi.org/10.3390/app122211476
Chicago/Turabian StyleGeorgi, Irini, Nikos Asoutis Didaras, Marios Nikolaidis, Tilemachos G. Dimitriou, Leonidas Charistos, Fani Hatjina, Grigoris D. Amoutzias, and Dimitris Mossialos. 2022. "The Impact of Vairimorpha (Nosema) ceranae Natural Infection on Honey Bee (Apis mellifera) and Bee Bread Microbiota" Applied Sciences 12, no. 22: 11476. https://doi.org/10.3390/app122211476
APA StyleGeorgi, I., Asoutis Didaras, N., Nikolaidis, M., Dimitriou, T. G., Charistos, L., Hatjina, F., Amoutzias, G. D., & Mossialos, D. (2022). The Impact of Vairimorpha (Nosema) ceranae Natural Infection on Honey Bee (Apis mellifera) and Bee Bread Microbiota. Applied Sciences, 12(22), 11476. https://doi.org/10.3390/app122211476










