Complexation of Amino Acids with Cadmium and Their Application for Cadmium-Contaminated Soil Remediation
Abstract
:1. Introduction
2. Experimental and Computational Section
2.1. Calculation Methods
2.2. Experimental Section
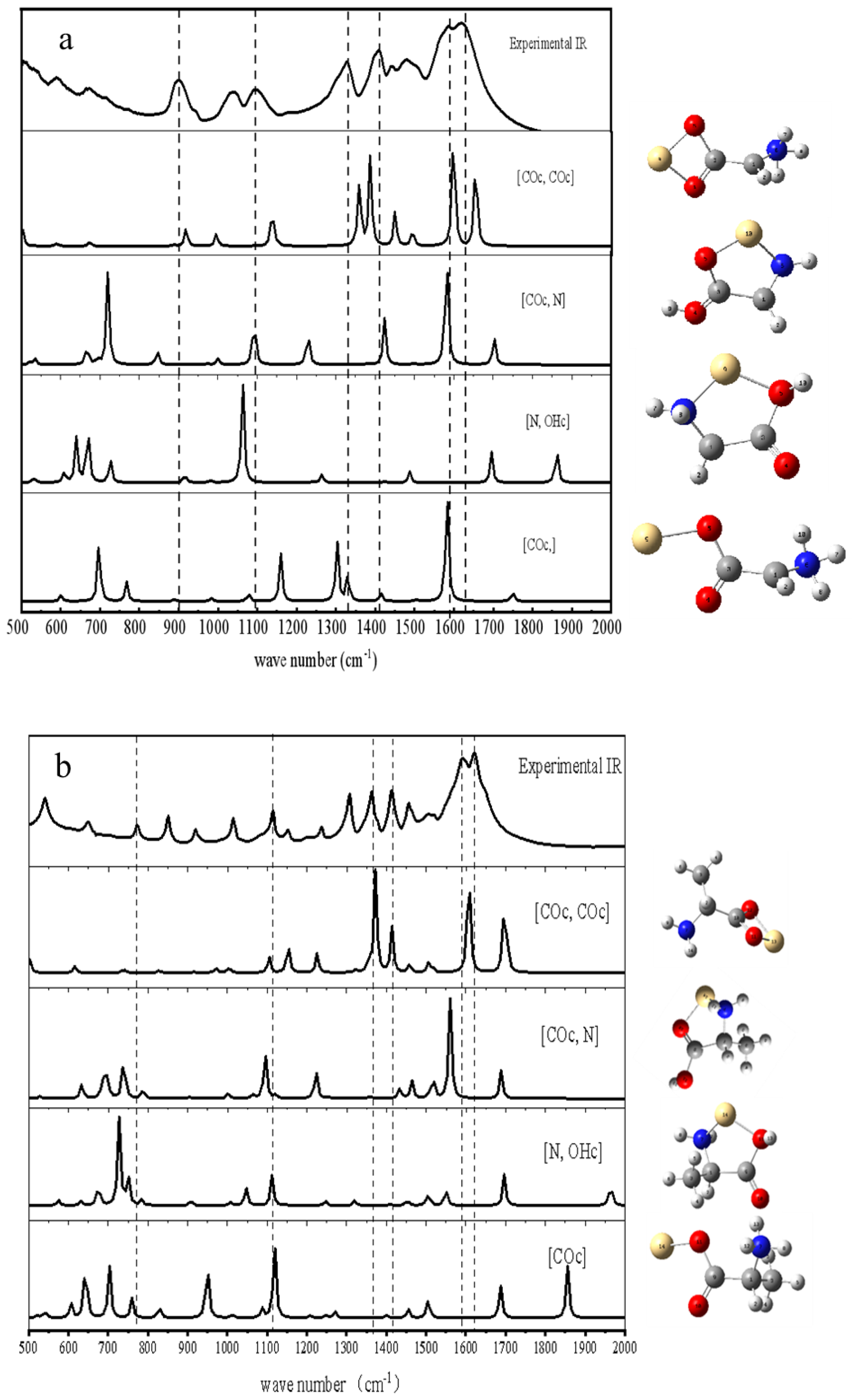
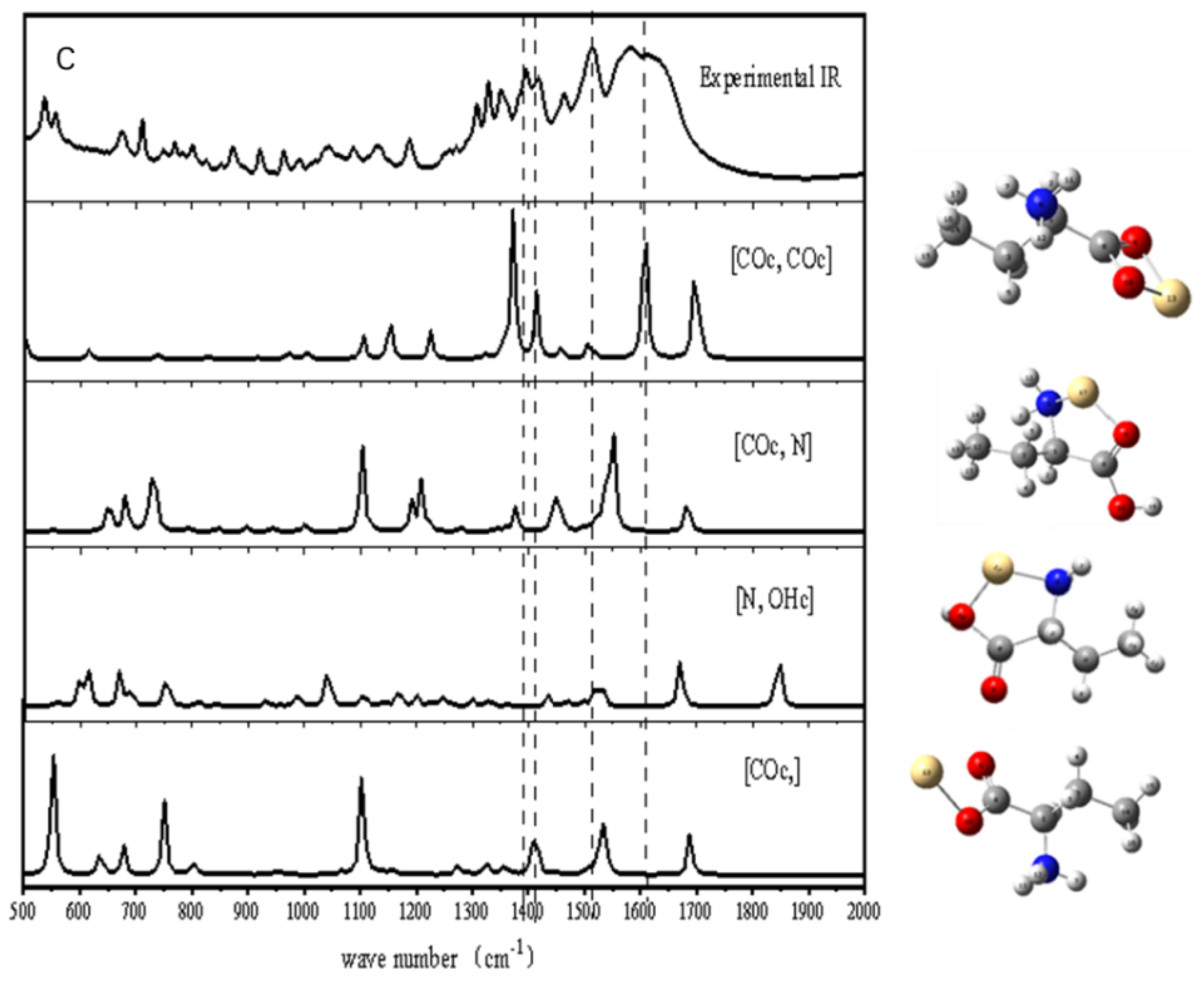
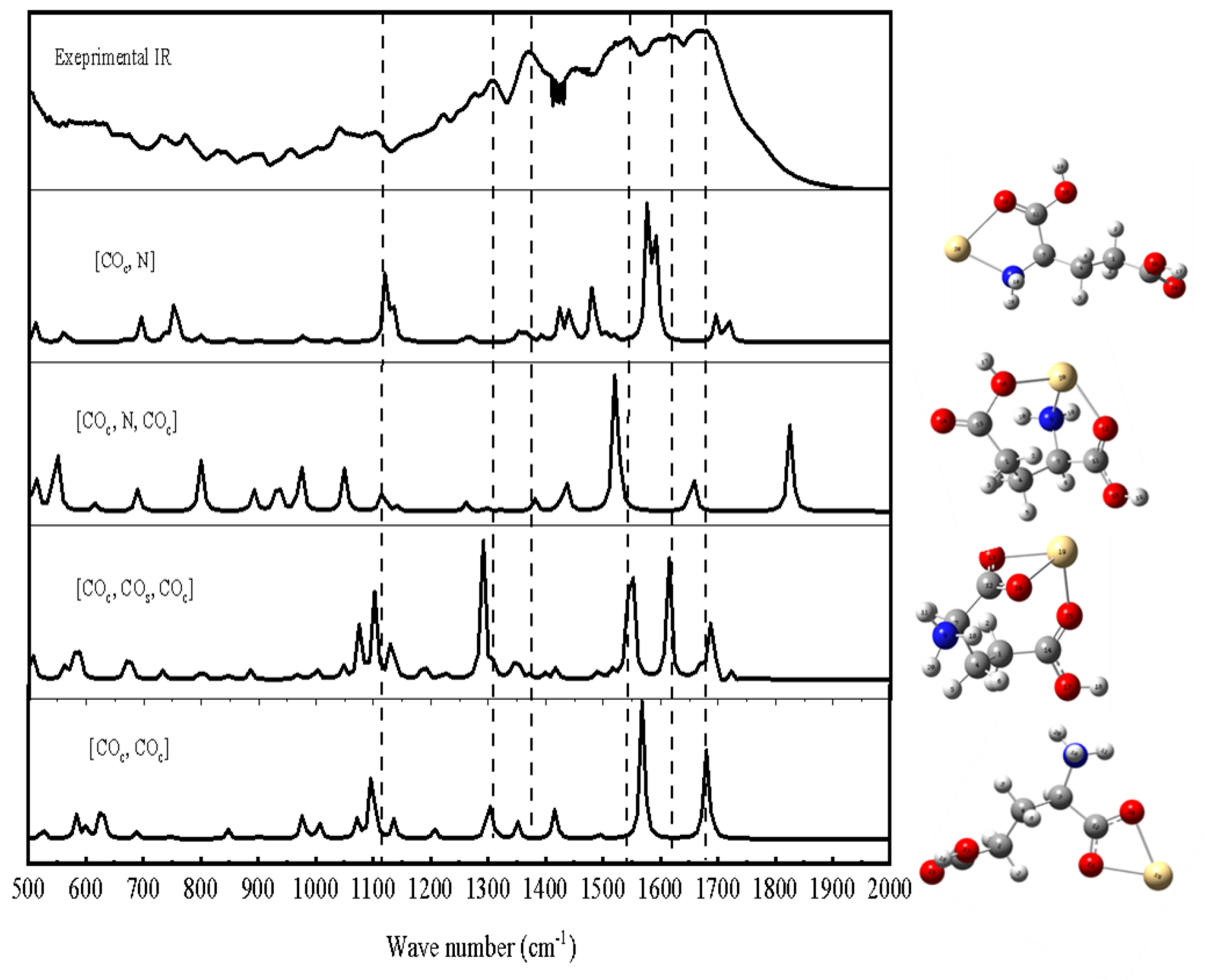
2.2.1. FTIR Detection of the Complexes
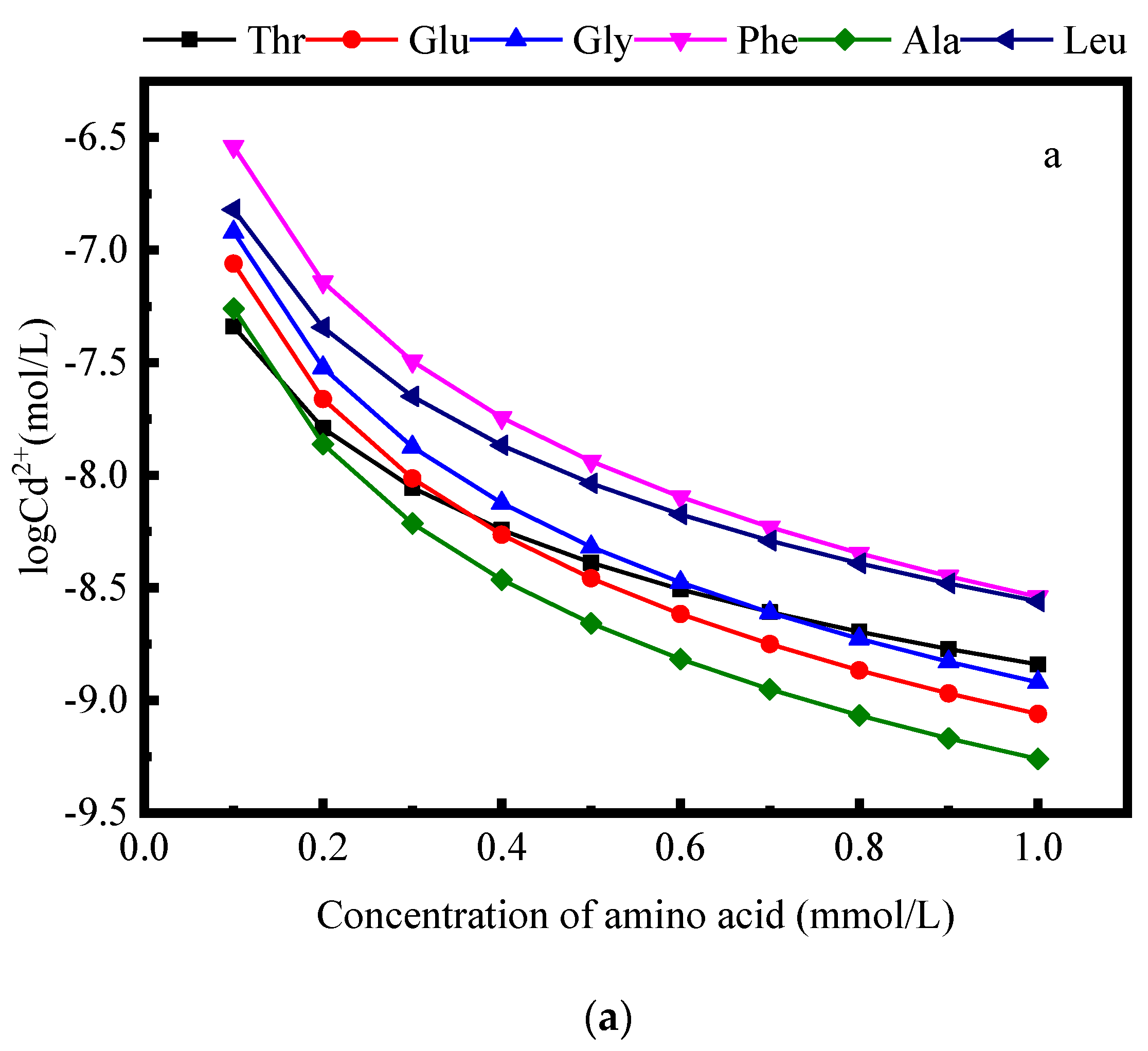
2.2.2. Effect of Amino Concentrations on Complexation
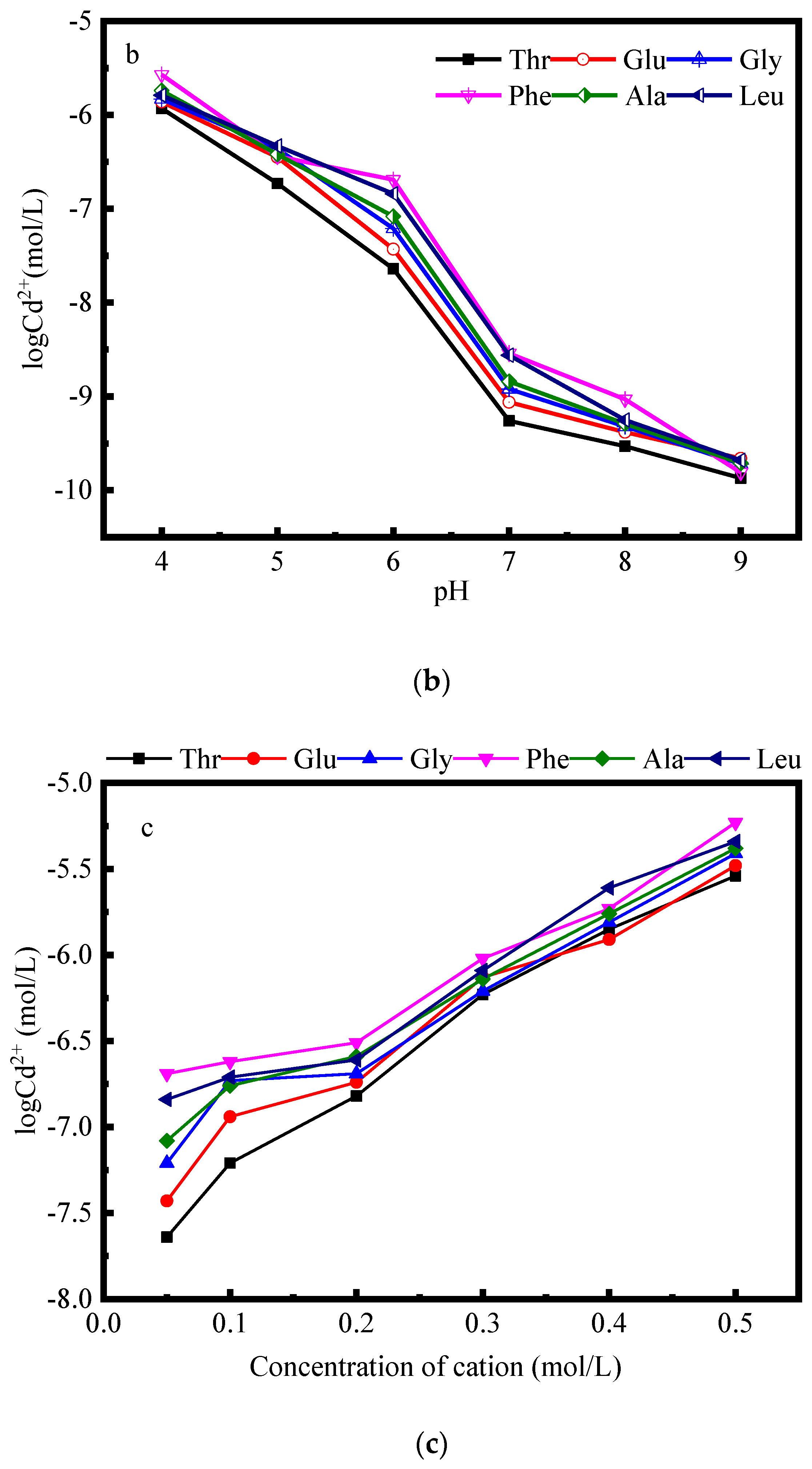
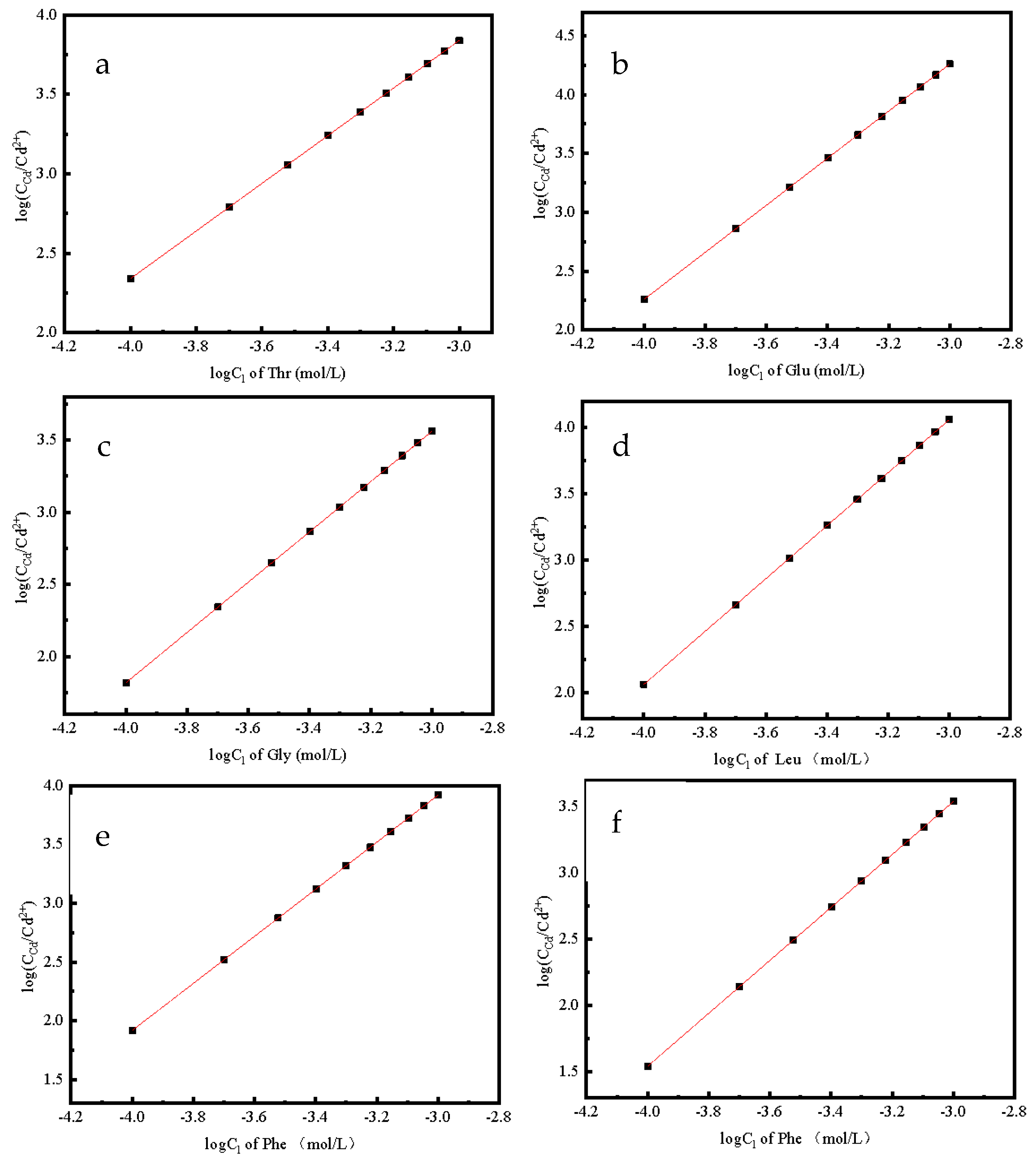
2.2.3. Effect of Cation Concentration on Complexation
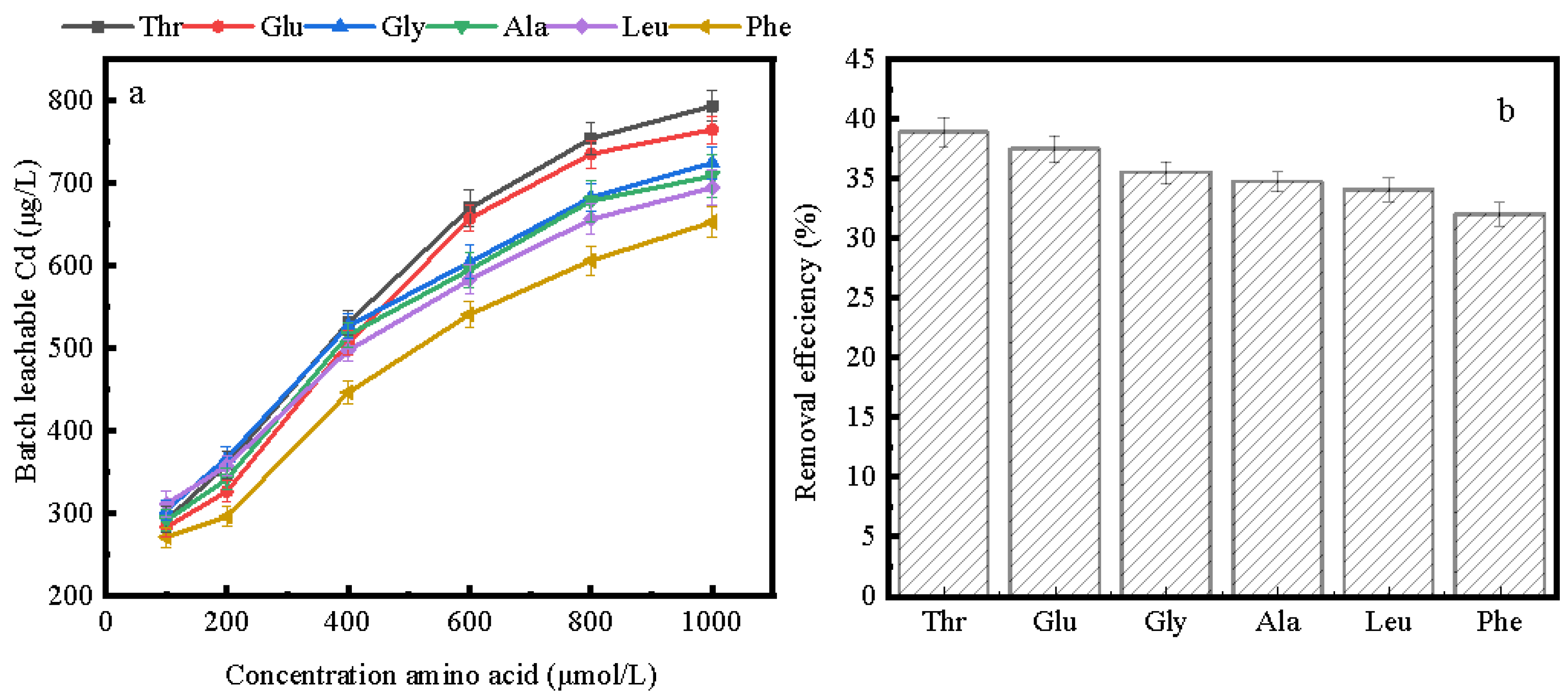
2.2.4. Batch Soil Experiments
3. Results and Discussion
3.1. The Complexes of Cadmium and Amino Acids
3.1.1. The Complexes of Gly, Leu, and Ala with Cd2+
3.1.2. The Complexes of Glu with Cd2+
3.1.3. The Complexes of Thr with Cd2+
3.1.4. The Complexes of Phe with Cd2+
3.1.5. The Theoretical Calculation of the Complex Energy
3.2. Pertinent Factors Affecting the Complex of Amino Acids and Cd
3.3. Removal of Cd from Contaminated Soil
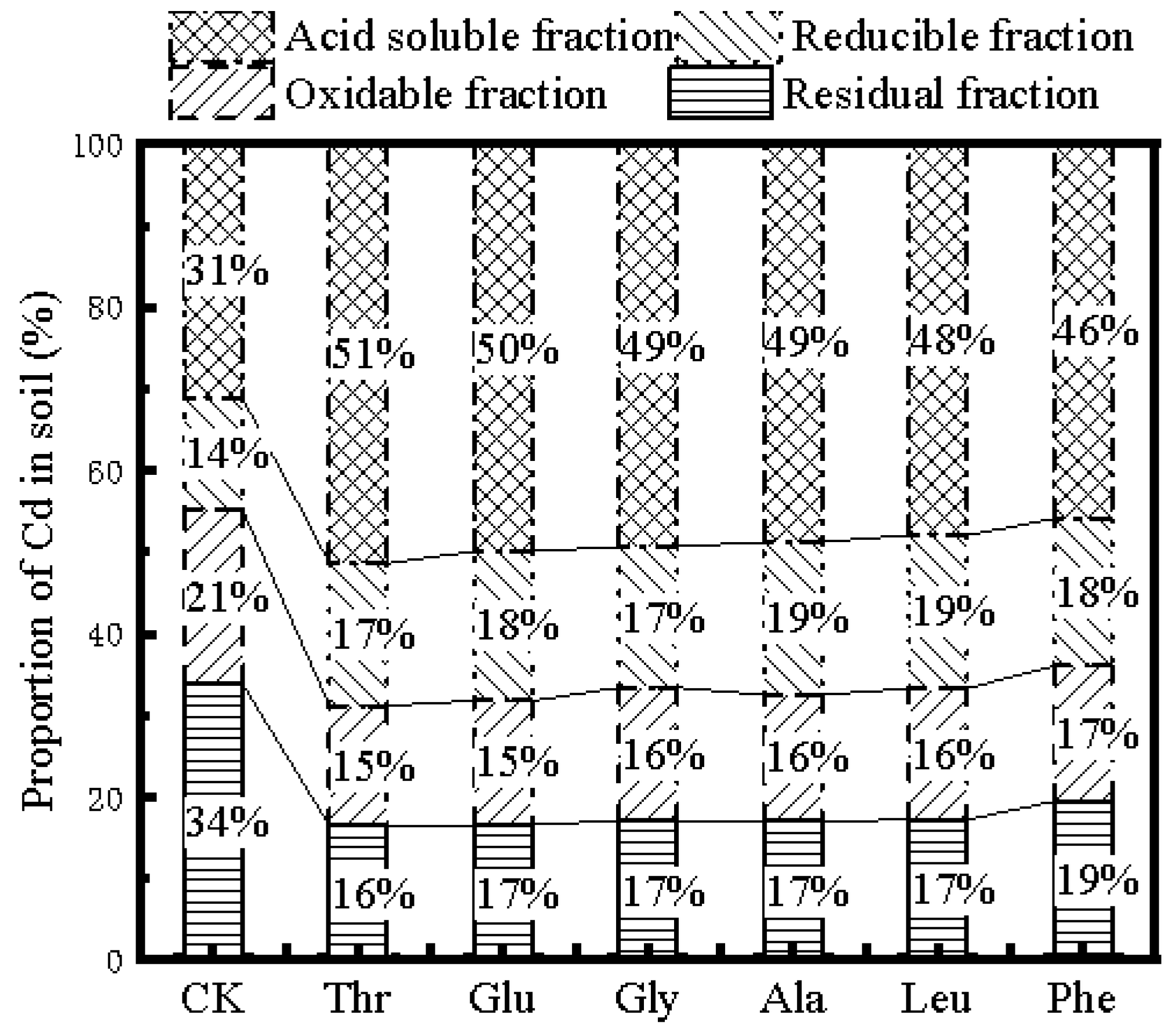
3.4. Distribution of Cd in the Soil Fraction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Y.; Ying, H.; Liu, Y. Heavy metal accumulation in iron plaque and growth of rice plants upon exposure to single and combined contamination by copper, cadmium and lead. Acta Ecol. Sin. 2009, 29, 320–326. [Google Scholar] [CrossRef]
- Zhao, F.; Liu, Y.; Hammouda, S.B.; Doshi, B.; Wang, S. MIL-101(Fe)/g-C3N4 for enhanced visible-light-driven photocatalysis toward simultaneous reduction of Cr(VI) and oxidation of bisphenol A in aqueous media. Appl. Catal. B Environ. 2020, 272, 119033. [Google Scholar] [CrossRef]
- Zhao, F.; Repo, E.; Yin, D.; Sillanp, M.E.T. Adsorption of Cd(II) and Pb(II) by a novel EGTA-modified chitosan material: Kinetics and isotherms. J. Colloid Interface 2013, 409, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, S.; Priya, T.; Khoo, K.S.; Hoang, T.K.; Ng, H.-S.; Munawaroh, H.S.H.; Karaman, C.; Orooji, Y.; Show, P.L. A critical review on various remediation approaches for heavy metal contaminants removal from contaminated soils. Chemosphere 2021, 287, 132369. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurti, G.S.R.; Megharaj, M.; Naidu, R. Bioavailability of cadmium−organic complexes to soil AlgaAn exception to the free ion model. J. Agric. Food Chem. 2004, 52, 3894–3899. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Z.; Li, A.; Cui, C. Enhanced electrokinetic remediation of heavy metals contaminated soil by biodegradable complexing agents. Environ. Pollut. 2021, 283, 117111. [Google Scholar] [CrossRef] [PubMed]
- Dolev, N.; Katz, Z.; Ludmer, Z.; Ullmann, A.; Goikhman, R. Natural amino acids as potential chelators for soil remediation. Environ. Res. 2020, 183, 109140. [Google Scholar] [CrossRef]
- Borggaard, O.K.; Holm, P.E.; Strobel, B.W. Potential of dissolved organic matter (DOM) to extract As, Cd, Co, Cr, Cu, Ni, Pb and Zn from polluted soils: A review. Geoderma 2019, 343, 235–246. [Google Scholar] [CrossRef]
- Hosseini, S.S.; Lakzian, A.; Halajnia, A.; Razavi, B.S. Optimization of EDTA and citric acid for risk assessment in the remediation of lead contaminated soil. Rhizosphere 2020, 17, 100277. [Google Scholar] [CrossRef]
- Yu, F.; Li, Y.; Li, F.; Li, C.; Liu, K. The effects of EDTA on plant growth and manganese (Mn) accumulation in Polygonum pubescens Blume cultured in unexplored soil, mining soil and tailing soil from the Pingle Mn mine, China. Ecotox. Environ. Safe 2019, 173, 235–242. [Google Scholar] [CrossRef]
- Vranova, V.; Rejsek, K.; Skene, K.R.; Janous, D.; Formanek, P. Methods of collection of plant root exudates in relation to plant metabolism and purpose: A review. J. Plant Nutr. Soil Sci. 2013, 176, 175–199. [Google Scholar] [CrossRef]
- Sun, L.; Cao, X.; Tan, C.; Deng, Y.; Bai, J. Analysis of the effect of cadmium stress on root exudates of Sedum plumbizincicola based on metabolomics. Ecotox. Environ. Safe 2020, 205, 111152. [Google Scholar] [CrossRef]
- Tao, Q.; Zhao, J.; Li, J.; Liu, Y.; Wang, C. Unique root exudate tartaric acid enhanced cadmium mobilization and uptake in Cd-hyperaccumulator Sedum alfredii. J. Hazard. Mater. 2019, 383, 121177. [Google Scholar] [CrossRef]
- Ubeynarayana, N.; Jeyakumar, P.; Bishop, P.; Pereira, R.C.; Anderson, C. Effect of soil cadmium on root organic acid secretion by forage crops. Environ. Pollut. 2021, 268, 115839. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shao, M.; Huang, M.; Sang, W.; Zheng, S.; Jiang, N.; Gao, Y. Enhanced remediation of heavy metals contaminated soils with EK-PRB using β-CD/hydrothermal biochar by waste cotton as reactive barrier. Chemosphere 2022, 286, 131470. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, S.; Khoshgoftarmanesh, A.H.; Afyuni, M.; Hadadzadeh, H.; Schulin, R. Zinc–amino acid complexes are more stable than free amino acids in saline and washed soils. Soil Biol. Biochem. 2013, 63, 73–79. [Google Scholar] [CrossRef]
- Rogiers, S.Y.; Holzapfel, B.P.; Scollary, G.R.; Canny, M.J.; Gourieroux, A.M. The amino acid distribution in rachis xylem sap and phloem exudate of Vitis vinifera ’Cabernet Sauvignon’ bunches. Plant Physiol. Biochem. 2016, 105, 45–54. [Google Scholar]
- Hua Changer, Z.S.; Wang, Z.; Wang, X.; Yang, J. Effect of nitrogen and phosphorus on the amino acids in root exudates and grains of rice during grain filling. Acta Agron. Sin. 2009, 34, 612–618. [Google Scholar]
- Jones, D.L.; Shannon, D.; Junvee-Fortune, T.; Farrar, J.F. Plant capture of free amino acids is maximized under high soil amino acid concentrations. Soil Biol. Biochem. 2005, 37, 179–181. [Google Scholar] [CrossRef]
- Karczewska, A.; Milko, K. Effects of chelating agents on copper, lead and zinc solubility in polluted soils and tailings produced by copper industry. Ecol. Chem. Eng. A 2010, 17, 395–403. [Google Scholar]
- Bell, J.; Samb, I.; Toullec, P.Y.; Michelet, V.; Leray, I. Synthesis and complexing properties of molecular probes linked with fluorescent phosphane oxide derivatives. J. Photochem. Photobiol. A Chem. 2016, 318, 25–32. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Metal-binding affinity and selectivity of nonstandard natural amino acid residues from DFT/CDM calculations. J. Phys. Chem. B 2009, 113, 11754–11764. [Google Scholar] [CrossRef]
- Hoyau, S.; Pélicier, J.-P.; Rogalewicz, F.O.; Hoppilliard, Y.; Ohanessian, G. Complexation of glycine by atomic metal cations in the gas phase. Eur. J. Mass Spectrom. 2001, 7, 303–311. [Google Scholar] [CrossRef]
- Nie, B.; Li, R.; Wu, Y.; Yuan, X.; Zhang, W. Theoretical calculation of the thermodynamic properties of 20 amino acid ionic liquids. J. Phys. Chem. B 2018, 122, 10548–10557. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, R.C.; Hopkinson, A.C.; Oomens, J.; Siu, C.K.; Siu, K.W.M.; Steill, J.D.; Verkerk, U.H.; Zhao, J. Conformation Switching in gas-phase complexes of histidine with alkaline earth ions. J. Phys. Chem. B 2009, 113, 10403–10408. [Google Scholar] [CrossRef]
- Frisch, Æ.; Frisch, M.J. Gaussian 98 User’s Reference; Gaussian: Pittsburgh, PA, USA, 1999. [Google Scholar]
- Boles, G.C.; Coates, R.A.; Berden, G.; Oomens, J.; Armentrout, P.B. Experimental and theoretical investigations of infrared multiple photon dissociation spectra of asparagine complexes with Zn2+ and Cd2+ and their deamidation processes. J. Phys. Chem. B 2016, 120, 12486–12500. [Google Scholar] [CrossRef]
- Fajković, H.; Rončević, S.; Nemet, I.; Prohić, E.; Leontićvazdar, D. Fractionation of metals by sequential extraction procedures (BCR and Tessier) in soil exposed to fire of wide temperature range. Geophys. Res. Abstr. 2017, 19, 424. [Google Scholar]
- Jockusch, R.A.; Price, W.D.; Williams, E.R. Structure of cationized arginine (Arg·M+, M = H, Li, Na, K, Rb, and Cs) in the gas phase: Further evidence for zwitterionic arginine. J. Phys. Chem. A 1999, 103, 9266–9274. [Google Scholar] [CrossRef] [Green Version]
- Armentrout, P.B.; Yang, B.; Rodgers, M.T. Metal cation dependence of interactions with amino acids: Bond energies of Rb+ and Cs+ to Met, Phe, Tyr, and Trp. J. Phys. Chem. B 2013, 117, 3771–3781. [Google Scholar] [CrossRef]
- Phillips, F.C. Manual of qualitative chemical analysis. J. Am. Chem. Soc. 2002, 20, 142–144. [Google Scholar] [CrossRef] [Green Version]
- Bowman, V.N.; Heaton, A.L.; Armentrout, P.B. Metal cation dependence of interactions with amino acids: Bond energies of Rb+ to Gly, Ser, Thr, and Pro. J. Phys. Chem. B 2010, 114, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.T.; Prell, J.S.; Steill, J.D.; Oomens, J.; Williams, E.R. Interactions of mono-and divalent metal ions with aspartic and glutamic acid investigated with IR photodissociation spectroscopy and theory. J. Phys. Chem. A 2008, 112, 10823–10830. [Google Scholar] [CrossRef] [PubMed]
- Remelli, M.; Nurchi, V.M.; Lachowicz, J.I.; Medici, S.; Zoroddu, M.A.; Peana, M. Competition between Cd(II) and other divalent transition metal ions during complex formation with amino acids, peptides, and chelating agents. Coord. Chem. Rev. 2016, 327, 55–69. [Google Scholar] [CrossRef]
- Rojo, H.; Gaona, X.; Rabung, T.; Polly, R.; Altmaier, M. Complexation of Nd(III)/Cm(III) with gluconate in alkaline NaCl and CaCl2 solutions: Solubility, TRLFS and DFT studies. Appl. Geochem. 2020, 126, 104864. [Google Scholar] [CrossRef]
- Beheshti, A.; Clegg, W.; Dale, S.; Hyvadi, R. Synthesis, crystal structures, and spectroscopic characterization of the neutral monomeric tetrahedral [M(Diap)2(OAc)2]center dot H2O complexes (M = Zn,Cd; Diap = 1,3-diazepane-2-thione; OAc = acetate) with N-H center dot center dot center dot O intermolecular hydrogen bonding interactions. Inorg. Chim. Acta 2007, 360, 2967–2972. [Google Scholar]
- Zhang, S.; Yu, T.; Sun, M.; Yu, H.; Zhang, Z.; Wang, S.; Jiang, H. Highly sensitive and selective fluorescence detection of copper (II) ion based on multi-ligand metal chelation. Talanta 2014, 126, 185–190. [Google Scholar] [CrossRef]
- Kumarasamy, K.; Devendhiran, T.; Lin, M.C.; Chiu, C.W. Synthesis and physical property studies of cyclometalated Pt(II) and Pd(II) complexes with tridentate ligands containing pyrazole and pyridine groups. Polyhedron 2020, 191, 114799. [Google Scholar] [CrossRef]
- Keskin, S.G.; Stanley, J.M.; Cowley, A.H. Synthesis, characterization and theoretical investigations of molybdenum carbonyl complexes with phosphorus/nitrogen/phosphorus ligand as bidentate and tridentate modes. Polyhedron 2017, 138, 206–217. [Google Scholar] [CrossRef]
- Fneich, B.N.; Das, A.; Kirschbaum, K.; Mason, M.R. Bidentate and tridentate coordination modes of bis(3-methylindolyl)-2-pyridylmethane in complexes of aluminum and gallium: Structural characterization of bridging N-indolide in a dialuminum complex-ScienceDirect. J. Organomet. Chem. 2018, 872, 12–23. [Google Scholar] [CrossRef]
- Aly, A.A.; Bräse, S.; Weis, P. Tridentate and bidentate copper complexes of [2.2]paracyclophanyl-substituted thiosemicarbazones, thiocarbazones, hydrazones and thioureas. J. Mol. Struct. 2018, 1178, 311–326. [Google Scholar] [CrossRef]
- Cao, J.S.; Wang, C.; Fang, F.; Lin, J.X. Removal of heavy metal Cu(II) in simulated aquaculture wastewater by modified palygorskite. Environ. Pollut. 2016, 219, 924–931. [Google Scholar] [CrossRef]
- Vacek, J.; Zatloukalová, M.; Vrba, J.; Vleeschouwer, F.D.; Proft, F.D.; Obluková, M.; Sokolová, R.; Pospíšil, J. Diferulate: A highly effective electron donor. J. Electroanal. Chem. 2020, 869, 113950. [Google Scholar] [CrossRef]
- Yan, B.; Niu, C.H.; Wang, J. Kinetics, electron-donor-acceptor interactions, and site energy distribution analyses of norfloxacin adsorption on pretreated barley straw. Chem. Eng. J. 2017, 330, 1211–1221. [Google Scholar] [CrossRef]
- Xie, X.; Yan, L.; Li, J.; Guan, L.; Chi, Z. Cadmium isotope fractionation during Cd-calcite coprecipitation: Insight from batch experiment. Sci. Total Environ. 2020, 760, 143330. [Google Scholar] [CrossRef] [PubMed]
- El-Haty, M.T.; Amrallah, A.H.; Mahmoud, R.A.; Ibrahim, A.A. pH-metric studies on ternary metal complexes of some amino acids and benzimidazole. Talanta 1995, 42, 1711. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, Z.; Wei, Z.; Spinney, R.; Sillanpää, M.; Tang, J.; Tam, M.; Xiao, R. Polyethylenimine-modified chitosan materials for the recovery of La (III) from leachates of bauxite residue. Chem. Eng. J. 2020, 388, 124307. [Google Scholar] [CrossRef]
- Murphy, J.M.; Gaertner, A.A.; Williams, T.; McMillen, C.D.; Powell, B.A.; Brumaghim, J.L. Stability constant determination of sulfur and selenium amino acids with Cu(II) and Fe(II). J. Inorg. Biochem. 2019, 195, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ondoo, E.; Bacchetta, G.; Lallena, A.M.; Navarro, F.B.; Jiménez, M. Use of BCR sequential extraction procedures for soils and plant metal transfer predictions in contaminated mine tailings in Sardinia. J. Geochem. Explor. 2017, 172, 133–141. [Google Scholar] [CrossRef]
- Xing, L.; Wen, J.; Yan, C.; Wang, Q.; Xue, Z. Improving the microenvironment of Cd-contaminated river sediments through humic substances washing and zeolite immobilization. Process. Saf. Environ. Prot. 2021, 146, 779–788. [Google Scholar] [CrossRef]
- Li, J.S.; Chen, Z.; Wang, Q.M.; Fang, L.; Xue, Q.; Cheeseman, C.R.; Donatello, S.; Liu, L.; Poon, C.S. Change in re-use value of incinerated sewage sludge ash due to chemical extraction of phosphorus. Waste Manag. 2018, 74, 404–412. [Google Scholar] [CrossRef]


| Molecule | M-L | RM-L (calc) | RM-L (expt) |
|---|---|---|---|
| [Cd(H2O)6]2+ | Cd-O | 2.26 | 2.27 (±0.04) |
| [Cd(NH3)6]2+ | Cd-N | 2.35 | 2.37 (±0.03) |
| Soil Components | Values |
|---|---|
| Organic matter | 4.88% |
| Clay (<0.002 mm) | 16.92% |
| Sand (2–0.02 mm) | 68.32% |
| Silt (0.02–0.002 mm) | 14.76% |
| pH | 6.7 |
| CEC (meq NH4+/100 g soil) | 21.41 |
| Total Cd | 20.33 mg/kg |
| Complexes | Structure | R Cd-Oc (Å) | R Cd-Xc (N, O) (Å) | R Cd-Ys (Å) | <OcCdXc (°) | <OCdYs(°) | <XcCdYs (°) |
|---|---|---|---|---|---|---|---|
| GlyCd2+ | [COc, COc] | 2.23 | 2.28 | 59.85 | |||
| [COc] | 2.18 | ||||||
| [Nc, OHc] | 2.23 | 2.16 | 80.26 | ||||
| [Nc, COc] | 2.28 | 2.16 | 80.30 | ||||
| AlaCd2+ | [COc, COc] | 2.28 | 2.23 | 59.93 | |||
| [COc] | 2.22 | ||||||
| [Nc, OHc] | 2.21 | 2.17 | 79.82 | ||||
| [Nc, COc] | 2.28 | 2.16 | 80.33 | ||||
| LeuCd2+ | [COc, COc] | 2.23 | 2.28 | 59.99 | |||
| [COc] | 2.18 | ||||||
| [Nc, OHc] | 2.22 | 2.17 | 49.87 | ||||
| [Nc, COc] | 2.25 | 2.17 | 48.60 | ||||
| GluCd2+ | [COc, COc, COs] | 2.21 | 2.31 | 2.28 | 59.42 | 88.50 | 87.34 |
| [COc, Nc, COs] | 2.26 | 2.17 | 2.3 | 115.00 | 98.59 | 76.73 | |
| [COc, COc] | 2.30 | 2.27 | 59.10 | ||||
| [Nc, COc] | 2.23 | 2.17 | 80.97 | ||||
| ThrCd2+ | [COc, Nc, COs] | 2.24 | 2.27 | 2.23 | 76.44 | 85.70 | 75.79 |
| [COc, COc] | 2.23 | 2.27 | 60.01 | ||||
| [COc, COc, COs] | 2.25 | 2.30 | 2.25 | 60.23 | 72.94 | 71.03 | |
| [N, CO] | 2.45 | 2.27 | 71.40 | ||||
| PheCd2+ | [COc, COc] | 2.20 | 2.54 | 48.87 | |||
| [N, CO] | 2.23 | 2.34 | 75.09 | ||||
| [N, P] | 2.20 | 2.33 | 37.53 |
| Amino Acid | ThrCd2+ | GluCd2+ | GlyCd2+ | AlaCd2+ | LeuCd2+ | PheCd2+ |
|---|---|---|---|---|---|---|
| Complex energy (kcal/mol) | −24.33 | −23.87 | −22.26 | −21.58 | −20.31 | −18.96 |
| Dissolving energy (kcal/mol) | −260.94 | −248.42 | −279.17 | −272.23 | −264.98 | −254.29 |
| Amino Acids | Thr | Glu | Gly | Ala | Leu | Phe |
|---|---|---|---|---|---|---|
| p | 0.67 | 0.5 | 0.57 | 0.5 | 0.5 | 0.5 |
| logβ | 5.56 | 5.13 | 5.05 | 5.03 | 4.96 | 4.77 |
| R2 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Yang, Z.; Huang, L.; Su, C. Complexation of Amino Acids with Cadmium and Their Application for Cadmium-Contaminated Soil Remediation. Appl. Sci. 2022, 12, 1114. https://doi.org/10.3390/app12031114
Yao W, Yang Z, Huang L, Su C. Complexation of Amino Acids with Cadmium and Their Application for Cadmium-Contaminated Soil Remediation. Applied Sciences. 2022; 12(3):1114. https://doi.org/10.3390/app12031114
Chicago/Turabian StyleYao, Wenbin, Zhihui Yang, Lei Huang, and Changqing Su. 2022. "Complexation of Amino Acids with Cadmium and Their Application for Cadmium-Contaminated Soil Remediation" Applied Sciences 12, no. 3: 1114. https://doi.org/10.3390/app12031114
APA StyleYao, W., Yang, Z., Huang, L., & Su, C. (2022). Complexation of Amino Acids with Cadmium and Their Application for Cadmium-Contaminated Soil Remediation. Applied Sciences, 12(3), 1114. https://doi.org/10.3390/app12031114







