LASSO-Cox Modeling of Survival Using High-Resolution CT-Based Radiomic Features in a Cohort of COVID-19 Patients and Its Generalizability to Standard Image Reconstruction
Abstract
:1. Introduction
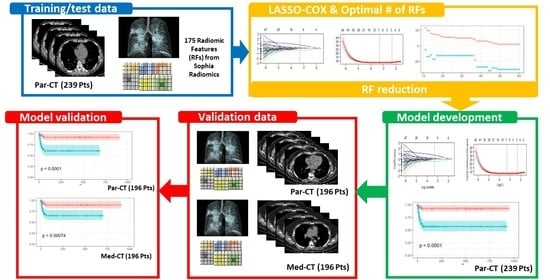
2. Materials and Methods
2.1. Patient Cohort
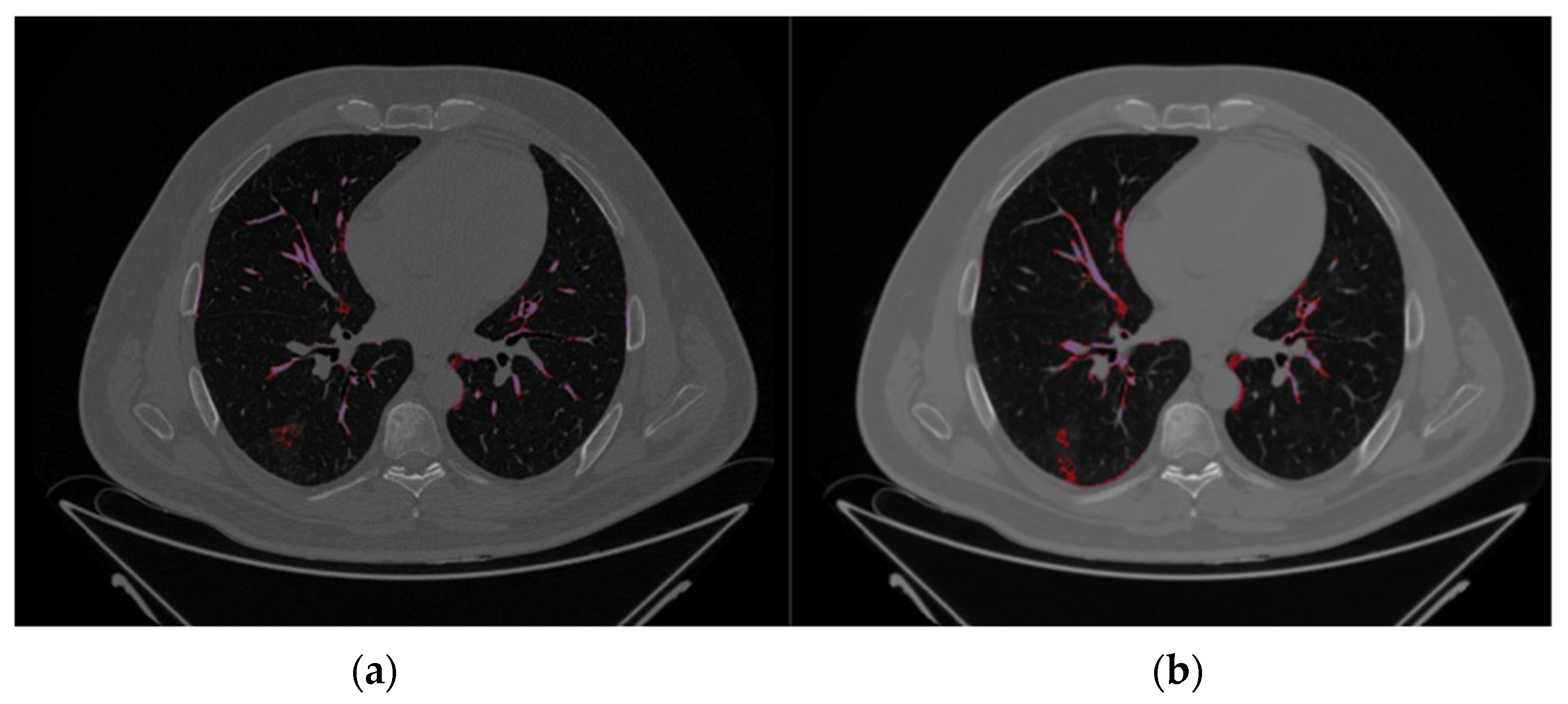
2.2. Image Segmentation and Feature Extraction
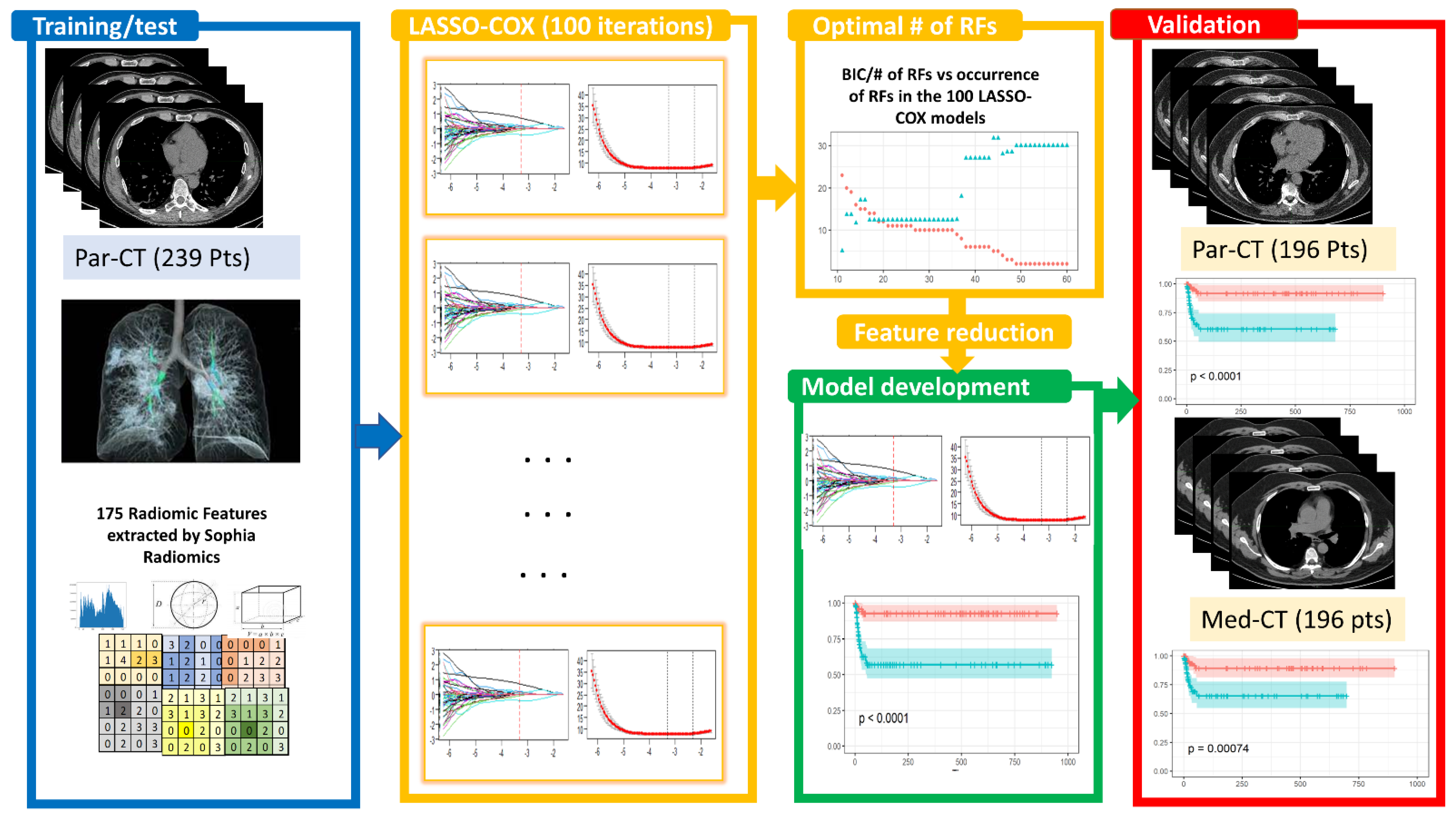
2.3. Predictive Model Building
2.3.1. Feature Selection
2.3.2. Bayesian Information Criterion (BIC)
2.3.3. ICC Analysis
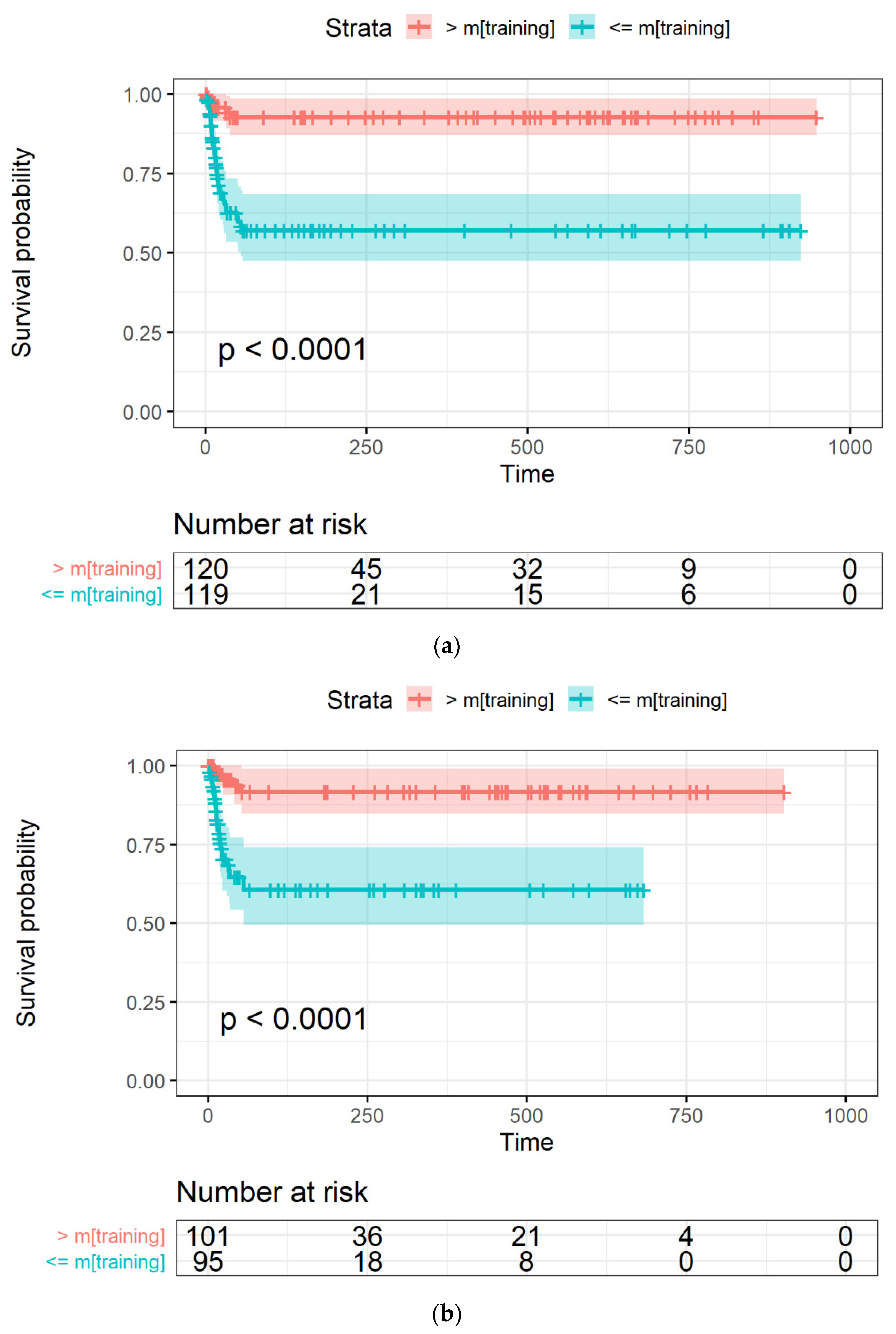
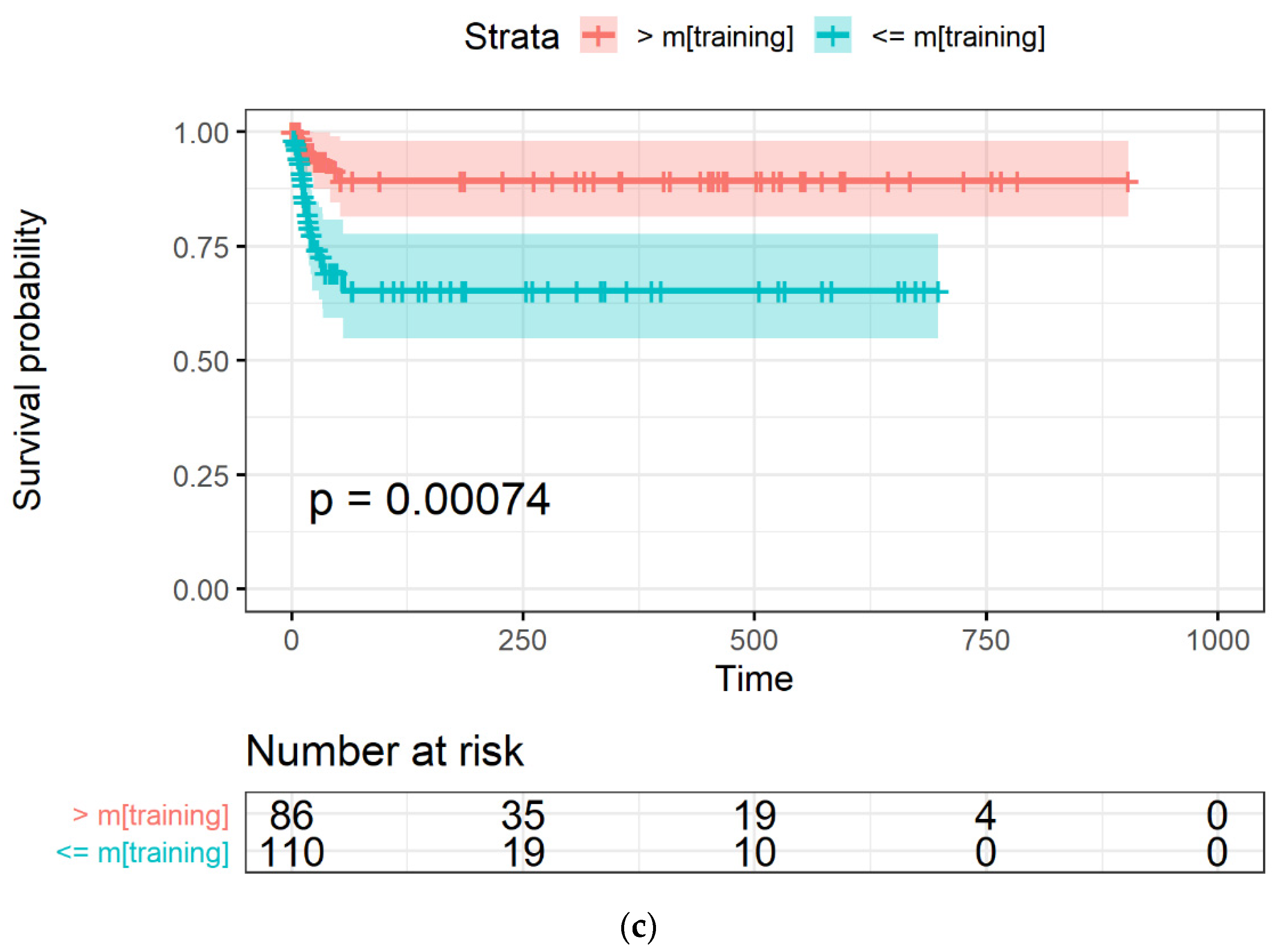
2.3.4. Model Evaluation through Survival Curves
2.3.5. Area under Curve (AUC)
3. Results
3.1. Patient Cohort
3.2. Image Reconstruction and VOI Delineation Results
3.3. LASSO-Cox for Feature Selection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grassi, R.; Belfiore, M.P.; Montanelli, A.; Patelli, G.; Urraro, F.; Giacobbe, G.; Fusco, R.; Granata, V.; Petrillo, A.; Sacco, P.; et al. COVID-19 pneumonia: Computer-aided quantification of healthy lung parenchyma, emphysema, ground glass and consolidation on chest computed tomography (CT). La Radiol. Med. 2020, 126, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Neri, E.; Miele, V.; Coppola, F.; Grassi, R. Use of CT and artificial intelligence in suspected or COVID-19 positive patients: Statement of the Italian Society of Medical and Interventional Radiology. La Radiol. Med. 2020, 125, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Manco, L.; Maffei, N.; Strolin, S.; Vichi, S.; Bottazzi, L.; Strigari, L. Basic of machine learning and deep learning in imaging for medical physicists. Phys. Medica 2021, 83, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Stancanello, J.; El Naqa, I. Beyond imaging: The promise of radiomics. Phys. Medica 2017, 38, 122–139. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging-“how-to” guide and critical reflection. Insights Imaging. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Lee, G.; Lee, H.Y.; Park, H.; Schiebler, M.L.; van Beek, E.J.; Ohno, Y.; Seo, J.B.; Leung, A. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur. J. Radiol. 2017, 86, 297–307. [Google Scholar] [CrossRef]
- Varghese, B.A.; Shin, H.; Desai, B.; Gholamrezanezhad, A.; Lei, X.; Perkins, M.; Oberai, A.; Nanda, N.; Cen, S.; Duddalwar, V. Predicting clinical outcomes in COVID-19 using radiomics on chest radiographs. Br. J. Radiol. 2021, 94, 20210221. [Google Scholar] [CrossRef]
- Spagnoli, L.; Morrone, M.F.; Giampieri, E.; Paolani, G.; Santoro, M.; Curti, N.; Coppola, F.; Ciccarese, F.; Vara, G.; Brandi, N.; et al. Outcome Prediction for SARS-CoV-2 Patients Using Machine Learning Modeling of Clinical, Radiological, and Radiomic Features Derived from Chest CT Images. Appl. Sci. 2022, 12, 4493. [Google Scholar] [CrossRef]
- Ke, Z.; Li, L.; Wang, L.; Liu, H.; Lu, X.; Zeng, F.; Zha, Y. Radiomics analysis enables fatal outcome prediction for hospitalized patients with coronavirus disease 2019 (COVID-19). Acta Radiol. 2021, 63, 319–327. [Google Scholar] [CrossRef]
- Xiao, F.; Sun, R.; Sun, W.; Xu, D.; Lan, L.; Li, H.; Xu, H. Radiomics analysis of chest CT to predict the overall survival for the severe patients of COVID-19 pneumonia. Phys. Med. Biol. 2021, 66, 105008. [Google Scholar] [CrossRef]
- Kao, Y.-S.; Lin, K.-T. A Meta-Analysis of Computerized Tomography-Based Radiomics for the Diagnosis of COVID-19 and Viral Pneumonia. Diagnostics 2021, 11, 991. [Google Scholar] [CrossRef] [PubMed]
- Cattabriga, A.; Cocozza, M.A.; Vara, G.; Coppola, F.; Golfieri, R. Lung CT Segmentation to Identify Consolidations and Ground Glass Areas for Quantitative Assesment of SARS-CoV Pneumonia. JoVE (J. Vis. Exp.) 2020, 166, e61737. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Mithun, S.; Jaiswar, V.; Sherkhane, U.B.; Purandare, N.C.; Prabhash, K.; Rangarajan, V.; Dekker, A.; Wee, L.; Traverso, A. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci. Rep. 2021, 11, 2055. [Google Scholar] [CrossRef]
- Bettinelli, A.; Marturano, F.; Avanzo, M.; Loi, E.; Menghi, E.; Mezzenga, E.; Pirrone, G.; Sarnelli, A.; Strigari, L.; Strolin, S.; et al. A Novel Benchmarking Approach to Assess the Agreement among Radiomic Tools. Radiology 2022, 303, 533–541. [Google Scholar] [CrossRef]
- Gideon, S. Estimating the Dimension of a Model. Ann. Stat. 1978, 6, 461–464. [Google Scholar]
- Kass, R.E.E. Bayes Factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Wang, S.; Li, L.; Qian, W.; Hu, Y.; Li, L.; Zhou, X.; Ma, H.; Li, H.; Wang, M.; et al. Radiomics Analysis of Computed Tomography helps predict poor prognostic outcome in COVID-19. Theranostics 2020, 10, 7231–7244. [Google Scholar] [CrossRef]
- Shiri, I.; Sorouri, M.; Geramifar, P.; Nazari, M.; Abdollahi, M.; Salimi, Y.; Khosravi, B.; Askari, D.; Aghaghazvini, L.; Hajianfar, G.; et al. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients. Comput. Biol. Med. 2021, 132, 104304. [Google Scholar] [CrossRef]
- Li, C.; Dong, D.; Li, L.; Gong, W.; Li, X.; Bai, Y.; Wang, M.; Hu, Z.; Zha, Y.; Tian, J. Classification of Severe and Critical Covid-19 Using Deep Learning and Radiomics. IEEE J. Biomed. Health Inform. 2020, 24, 3585–3594. [Google Scholar] [CrossRef]
- Ferreira Junior, J.R.; Cardona Cardenas, D.A.; Moreno, R.A.; de Sá Rebelo, M.F.; Krieger, J.E.; Gutierrez, M.A. Novel Chest Radiographic Biomarkers for COVID-19 Using Radiomic Features Associated with Diagnostics and Outcomes. J. Digit. Imaging 2021, 34, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Yang, P.; Xie, Y.; Woodruff, H.C.; Rao, X.; Guiot, J.; Frix, A.-N.; Louis, R.; Moutschen, M.; Li, J.; et al. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: An international multicentre study. Eur. Respir. J. 2020, 56, 2001104. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Strolin, S.; Paolani, G.; Della Gala, G.; Bartoloni, A.; Giacometti, C.; Ammendolia, I.; Morganti, A.G.; Strigari, L. Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond. Appl. Sci. 2022, 12, 3223. [Google Scholar] [CrossRef]


| Feature Name | Cox-Coef. | Hazard Ratio | ICC [Range] | p-Value (ICC) |
|---|---|---|---|---|
| Area density enclosing ellipsoid | −0.30425 | 0.7377 | 0.87 [0.82, 0.90] | <0.0001 |
| Cluster shade | −0.32886 | 0.7197 | 0.83 [0.79, 0.87] | <0.0001 |
| Intensity histogram quartile coefficient of dispersion | 0.65043 | 1.9163 | 0.85 [0.81, 0.89] | <0.0001 |
| Min value | −0.12927 | 0.8787 | 0.17 [0.03, 0.30] | 0.0081 |
| Normalized zone distance non-uniformity | 0.32209 | 1.3800 | 0.52 [0.41, 0.61] | 0.1800 |
| Ref. | Patients [Train/Test/Validation] | Follow-up Length (Days) | Segmentation Type (Tool) | Predictors | Modelling | Outcome | Performance |
|---|---|---|---|---|---|---|---|
| [7] | 167 [NS/NS/NA] | NS | Manual (ITK-Snap) 2D extraction on CXR | RFs from lesion only in CXRs | Adaboost | Death | AUC = 0.71 |
| [9] | 96 [66/30/NA] | 62 | Semi-automatic (LungSegmentation Kit GE) | Demographics, Laboratory tests and RFs | Lasso-Cox Proportional Hazard | OS, death | AUCtest = 0.871 |
| [18] | EarlyCT 317 [212/105/NA] LateCT 175 [139/36/NA] | ~30 | Automatic DenseNet121-FPN | Demographics, Comorbidities, RFs | Lasso-Cox Proportional Hazard | Poor outcome | AUCtest,early = 0.816 AUCtest,late = 0.976 |
| [19] | 152 [106/46/NA] | NS | Manual by radiologist with 3d Slicer | Laboratory tests, radiological score, RFs | XGBoost | Death | AUCcombined = 0.95 |
| This study | 435 [167/72/196] | 948 | Semi-automatic (Sophia Radiomics DDM) | RFs | Lasso-Cox Proportional Hazard | OS, death | AUCPar-CT = 0.764 AUCMed-CT = 0.748 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paolani, G.; Spagnoli, L.; Morrone, M.F.; Santoro, M.; Coppola, F.; Strolin, S.; Golfieri, R.; Strigari, L. LASSO-Cox Modeling of Survival Using High-Resolution CT-Based Radiomic Features in a Cohort of COVID-19 Patients and Its Generalizability to Standard Image Reconstruction. Appl. Sci. 2022, 12, 12065. https://doi.org/10.3390/app122312065
Paolani G, Spagnoli L, Morrone MF, Santoro M, Coppola F, Strolin S, Golfieri R, Strigari L. LASSO-Cox Modeling of Survival Using High-Resolution CT-Based Radiomic Features in a Cohort of COVID-19 Patients and Its Generalizability to Standard Image Reconstruction. Applied Sciences. 2022; 12(23):12065. https://doi.org/10.3390/app122312065
Chicago/Turabian StylePaolani, Giulia, Lorenzo Spagnoli, Maria Francesca Morrone, Miriam Santoro, Francesca Coppola, Silvia Strolin, Rita Golfieri, and Lidia Strigari. 2022. "LASSO-Cox Modeling of Survival Using High-Resolution CT-Based Radiomic Features in a Cohort of COVID-19 Patients and Its Generalizability to Standard Image Reconstruction" Applied Sciences 12, no. 23: 12065. https://doi.org/10.3390/app122312065
APA StylePaolani, G., Spagnoli, L., Morrone, M. F., Santoro, M., Coppola, F., Strolin, S., Golfieri, R., & Strigari, L. (2022). LASSO-Cox Modeling of Survival Using High-Resolution CT-Based Radiomic Features in a Cohort of COVID-19 Patients and Its Generalizability to Standard Image Reconstruction. Applied Sciences, 12(23), 12065. https://doi.org/10.3390/app122312065