A Deep Learning Approach to Upscaling “Low-Quality” MR Images: An In Silico Comparison Study Based on the UNet Framework
Abstract
1. Introduction
- Two different acquisition scenarios: (a) the With-Priors that mimic studies that entail the collection of both an LQ image and its complementary high quality prior to upscale the image to a corresponding HQ target, (b) the WithOut-Priors that mimic studies that entail the collection of only a single LQ Image to upscale the image to a corresponding HQ target.
- Creation of synthetic training and testing data so we have the same set of LQ images, its complementary HQ prior, and an HQ image (ground truth). In addition, hyperintense lesions of random intensity, size, and position were added to increase the variability in the images.
- The collected LQ Images were truncated to three smaller matrix sizes to mimic studies where the acquisition matrix sizes are small. We train the networks to upscale these images in two acquisition scenarios.
- An extensive analysis of the quality of the upscaled images obtained from different UNet was performed using various indices and statistical tests using a mixed effects model.
2. Materials and Methods
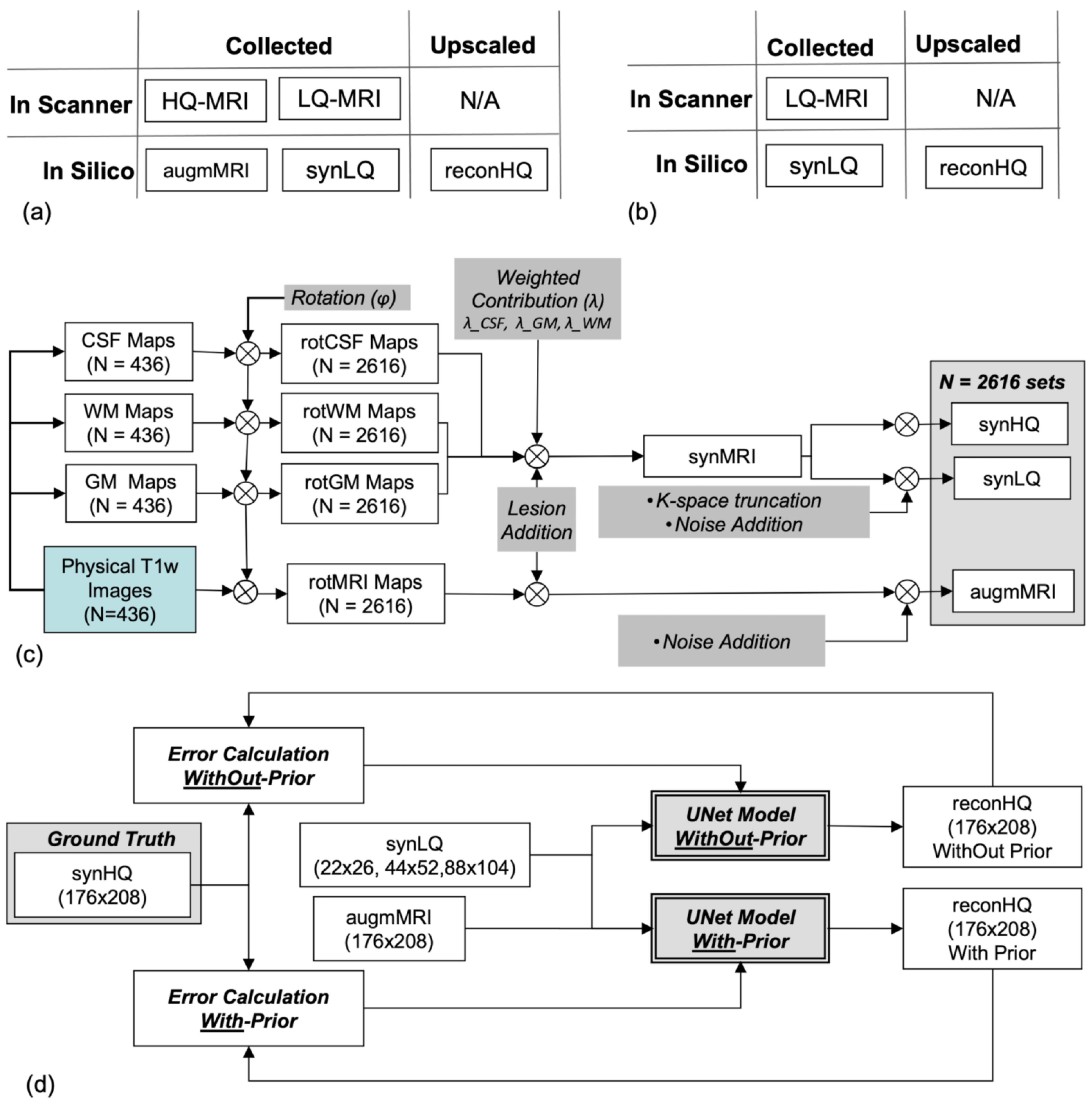
2.1. Training and Testing Dataset
2.2. UNet Architectures
2.3. Dense UNet (DUNet)
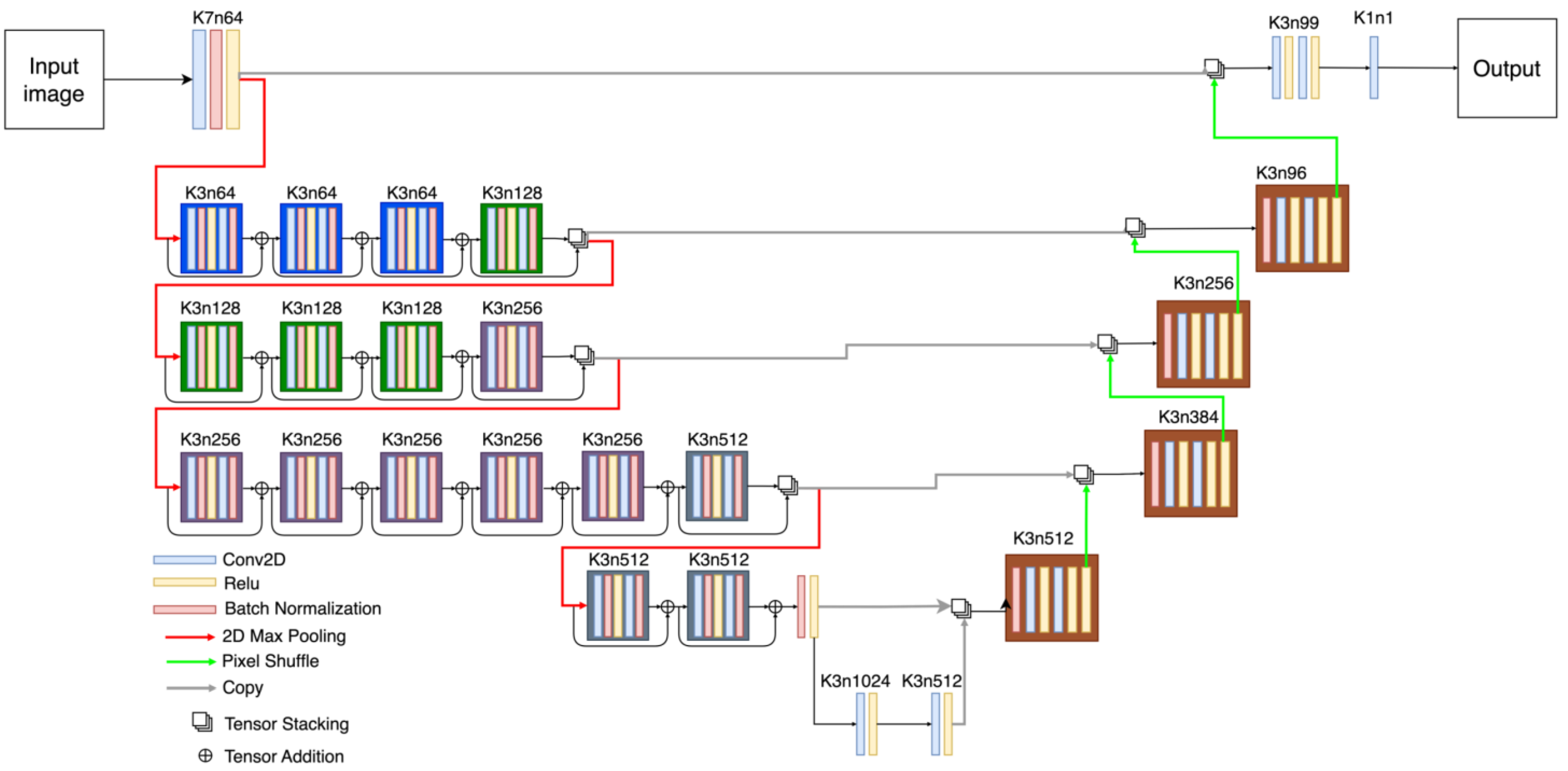
2.4. Robust UNet (RUNet)
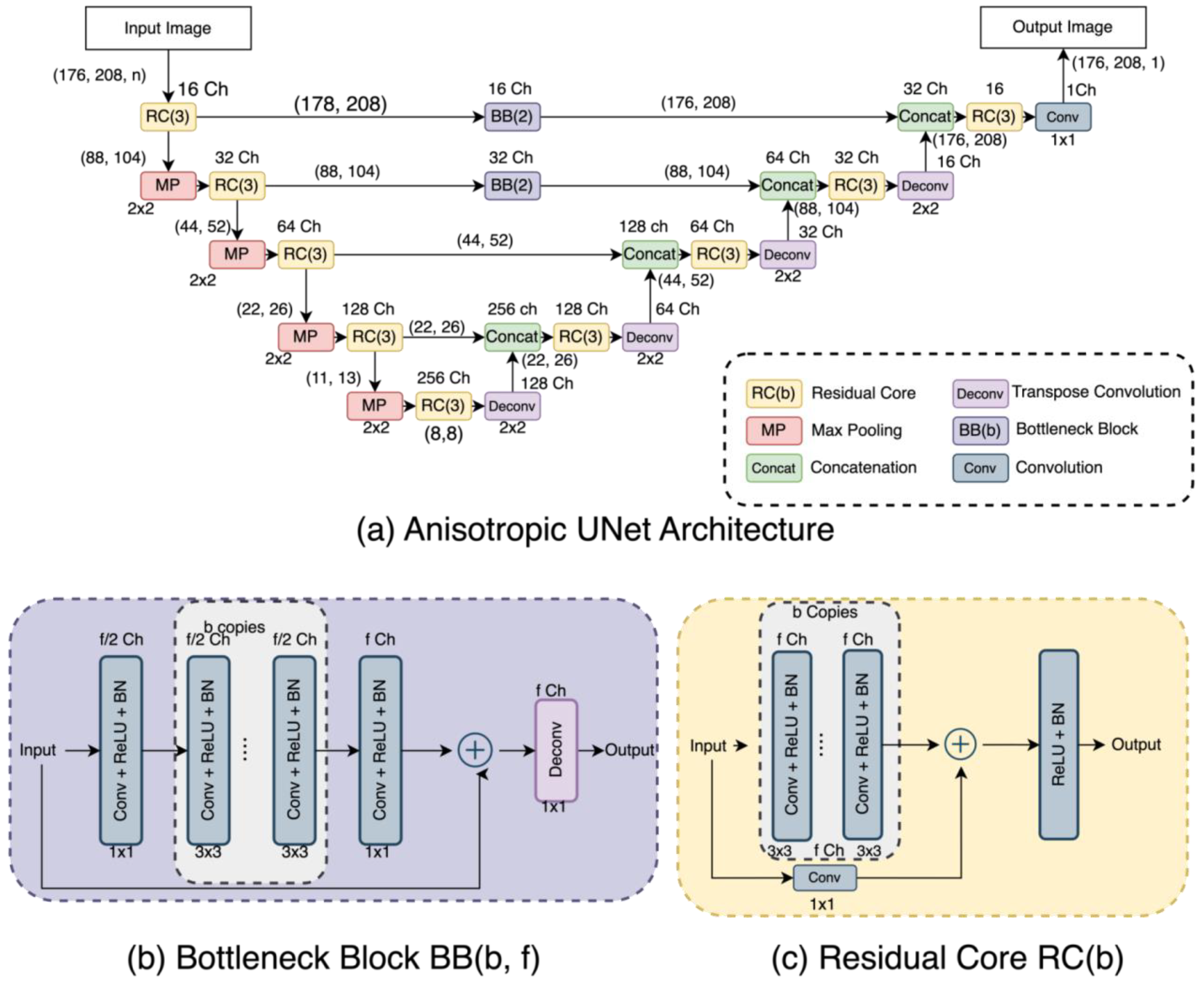
2.5. Anisotropic UNet (AUNet)
2.6. Network Implementation and Training
2.7. Data Analysis and Statistics
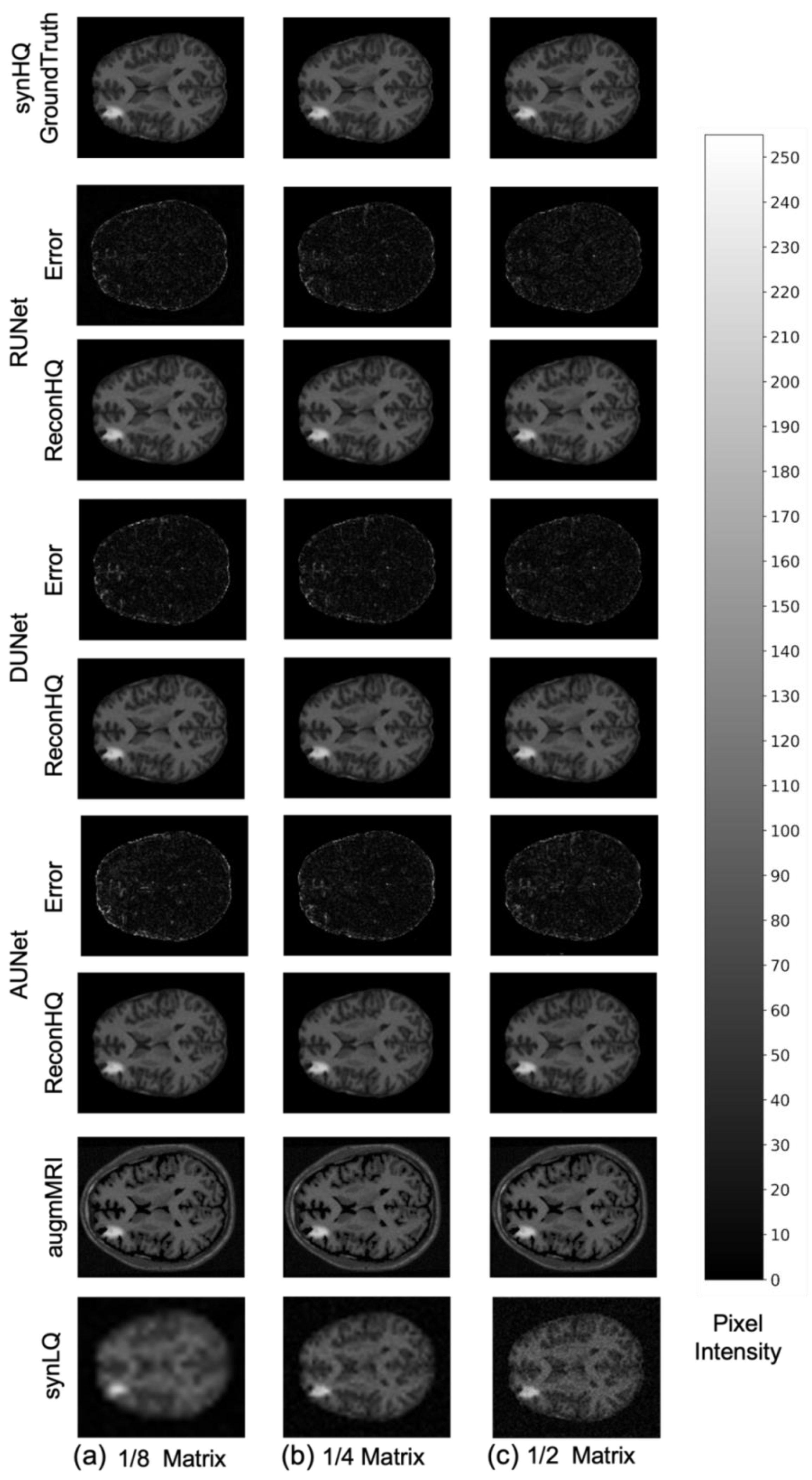
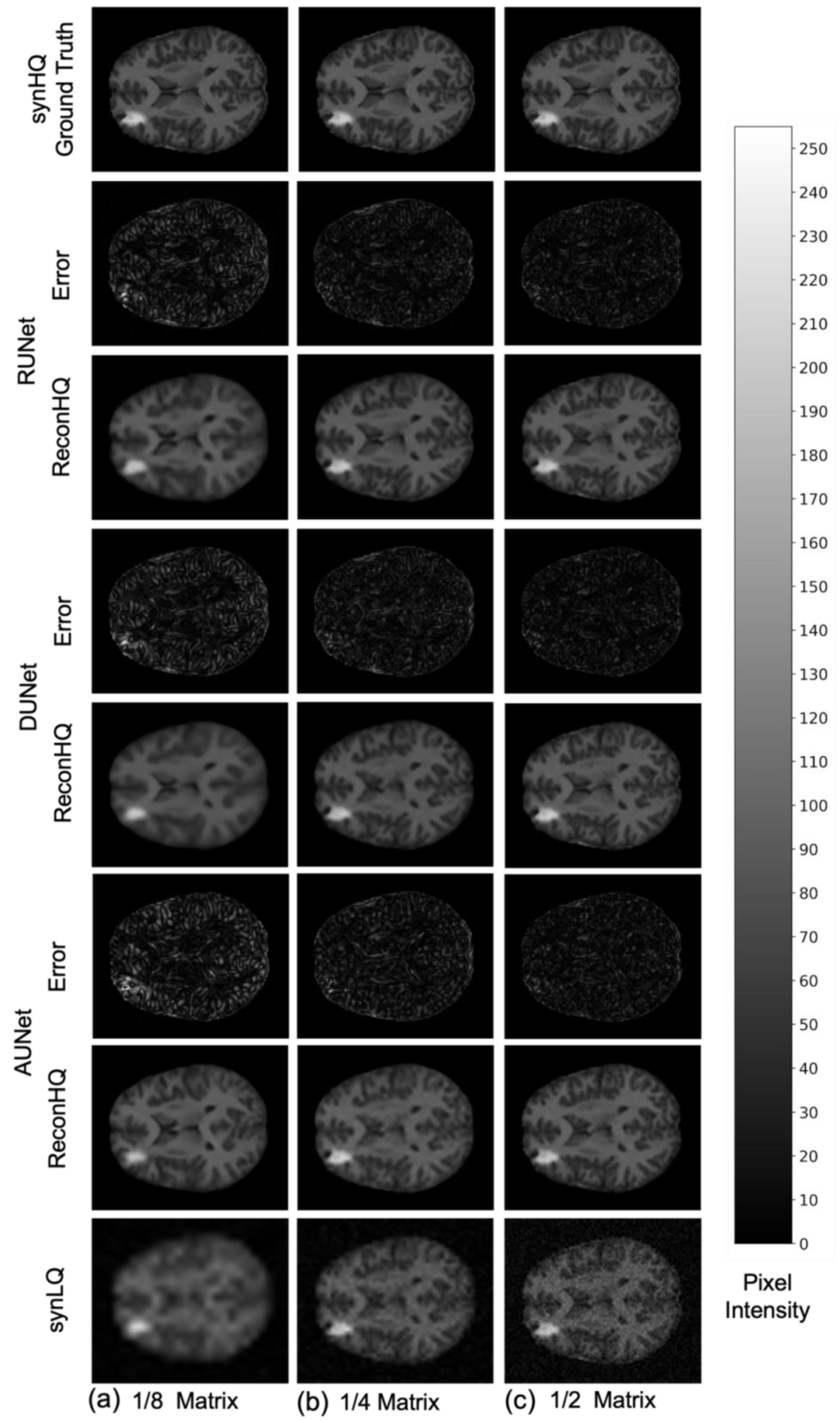
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parker, D.L.; Gullberg, G.T. Signal-to-Noise Efficiency in Magnetic Resonance Imaging. Med. Phys. 1990, 17, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Constable, R.T.; Henkelman, R.M. Contrast, Resolution, and Detectability in MR Imaging. J. Comput. Assist. Tomogr. 1991, 15, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Macovski, A. Noise in MRI. Magn. Reson. Med. 1996, 36, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Plenge, E.; Poot, D.H.J.; Bernsen, M.; Kotek, G.; Houston, G.; Wielopolski, P.; van der Weerd, L.; Niessen, W.J.; Meijering, E. Super-Resolution Methods in MRI: Can They Improve the Trade-off between Resolution, Signal-to-Noise Ratio, and Acquisition Time? Magn. Reson. Med. 2012, 68, 1983–1993. [Google Scholar] [CrossRef]
- Peters, D.C.; Korosec, F.R.; Grist, T.M.; Block, W.F.; Holden, J.E.; Vigen, K.K.; Mistretta, C.A. Undersampled Projection Reconstruction Applied to MR Angiography. Magn. Reson. Med. 2000, 43, 91–101. [Google Scholar] [CrossRef]
- Lustig, M.; Donoho, D.; Pauly, J.M. Sparse MRI: The Application of Compressed Sensing for Rapid MR Imaging. Magn. Reson. Med. 2007, 58, 1182–1195. [Google Scholar] [CrossRef]
- Heidemann, R.M.; Özsarlak, Ö.; Parizel, P.M.; Michiels, J.; Kiefer, B.; Jellus, V.; Müller, M.; Breuer, F.; Blaimer, M.; Griswold, M.A.; et al. A Brief Review of Parallel Magnetic Resonance Imaging. Eur. Radiol. 2003, 13, 2323–2337. [Google Scholar] [CrossRef]
- Pruessmann, K.P. Encoding and Reconstruction in Parallel MRI. NMR Biomed. 2006, 19, 288–299. [Google Scholar] [CrossRef]
- Brateman, L. Chemical Shift Imaging: A Review. Am. J. Roentgenol. 1986, 146, 971–980. [Google Scholar] [CrossRef]
- Marques, J.P.; Simonis, F.F.J.; Webb, A.G. Low-field MRI: An MR Physics Perspective. J. Magn. Reson. Imaging 2019, 49, 1528–1542. [Google Scholar] [CrossRef]
- Hu, R.; Kleimaier, D.; Malzacher, M.; Hoesl, M.A.U.; Paschke, N.K.; Schad, L.R. X-nuclei Imaging: Current State, Technical Challenges, and Future Directions. J. Magn. Reson. Imaging 2020, 51, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.-H.; Ducournau, A.; Fablet, R.; Rousseau, F. Brain MRI Super-Resolution Using Deep 3D Convolutional Networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 197–200. [Google Scholar]
- Chen, Y.; Shi, F.; Christodoulou, A.G.; Xie, Y.; Zhou, Z.; Li, D. Efficient and accurate MRI super-resolution using a generative adversarial network and 3D multi-level densely connected network. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); LNCS; Springer: Cham, Switzerland, 2018; Volume 11070. [Google Scholar]
- van Reeth, E.; Tham, I.W.K.; Tan, C.H.; Poh, C.L. Super-Resolution in Magnetic Resonance Imaging: A Review. Concepts Magn. Reson. Part A 2012, 40A, 306–325. [Google Scholar] [CrossRef]
- Cherukuri, V.; Guo, T.; Schiff, S.J.; Monga, V. Deep MR Brain Image Super-Resolution Using Spatio-Structural Priors. IEEE Trans. Image Process. 2020, 29, 1368–1383. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Iwamoto, Y.; Lin, L.; Xu, R.; Tong, R.; Chen, Y.-W. VolumeNet: A Lightweight Parallel Network for Super-Resolution of MR and CT Volumetric Data. IEEE Trans. Image Process. 2021, 30, 4840–4854. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Taviani, V.; Malkiel, I.; Cheng, J.Y.; Tamir, J.I.; Shaikh, J.; Chang, S.T.; Hardy, C.J.; Pauly, J.M.; Vasanawala, S.S. Variable-Density Single-Shot Fast Spin-Echo MRI with Deep Learning Reconstruction by Using Variational Networks. Radiology 2018, 289, 366–373. [Google Scholar] [CrossRef]
- Pizurica, A.; Wink, A.; Vansteenkiste, E.; Philips, W.; Roerdink, B.J. A Review of Wavelet Denoising in MRI and Ultrasound Brain Imaging. Curr. Med. Imaging Rev. 2006, 2, 247–260. [Google Scholar] [CrossRef]
- Zhang, K.; Zuo, W.; Chen, Y.; Meng, D.; Zhang, L. Beyond a Gaussian Denoiser: Residual Learning of Deep CNN for Image Denoising. IEEE Trans. Image Process. 2017, 26, 3142–3155. [Google Scholar] [CrossRef]
- Fan, L.; Zhang, F.; Fan, H.; Zhang, C. Brief Review of Image Denoising Techniques. Vis. Comput. Ind. Biomed. Art 2019, 2, 7. [Google Scholar] [CrossRef]
- Iqbal, Z.; Nguyen, D.; Hangel, G.; Motyka, S.; Bogner, W.; Jiang, S. Super-Resolution 1H Magnetic Resonance Spectroscopic Imaging Utilizing Deep Learning. Front. Oncol. 2019, 9, 1010. [Google Scholar] [CrossRef]
- Hu, X.; Naiel, M.A.; Wong, A.; Lamm, M.; Fieguth, P. RUNet: A Robust UNet Architecture for Image Super-Resolution. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Long Beach, CA, USA, 16–17 June 2019. [Google Scholar]
- Lin, H.; Figini, M.; Tanno, R.; Blumberg, S.B.; Kaden, E.; Ogbole, G.; Brown, B.J.; D’Arco, F.; Carmichael, D.W.; Lagunju, I.; et al. Deep learning for low-field to high-field mr: Image quality transfer with probabilistic decimation simulator. In International Workshop on Machine Learning for Medical Image Reconstruction; Springer: Cham, Switzerland, 2019; pp. 58–70. [Google Scholar]
- Masutani, E.M.; Bahrami, N.; Hsiao, A. Deep Learning Single-Frame and Multiframe Super-Resolution for Cardiac MRI. Radiology 2020, 295, 552–561. [Google Scholar] [CrossRef]
- Chatterjee, S.; Sarasaen, C.; Rose, G.; Nürnberger, A.; Speck, O. DDoS-UNet: Incorporating Temporal Information Using Dynamic Dual-Channel UNet for Enhancing Super-Resolution of Dynamic MRI. arXiv 2022, arXiv:2202.05355. [Google Scholar]
- Chatterjee, S.; Sciarra, A.; Dunnwald, M.; Mushunuri, R.V.; Podishetti, R.; Rao, R.N.; Gopinath, G.D.; Oeltze-Jafra, S.; Speck, O.; Nurnberger, A. ShuffleUNet: Super Resolution of Diffusion-Weighted MRIs Using Deep Learning. In Proceedings of the 2021 29th European Signal Processing Conference (EUSIPCO), Dublin, Ireland, 23–27 August 2021; pp. 940–944. [Google Scholar]
- Ding, P.L.K.; Li, Z.; Zhou, Y.; Li, B. Deep Residual Dense U-Net for Resolution Enhancement in Accelerated MRI Acquisition. In Proceedings of the Medical Imaging 2019: Image Processing, San Diego, California, USA, 19–21 February 2019; p. 14. [Google Scholar]
- Nasrin, S.; Alom, M.Z.; Burada, R.; Taha, T.M.; Asari, V.K. Medical Image Denoising with Recurrent Residual U-Net (R2U-Net) Base Auto-Encoder. In Proceedings of the 2019 IEEE National Aerospace and Electronics Conference (NAECON), Dayton, OH, USA, 15–19 July 2019; pp. 345–350. [Google Scholar]
- Koonjoo, N.; Zhu, B.; Bagnall, G.C.; Bhutto, D.; Rosen, M.S. Boosting the Signal-to-Noise of Low-Field MRI with Deep Learning Image Reconstruction. Sci Rep. 2021, 11, 8248. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, D.; Bozorgtabar, B.; Garnavi, R. Image Super-Resolution Using Progressive Generative Adversarial Networks for Medical Image Analysis. Comput. Med. Imaging Graph. 2019, 71, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, I.; Vilaplana, V. Brain MRI Super-Resolution Using 3D Generative Adversarial Networks. arXiv 2018, arXiv:1812.11440. [Google Scholar]
- Lyu, Q.; Shan, H.; Wang, G. MRI Super-Resolution With Ensemble Learning and Complementary Priors. IEEE Trans. Comput. Imaging 2020, 6, 615–624. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016, Athens, Greece, 17–21 October 2016; Springer: Cham, Switzerland, 2016; pp. 424–432. [Google Scholar]
- Sharma, R.; Eick, C.F.; Tsekos, N.V. Myocardial Infarction Segmentation in Late Gadolinium Enhanced MRI Images Using Data Augmentation and Chaining Multiple U-Net. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, Ohio, USA, 26–28 October 2020; pp. 975–980. [Google Scholar]
- Soomro, T.A.; Afifi, A.J.; Gao, J.; Hellwich, O.; Paul, M.; Zheng, L. Strided U-Net Model: Retinal Vessels Segmentation Using Dice Loss. In Proceedings of the 2018 Digital Image Computing: Techniques and Applications (DICTA), Canberra, Australia, 10–13 December 2018; pp. 1–8. [Google Scholar]
- Chen, C.; Zhou, K.; Wang, Z.; Xiao, R. Generative Consistency for Semi-Supervised Cerebrovascular Segmentation from TOF-MRA. IEEE Trans. Med. Imaging 2022, 1. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, K.; Zha, M.; Qu, X.; Guo, X.; Chen, H.; Wang, Z.; Xiao, R. An Effective Deep Neural Network for Lung Lesions Segmentation from COVID-19 CT Images. IEEE Trans. Industr Inform. 2021, 17, 6528–6538. [Google Scholar] [CrossRef]
- Steeden, J.A.; Quail, M.; Gotschy, A.; Mortensen, K.H.; Hauptmann, A.; Arridge, S.; Jones, R.; Muthurangu, V. Rapid Whole-Heart CMR with Single Volume Super-Resolution. J. Cardiovasc. Magn. Reson. 2020, 22, 56. [Google Scholar] [CrossRef]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. No New-Net. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 234–244. [Google Scholar]
- Zaaraoui, W.; Konstandin, S.; Audoin, B.; Nagel, A.M.; Rico, A.; Malikova, I.; Soulier, E.; Viout, P.; Confort-Gouny, S.; Cozzone, P.J.; et al. Distribution of Brain Sodium Accumulation Correlates with Disability in Multiple Sclerosis: A Cross-Sectional 23Na MR Imaging Study. Radiology 2012, 264, 859–867. [Google Scholar] [CrossRef]
- O’Reilly, T.; Webb, A.G. In Vivo T1 and T2 Relaxation Time Maps of Brain Tissue, Skeletal Muscle, and Lipid Measured in Healthy Volunteers at 50 MT. Magn. Reson. Med. 2022, 87, 884–895. [Google Scholar] [CrossRef]
- Pinheiro, J.C.; Bates, D.M. Mixed-Effects Models in S and S-PLUS; Springer: New York, NY, USA, 2000; ISBN 0-387-98957-9. [Google Scholar]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A Review of Medical Image Data Augmentation Techniques for Deep Learning Applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical Image Synthesis for Data Augmentation and Anonymization Using Generative Adversarial Networks. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Cham, Switzerland, 2018; pp. 1–11. [Google Scholar]
- Lei, Y.; Dong, X.; Tian, Z.; Liu, Y.; Tian, S.; Wang, T.; Jiang, X.; Patel, P.; Jani, A.B.; Mao, H.; et al. CT Prostate Segmentation Based on Synthetic MRI-aided Deep Attention Fully Convolution Network. Med. Phys. 2020, 47, 530–540. [Google Scholar] [CrossRef]
- Liu, Y.; Lei, Y.; Fu, Y.; Wang, T.; Zhou, J.; Jiang, X.; McDonald, M.; Beitler, J.J.; Curran, W.J.; Liu, T.; et al. Head and Neck Multi-organ Auto-segmentation on CT Images Aided by Synthetic MRI. Med. Phys. 2020, 47, 4294–4302. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Wang, T.; Tian, S.; Dong, X.; Jani, A.B.; Schuster, D.; Curran, W.J.; Patel, P.; Liu, T.; Yang, X. Male Pelvic Multi-Organ Segmentation Aided by CBCT-Based Synthetic MRI. Phys. Med. Biol. 2020, 65, 035013. [Google Scholar] [CrossRef] [PubMed]
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-Sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; ben Slima, M.; ben Hamida, A.; Mhiri, C.; Ben Mahfoudh, K. Denoising and Contrast-Enhancement Approach of Magnetic Resonance Imaging Glioblastoma Brain Tumors. J. Med. Imaging 2019, 6, 044002. [Google Scholar] [CrossRef]
- Kleesiek, J.; Morshuis, J.N.; Isensee, F.; Deike-Hofmann, K.; Paech, D.; Kickingereder, P.; Köthe, U.; Rother, C.; Forsting, M.; Wick, W.; et al. Can Virtual Contrast Enhancement in Brain MRI Replace Gadolinium? Investig. Radiol. 2019, 54, 653–660. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on Machine Learning, Lille, France, 6–11 July 2015. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified Linear Units Improve Restricted Boltzmann Machines. In Proceedings of the International Conference on Machine Learning, Haifa, Israel, 21–24 June 2010. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Shi, W.; Caballero, J.; Huszar, F.; Totz, J.; Aitken, A.P.; Bishop, R.; Rueckert, D.; Wang, Z. Real-Time Single Image and Video Super-Resolution Using an Efficient Sub-Pixel Convolutional Neural Network. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 1874–1883. [Google Scholar]
- Ledig, C.; Theis, L.; Huszar, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 105–114. [Google Scholar]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image Super-Resolution Using Deep Convolutional Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Dalmaz, O.; Yurt, M.; Cukur, T. ResViT: Residual Vision Transformers for Multimodal Medical Image Synthesis. IEEE Trans. Med. Imaging 2022, 41, 2598–2614. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Cao, J.; Sun, G.; Zhang, K.; van Gool, L.; Timofte, R. SwinIR: Image Restoration Using Swin Transformer. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW), Montreal, BC, Canada, 11–17 October 2021; pp. 1833–1844. [Google Scholar]
- Yang, F.; Yang, H.; Fu, J.; Lu, H.; Guo, B. Learning Texture Transformer Network for Image Super-Resolution. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 5790–5799. [Google Scholar]







| Acquisition Scenario | Number of Epochs | Upscaling Method | Mean Square Error (MSE) | Mean Intensity Error (MIE) | Mean Structural Similarity (SSIM) | Peak Signal to Noise Ratio (PSNR) |
|---|---|---|---|---|---|---|
| With-Prior | 100 Epochs | RUNet | 9.51 (3.00) | 1.74 (0.28) | 96.44 (0.48) | 39.17 (1.12) |
| DUNet | 10.72 (3.78) | 1.77 (0.38) | 97.62 (0.41) | 38.61 (1.46) | ||
| AUNet | 15.90 (4.68) | 2.10 (0.38) | 96.17 (0.70)) | 37.29 (1.25) | ||
| 1000 Epochs | RUNet | 8.67 (2.53) | 1.62 (0.29) | 97.45 (0.37) | 39.54 (1.20) | |
| DUNet | 8.13 (2.54) | 1.46 (0.24) | 97.99 (0.35) | 39.90 (1.12) | ||
| AUNet | 10.71 (4.60) | 1.75 (0.46) | 97.78 (0.42) | 38.67 (1.71) | ||
| WithOut-Prior | 100 Epochs | Runet | 65.73 (19.08) | 4.57 (0.72) | 83.69 (1.49) | 33.11 (0.63) |
| DUNet | 66.07 (20.58) | 4.50 (0.78) | 84.21 (1.53) | 33.11 (0.70) | ||
| AUNet | 76.26 (20.88) | 4.73 (0.75) | 83.08 (1.55) | 33.04 (0.65) | ||
| 1000 Epochs | RUNet | 66.64 (23.17) | 4.54 (0.86) | 84.16 (1.53) | 33.12 (0.72) | |
| DUNet | 65.06 (21.57) | 4.45 (0.81) | 84.77 (1.51) | 33.15 (0.71) | ||
| AUNet | 75.25 (21.76) | 4.68 (0.79) | 83.26 (1.39) | 33.10 (0.70) |
| Acquisition Scenario | Number of Epochs | Upscaling Method | Mean Square Error (MSE) | Mean Intensity Error (MIE) | Mean Structural Similarity (SSIM) | Peak Signal to Noise Ratio (PSNR) |
|---|---|---|---|---|---|---|
| With-Prior | 100 Epochs | RUNet | 9.20 (3.03) | 1.58 (0.28) | 97.54 (0.39) | 39.33 (1.18) |
| DUNet | 9.24 (3.82) | 1.62 (0.39) | 97.87 (0.33) | 39.22 (1.53) | ||
| AUNet | 11.76 (4.28) | 1.80 (0.39) | 97.24 (0.41) | 38.40 (1.38) | ||
| 1000 Epochs | RUNet | 6.36 (1.92) | 1.29 (0.22) | 98.25 (0.25) | 40.86 (1.15) | |
| DUNet | 6.73 (1.91) | 1.33 (0.21) | 98.26 (0.24) | 40.58 (1.10) | ||
| AUNet | 8.39 (3.88) | 1.56 (0.43) | 98.17 (0.29) | 39.58 (1.76) | ||
| WithOut-Prior | 100 Epochs | Runet | 27.38 (10.41) | 2.90 (0.58) | 93.24 (0.73) | 35.09 (1.04) |
| DUNet | 29.81 (13.66) | 3.01 (0.70) | 92.93 (0.79) | 34.85 (1.13) | ||
| AUNet | 40.07 (15.44) | 3.57 (0.77) | 91.67 (0.79) | 33.91 (0.99) | ||
| 1000 Epochs | RUNet | 25.22 (10.51) | 2.73 (0.61) | 94.12 (0.64) | 35.43 (1.81) | |
| DUNet | 27.05 (12.04) | 2.84 (0.64) | 93.86 (0.70) | 35.16 (1.13) | ||
| AUNet | 26.43 (10.60) | 2.81 (0.63) | 94.00 (0.67) | 35.23 (1.21) |
| Acquisition Scenario | Number of Epochs | Upscaling Method | Mean Square Error (MSE) | Mean Intensity Error (MIE) | Mean Structural Similarity (SSIM) | Peak Signal to Noise Ratio (PSNR) |
|---|---|---|---|---|---|---|
| With-Prior | 100 Epochs | RUNet | 7.80 (2.02) | 1.47 (0.21) | 97.79 (0.36) | 39.84 (1.02) |
| DUNet | 8.54 (0.89) | 1.57 (0.37) | 97.98 (0.30) | 39.47 (1.53) | ||
| AUNet | 14.17 (5.77) | 2.01 (0.49) | 96.76 (0.64) | 37.60 (1.57) | ||
| 1000 Epochs | RUNet | 6.51 (1.64) | 1.34 (0.20) | 98.08 (0.25) | 40.59 (1.02) | |
| DUNet | 6.35 (2.35) | 1.32 (0.28) | 98.40 (0.22) | 40.73 (1.34) | ||
| AUNet | 8.35 (4.06) | 1.53 (0.44) | 98.17 (0.27) | 39.73 (1.76) | ||
| WithOut-Prior | 100 Epochs | Runet | 21.33 (9.22) | 2.54 (0.56) | 94.77 (0.52) | 35.81 (1.18) |
| DUNet | 21.59 (10.89) | 2.55 (0.64) | 95.00 (0.54) | 35.82 (1.29) | ||
| AUNet | 24.32 (10.69) | 2.72 (0.64) | 94.45 (0.56) | 35.41 (1.23) | ||
| 1000 Epochs | RUNet | 19.63 (9.25) | 2.42 (0.55) | 95.34 (0.47) | 36.14 (1.21) | |
| DUNet | 20.39 (10.30) | 2.47 (0.63) | 95.50 (0.50) | 36.04 (1.33) | ||
| AUNet | 19.61 (9.38) | 2.42 (0.60) | 95.57 (0.47) | 36.15 (1.33) |
| Acquisition Scenario | Response Variable | UNets | Matrix Size | UNets × Matrix Size |
|---|---|---|---|---|
| With-Prior | log(MSE) | 2516 | 476 | 124 |
| log(MIE) | 1191 | 430 | 70 | |
| SSIM | 8239 | 5607 | 970 | |
| PSNR | 1585 | 254 | 88 | |
| WithOut-Prior | log(MSE) | 495 | 12,133 | 59 |
| log(MIE) | 295 | 7806 | 66 | |
| SSIM | 931 | 114,955 | 139 | |
| PSNR | 242 | 4674 | 81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, R.; Tsiamyrtzis, P.; Webb, A.G.; Seimenis, I.; Loukas, C.; Leiss, E.; Tsekos, N.V. A Deep Learning Approach to Upscaling “Low-Quality” MR Images: An In Silico Comparison Study Based on the UNet Framework. Appl. Sci. 2022, 12, 11758. https://doi.org/10.3390/app122211758
Sharma R, Tsiamyrtzis P, Webb AG, Seimenis I, Loukas C, Leiss E, Tsekos NV. A Deep Learning Approach to Upscaling “Low-Quality” MR Images: An In Silico Comparison Study Based on the UNet Framework. Applied Sciences. 2022; 12(22):11758. https://doi.org/10.3390/app122211758
Chicago/Turabian StyleSharma, Rishabh, Panagiotis Tsiamyrtzis, Andrew G. Webb, Ioannis Seimenis, Constantinos Loukas, Ernst Leiss, and Nikolaos V. Tsekos. 2022. "A Deep Learning Approach to Upscaling “Low-Quality” MR Images: An In Silico Comparison Study Based on the UNet Framework" Applied Sciences 12, no. 22: 11758. https://doi.org/10.3390/app122211758
APA StyleSharma, R., Tsiamyrtzis, P., Webb, A. G., Seimenis, I., Loukas, C., Leiss, E., & Tsekos, N. V. (2022). A Deep Learning Approach to Upscaling “Low-Quality” MR Images: An In Silico Comparison Study Based on the UNet Framework. Applied Sciences, 12(22), 11758. https://doi.org/10.3390/app122211758






