A Shallow Domain Knowledge Injection (SDK-Injection) Method for Improving CNN-Based ECG Pattern Classification
Abstract
:Featured Application
Abstract
1. Introduction
- We propose a method to further improve the accuracy of the existing parameter-optimized CNN by using accumulated medical domain knowledge, which could not be utilized due to the inexplicability of deep learning.
- By utilizing shallow domain knowledge that can be acquired even by non-medical experts, it is possible to further improve the existing accuracy while minimizing the cost of introducing medical experts.
- By utilizing the existing CNN parameters that have already been optimized, the accuracy can be further improved without the cost of an additional parameter optimization process.
- By applying the proposed method to the single ECG-based analysis, the original accuracy can be improved to a level similar to, or higher than, the accuracy of the multi-ECG-based analysis. As a result, it can be applied to smart hospital or healthcare services using wearable devices, such as smart watches, that measure just a single ECG stream.
2. Related Work
2.1. Deep and Shallow Domain Knowledge for the ECG Pattern Classification
2.2. Automatic ECG Diagnosis and ECG Pattern Classification
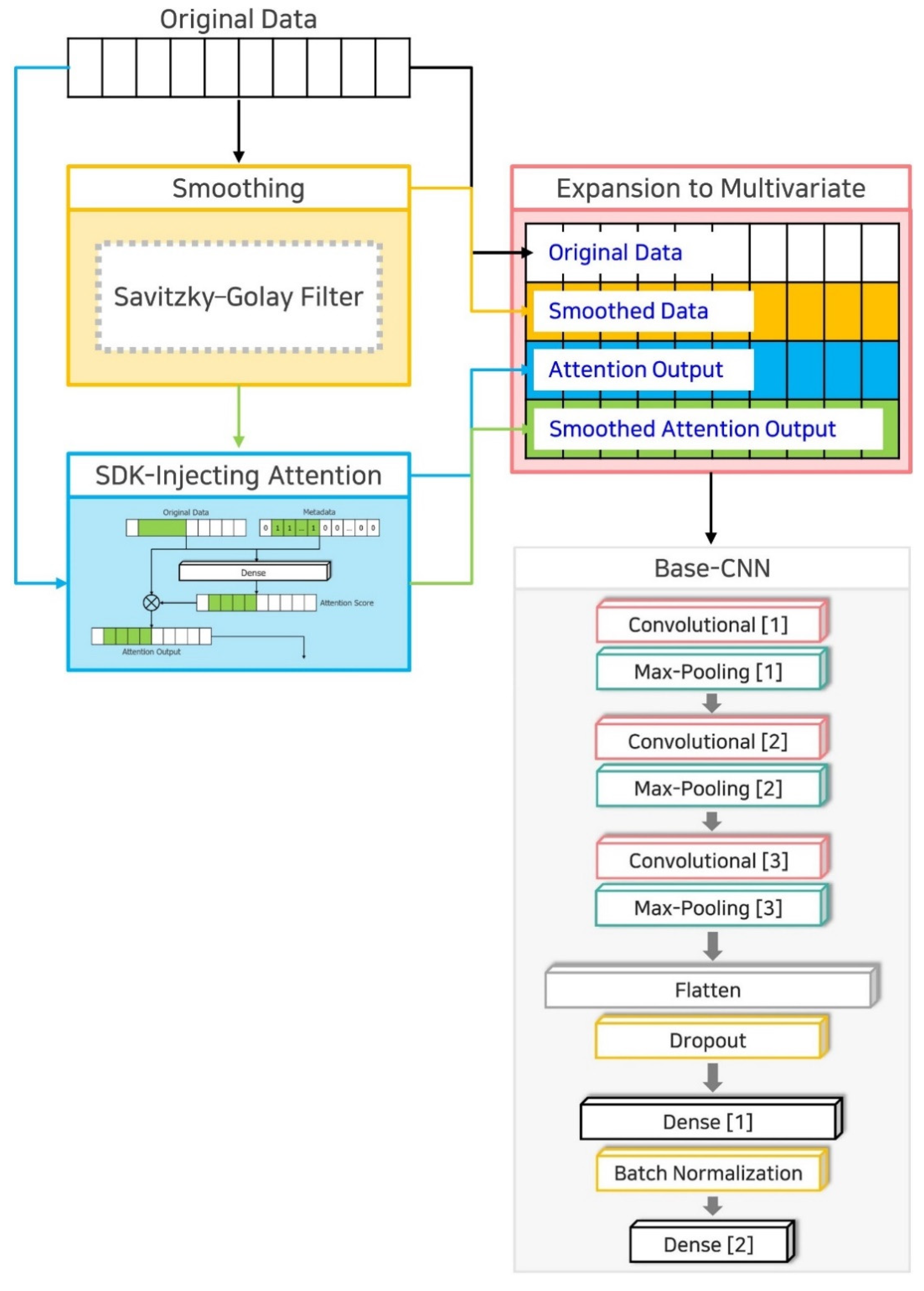
3. SDK-Injection Method
- Technique 1: Curve fitting-based smoothing;
- Technique 2: SDK-Injecting attention;
- Technique 3: Expansion to multivariate.
3.1. Technique 1: Curve Fitting-Based Smoothing
3.2. Technique 2: SDK-Injecting Attention
3.3. Technique 3: Expansion to Multivariate
4. Performance Evaluation Results and Discussion
4.1. Dataset and Environment
4.2. Experiments and Results
- ;
- ;
- ;
- ;
- ;
- .
4.2.1. Result of Experiment 1
4.2.2. Result of Experiment 2
4.2.3. Result of Experiment 3
4.2.4. Result of Experiment 4
4.2.5. Result of Experiment 5
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moses, D. A Survey of Data Mining Algorithms Used in Cardiovascular Disease Diagnosis from Multi-Lead ECG Data. Kuwait J. Sci. 2015, 42, 206–235. [Google Scholar]
- Glushica, B.; Aleksovski, B.; Kuhar, A. Automatic Detection of Characteristic Segments of a Recorded ECG Signal with Noise Handling Methods. IOP Conf. Ser. Mater. Sci. Eng. 2021, 1032, 012047. [Google Scholar] [CrossRef]
- Zifan, A.; Saberi, S.; Moradi, M.H.; Towhidkhah, F. Automated ECG Segmentation Using Piecewise Derivative Dynamic Time Warping. Int. J. Biol. Med. Sci. 2006, 6, 181–185. [Google Scholar]
- Beraza, I.; Romero, I. Comparative Study of Algorithms for ECG Segmentation. Biomed. Signal Process. Control 2017, 34, 166–173. [Google Scholar] [CrossRef]
- Luz, E.J.; Da, S.; Schwartz, W.R.; Cámara-Chávez, G.; Menotti, D. ECG-Based Heartbeat Classification for Arrhythmia Detection: A Survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Barman, T.; Ghongade, R.; Ratnaparkhi, A. Rough Set Based Segmentation and Classification Model for ECG. In Proceedings of the 2016 Conference on Advances in Signal Processing (CASP), Pune, India, 9–11 June 2016; pp. 18–23. [Google Scholar]
- Rai, H.M.; Chatterjee, K. Hybrid CNN-LSTM Deep Learning Model and Ensemble Technique for Automatic Detection of Myocardial Infarction Using Big ECG Data. Appl. Intell. 2021, 1–19. [Google Scholar] [CrossRef]
- Essa, E.; Xie, X. An Ensemble of Deep Learning-Based Multi-Model for ECG Heartbeats Arrhythmia Classification. IEEE Access 2021, 9, 103452–103464. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated Atrial Fibrillation Detection Using a Hybrid CNN-LSTM Network on Imbalanced ECG Datasets. Biomed. Signal Process. Control 2021, 63, 102194. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J. Application of Heartbeat-Attention Mechanism for Detection of Myocardial Infarction Using 12-Lead ECG Records. Appl. Sci. 2019, 9, 3328. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Huang, R.; Yu, F.; Huang, X.; Cui, L. EasiECG: A Novel Inter-Patient Arrhythmia Classification Method Using ECG Waves. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 1–8. [Google Scholar]
- Jiang, M.; Gu, J.; Li, Y.; Wei, B.; Zhang, J.; Wang, Z.; Xia, L. HADLN: Hybrid Attention-Based Deep Learning Network for Automated Arrhythmia Classification. Front. Physiol. 2021, 12, 683025. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Lu, C.; Sun, Y.; Yang, M.; Liu, C.; Ou, C. Automatic ECG Classification Using Continuous Wavelet Transform and Convolutional Neural Network. Entropy 2021, 23, 119. [Google Scholar] [CrossRef] [PubMed]
- Couceiro, R.; Carvalho, P.; Henriques, J.; Antunes, M.; Harris, M.; Habetha, J. Detection of Atrial Fibrillation Using Model-Based ECG Analysis. In Proceedings of the 2008 19th International Conference on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008; pp. 1–5. [Google Scholar]
- Chen, X.; Cheng, Z.; Wang, S.; Lu, G.; Xv, G.; Liu, Q.; Zhu, X. Atrial Fibrillation Detection Based on Multi-Feature Extraction and Convolutional Neural Network for Processing ECG Signals. Comput. Methods Programs Biomed. 2021, 202, 106009. [Google Scholar] [CrossRef] [PubMed]
- Pouyanfar, S.; Sadiq, S.; Yan, Y.; Tian, H.; Tao, Y.; Reyes, M.P.; Shyu, M.-L.; Chen, S.-C.; Iyengar, S.S. A Survey on Deep Learning: Algorithms, Techniques, and Applications. ACM Comput. Surv. 2019, 51, 1–36. [Google Scholar] [CrossRef]
- Holzinger, A. From Machine Learning to Explainable AI. In Proceedings of the 2018 World Symposium on Digital Intelligence for Systems and Machines (DISA), Košice, Slovakia, 23–25 August 2018; pp. 55–66. [Google Scholar]
- Lastre-Domínguez, C.; Shmaliy, Y.S.; Ibarra-Manzano, O.; Munoz-Minjares, J.; Morales-Mendoza, L.J. ECG Signal Denoising and Features Extraction Using Unbiased FIR Smoothing. BioMed. Res. Int. 2019, 2019, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, S.R.; Seelamantula, C.S. On the Selection of Optimum Savitzky-Golay Filters. IEEE Trans. Signal Process. 2013, 61, 380–391. [Google Scholar] [CrossRef]
- Awal, M.A.; Mostafa, S.S.; Ahmad, M. Performance Analysis of Savitzky-Golay Smoothing Filter Using ECG Signal. Int. J. Comput. Inf. Technol. 2011, 1, 24. [Google Scholar]
- Hussain, I.; Park, S.J. Big-ECG: Cardiographic Predictive Cyber-Physical System for Stroke Management. IEEE Access 2021, 9, 123146–123164. [Google Scholar] [CrossRef]
- Tseng, K.-K.; Wang, C.; Xiao, T.; Chen, C.-M.; Hassan, M.M.; de Albuquerque, V.H.C. Sliding Large Kernel of Deep Learning Algorithm for Mobile Electrocardiogram Diagnosis. Comput. Electr. Eng. 2021, 96, 107521. [Google Scholar] [CrossRef]
- Sereda, I.; Alekseev, S.; Koneva, A.; Kataev, R.; Osipov, G. ECG Segmentation by Neural Networks: Errors and Correction. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–7. [Google Scholar]
- Sodmann, P.; Vollmer, M. ECG Segmentation Using a Neural Network as the Basis for Detection of Cardiac Pathologies. In Proceedings of the 2020 Computing in Cardiology, Rimini, Italy, 13–16 September 2020; pp. 1–4. [Google Scholar]
- Wang, S.; Li, R.; Wang, X.; Shen, S.; Zhou, B.; Wang, Z. Multiscale Residual Network Based on Channel Spatial Attention Mechanism for Multilabel ECG Classification. J. Healthc. Eng. 2021, 2021, e6630643. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Li, Z. Recognition of ECG Signals by Convolutional Neural Network Based on Attentional Mechanism. In Proceedings of the 2021 4th International Conference on Advanced Electronic Materials, Computers and Software Engineering (AEMCSE), Changsha, China, 26–28 March 2021; pp. 124–127. [Google Scholar]
- Ullah, A.; Tu, S.; Mehmood, R.M.; Ehatisham-ul-haq, M. A Hybrid Deep CNN Model for Abnormal Arrhythmia Detection Based on Cardiac ECG Signal. Sensors 2021, 21, 951. [Google Scholar] [CrossRef] [PubMed]
- Time Series Classification Website. Available online: http://www.timeseriesclassification.com/dataset.php (accessed on 6 September 2021).
- PhysioNet Databases. Available online: https://www.physionet.org/about/database/ (accessed on 6 September 2021).











| No. | Dataset | Size (Train) | Size (Test) | Pattern Length | No. of Classes | Class Ratio (Train) | Class Ratio (Test) | Standard Deviation (Class Ratio) | Target Disease |
|---|---|---|---|---|---|---|---|---|---|
| 1 | AF-D1 | 15 | 15 | 640 | 3 | 1 | 1 | 0 | Atrial Fibrillation |
| 1 | 1 | ||||||||
| 1 | 1 | ||||||||
| 2 | AF-D2 | 15 | 15 | 640 | 3 | 1 | 1 | 0 | Atrial Fibrillation |
| 1 | 1 | ||||||||
| 1 | 1 | ||||||||
| 3 | ECG200 | 100 | 100 | 96 | 2 | 1 | 1 | 0.71 | ischemia |
| 2.23 | 1.78 | ||||||||
| 4 | ECG5000 | 500 | 4500 | 140 | 5 | 146 | 119.41 | 58.82 | Congestive Heart Failure |
| 88.5 | 72.27 | ||||||||
| 5 | 3.91 | ||||||||
| 9.5 | 7.95 | ||||||||
| 1 | 1 | ||||||||
| 5 | TwoLead | 23 | 1139 | 82 | 2 | 1.09 | 1 | 0.03 | Non-disease |
| 1 | 1 |
| Metric | Dataset | Base-CNN | SDK-Injection | Increasement by SDK-Injection |
|---|---|---|---|---|
| Accuracy (%) | AF-D1 | 26.67 | 46.67 | 20.00 |
| AF-D2 | 40.00 | 60.00 | 20.00 | |
| ECG200 | 92.00 | 95.00 | 3.00 | |
| ECG5000 | 93.76 | 94.38 | 0.62 | |
| TwoLead | 69.89 | 85.25 | 15.36 | |
| Precision | AF-D1 | 0.30 | 0.47 | 0.17 |
| AF-D2 | 0.33 | 0.70 | 0.37 | |
| ECG200 | 0.92 | 0.95 | 0.03 | |
| ECG5000 | 0.70 | 0.75 | 0.05 | |
| TwoLead | 0.72 | 0.86 | 0.14 | |
| Sensitivity | AF-D1 | 0.27 | 0.47 | 0.20 |
| AF-D2 | 0.40 | 0.60 | 0.20 | |
| ECG200 | 0.92 | 0.95 | 0.03 | |
| ECG5000 | 0.52 | 0.58 | 0.06 | |
| TwoLead | 0.70 | 0.86 | 0.16 | |
| Specificity | AF-D1 | 0.63 | 0.73 | 0.10 |
| AF-D2 | 0.70 | 0.80 | 0.10 | |
| ECG200 | 0.92 | 0.95 | 0.03 | |
| ECG5000 | 0.98 | 0.98 | 0.00 | |
| TwoLead | 0.70 | 0.86 | 0.16 | |
| F1-Score | AF-D1 | 0.28 | 0.46 | 0.18 |
| AF-D2 | 0.35 | 0.60 | 0.25 | |
| ECG200 | 0.92 | 0.95 | 0.03 | |
| ECG5000 | 0.56 | 0.63 | 0.07 | |
| TwoLead | 0.70 | 0.86 | 0.16 |
| Method | Dataset | Class | Accuracy (%) | Precision | Sensitivity | Specificity | F1-Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Per Class | Average | Per Class | Average | Per Class | Average | Per Class | Average | ||||
| Base- CNN | AF-D1 | n | 26.67 | 0.50 | 0.30 | 0.40 | 0.27 | 0.80 | 0.63 | 0.44 | 0.28 |
| s | 0.14 | 0.20 | 0.40 | 0.17 | |||||||
| t | 0.25 | 0.20 | 0.70 | 0.22 | |||||||
| AF-D2 | n | 40.00 | 0.38 | 0.33 | 0.60 | 0.40 | 0.50 | 0.70 | 0.46 | 0.35 | |
| s | 0.00 | 0.00 | 0.80 | NaN | |||||||
| t | 0.60 | 0.60 | 0.80 | 0.60 | |||||||
| ECG200 | −1 | 92.00 | 0.89 | 0.92 | 0.89 | 0.92 | 0.94 | 0.92 | 0.89 | 0.92 | |
| 1 | 0.94 | 0.94 | 0.89 | 0.94 | |||||||
| ECG5000 | 1 | 93.76 | 0.97 | 0.70 | 1.00 | 0.52 | 0.96 | 0.98 | 0.98 | 0.56 | |
| 2 | 0.90 | 0.96 | 0.94 | 0.93 | |||||||
| 3 | 0.43 | 0.07 | 1.00 | 0.12 | |||||||
| 4 | 0.68 | 0.33 | 0.99 | 0.45 | |||||||
| 5 | 0.50 | 0.23 | 1.00 | 0.31 | |||||||
| TwoLead | 1 | 69.89 | 0.65 | 0.72 | 0.84 | 0.70 | 0.56 | 0.70 | 0.74 | 0.70 | |
| 2 | 0.78 | 0.56 | 0.84 | 0.65 | |||||||
| SDK- Injection | AF-D1 | n | 46.67 | 0.40 | 0.47 | 0.40 | 0.47 | 0.70 | 0.73 | 0.40 | 0.46 |
| s | 0.50 | 0.40 | 0.80 | 0.44 | |||||||
| t | 0.50 | 0.60 | 0.70 | 0.55 | |||||||
| AF-D2 | n | 60.00 | 0.50 | 0.70 | 0.80 | 0.60 | 0.60 | 0.80 | 0.62 | 0.60 | |
| s | 1.00 | 0.40 | 1.00 | 0.57 | |||||||
| t | 0.60 | 0.60 | 0.80 | 0.60 | |||||||
| ECG200 | −1 | 95.00 | 0.94 | 0.95 | 0.92 | 0.95 | 0.97 | 0.95 | 0.93 | 0.95 | |
| 1 | 0.95 | 0.97 | 0.92 | 0.96 | |||||||
| ECG5000 | 1 | 94.38 | 0.97 | 0.75 | 1.00 | 0.58 | 0.96 | 0.98 | 0.99 | 0.63 | |
| 2 | 0.93 | 0.95 | 0.96 | 0.94 | |||||||
| 3 | 0.60 | 0.41 | 0.99 | 0.49 | |||||||
| 4 | 0.66 | 0.39 | 0.99 | 0.49 | |||||||
| 5 | 0.60 | 0.14 | 1.00 | 0.22 | |||||||
| TwoLead | 1 | 85.25 | 0.82 | 0.86 | 0.91 | 0.86 | 0.80 | 0.86 | 0.86 | 0.86 | |
| 2 | 0.90 | 0.80 | 0.91 | 0.85 | |||||||
| Metric | Dataset | Base-CNN (Univariate) | Base-CNN with Multivariate (AF-D1, AF-D2) | SDK-Injection (Univariate) |
|---|---|---|---|---|
| Accuracy (%) | AF-D1 | 26.67 | 46.67 | 46.67 |
| AF-D2 | 40.00 | 60.00 | ||
| Precision | AF-D1 | 0.30 | 0.48 | 0.47 |
| AF-D2 | 0.33 | 0.70 | ||
| Sensitivity | AF-D1 | 0.27 | 0.47 | 0.47 |
| AF-D2 | 0.40 | 0.60 | ||
| Specificity | AF-D1 | 0.63 | 0.73 | 0.73 |
| AF-D2 | 0.70 | 0.80 | ||
| F1-Score | AF-D1 | 0.28 | 0.45 | 0.46 |
| AF-D2 | 0.35 | 0.60 |
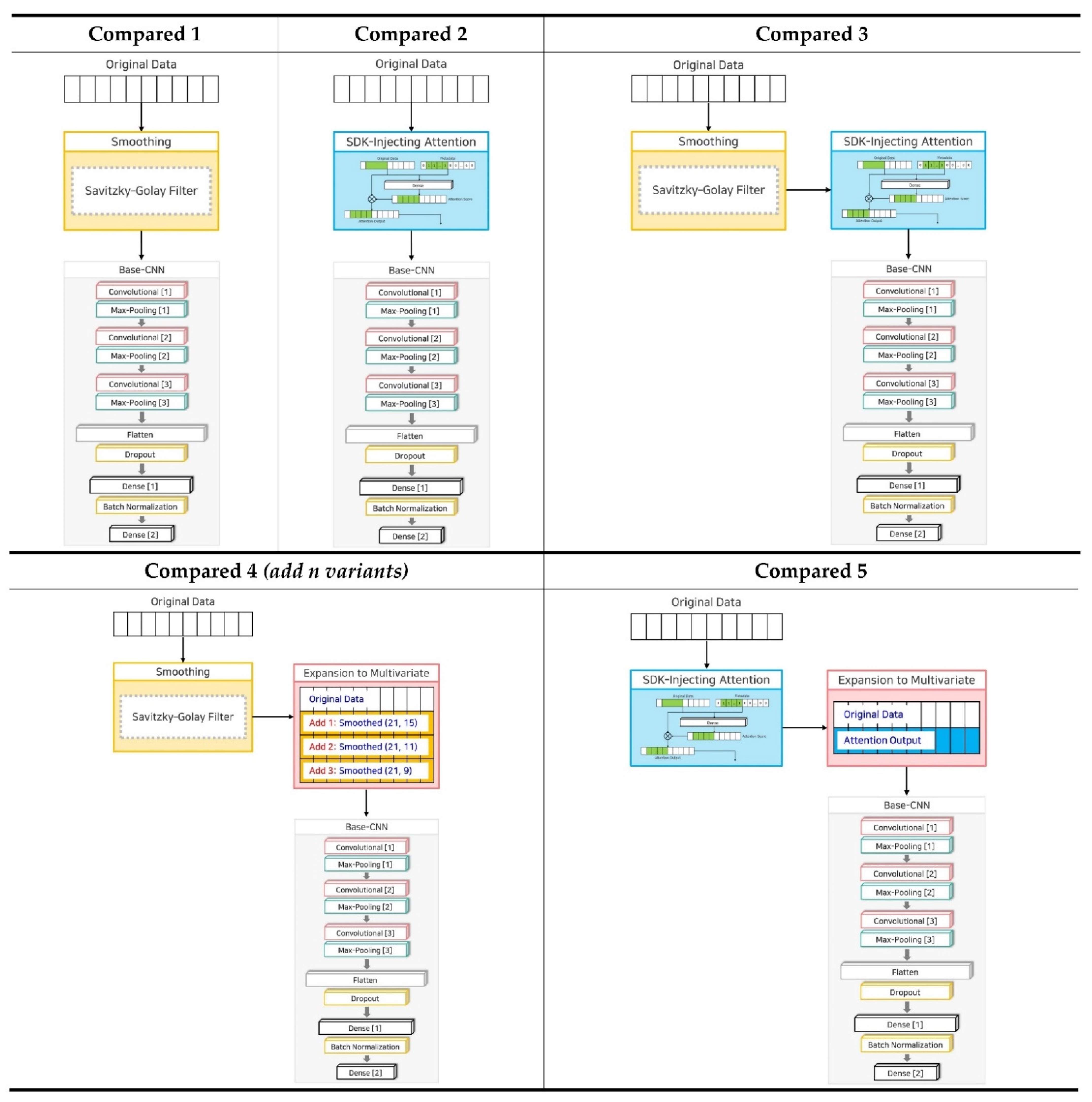
| No. | Name | Technique 1 | Technique 2 | Technique 3 | Variants |
|---|---|---|---|---|---|
| 1 | Base-CNN | X | X | X | 1: Original Data |
| 2 | SDK-Injection | O | O | O | 1: Original Data 2: Smoothed Data (21 window size, 13 order) 3: Attention Output 4: Smoothed Attention Output |
| 3 | Compared 1 | O | X | X | 1: Original Data |
| 4 | Compared 2 | X | O | X | 1: Original Data |
| 5 | Compared 3 | O | O | X | 1: Original Data |
| 6 | Compared 4 (add 1 variant) | O | X | O | 1: Original Data 2: Smoothed Data (21 window size, 15 order) |
| 7 | Compared 4 (add 2 variants) | O | X | O | 1: Original Data 2: Smoothed Data (21 window size, 15 order) 3: Smoothed Data (21 window size, 11 order) |
| 8 | Compared 4 (add 3 variants) | O | X | O | 1: Original Data 2: Smoothed Data (21 window size, 15 order) 3: Smoothed Data (21 window size, 11 order) 4: Smoothed Data (21 window size, 9 order) |
| 9 | Compared 5 | X | O | O | 1: Original Data 2: Attention Output |
| Metric | Dataset | Base-CNN | SDK- Injection | Compared 1 | Compared 2 | Compared 3 | Compared 4 | Compared 5 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Add 1 Variants | Add 2 Variants | Add 3 Variants | ||||||||
| Accuracy (%) | AF-D1 | 26.67 | 46.67 | 40.00 | 33.33 | 33.33 | 26.67 | 26.67 | 26.67 | 26.67 |
| AF-D2 | 40.00 | 60.00 | 46.67 | 40.00 | 33.33 | 40.00 | 33.33 | 40.00 | 46.67 | |
| ECG200 | 92.00 | 95.00 | 92.00 | 93.00 | 92.00 | 93.00 | 93.00 | 93.00 | 93.00 | |
| ECG5000 | 93.76 | 94.38 | 94.16 | 94.20 | 94.11 | 94.33 | 94.24 | 94.13 | 94.18 | |
| TwoLead | 69.89 | 85.25 | 69.53 | 72.52 | 70.41 | 65.67 | 66.64 | 67.95 | 77.88 | |
| Precision | AF-D1 | 0.30 | 0.47 | 0.33 | 0.22 | 0.28 | 0.21 | 0.17 | 0.17 | 0.17 |
| AF-D2 | 0.33 | 0.70 | 0.31 | 0.44 | 0.25 | 0.30 | 0.22 | 0.37 | 0.48 | |
| ECG200 | 0.92 | 0.95 | 0.92 | 0.93 | 0.92 | 0.94 | 0.93 | 0.94 | 0.92 | |
| ECG5000 | 0.70 | 0.75 | 0.70 | 0.68 | 0.64 | 0.73 | 0.72 | 0.68 | 0.69 | |
| TwoLead | 0.72 | 0.86 | 0.71 | 0.78 | 0.74 | 0.66 | 0.69 | 0.68 | 0.79 | |
| Sensitivity | AF-D1 | 0.27 | 0.47 | 0.40 | 0.33 | 0.33 | 0.27 | 0.27 | 0.27 | 0.27 |
| AF-D2 | 0.40 | 0.60 | 0.47 | 0.40 | 0.33 | 0.40 | 0.33 | 0.40 | 0.47 | |
| ECG200 | 0.92 | 0.95 | 0.91 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 0.93 | |
| ECG5000 | 0.52 | 0.58 | 0.57 | 0.58 | 0.56 | 0.57 | 0.58 | 0.55 | 0.56 | |
| TwoLead | 0.70 | 0.86 | 0.70 | 0.77 | 0.72 | 0.66 | 0.67 | 0.68 | 0.79 | |
| Specificity | AF-D1 | 0.63 | 0.73 | 0.70 | 0.67 | 0.67 | 0.63 | 0.63 | 0.63 | 0.63 |
| AF-D2 | 0.70 | 0.80 | 0.73 | 0.70 | 0.67 | 0.70 | 0.67 | 0.70 | 0.73 | |
| ECG200 | 0.92 | 0.95 | 0.91 | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | 0.93 | |
| ECG5000 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 | |
| TwoLead | 0.70 | 0.86 | 0.70 | 0.77 | 0.72 | 0.66 | 0.67 | 0.68 | 0.79 | |
| F1-Score | AF-D1 | 0.28 | 0.46 | 0.35 | 0.26 | 0.25 | 0.23 | 0.20 | 0.20 | 0.20 |
| AF-D2 | 0.35 | 0.60 | 0.37 | 0.41 | 0.29 | 0.32 | 0.25 | 0.36 | 0.45 | |
| ECG200 | 0.92 | 0.95 | 0.92 | 0.93 | 0.92 | 0.93 | 0.93 | 0.93 | 0.92 | |
| ECG5000 | 0.56 | 0.63 | 0.61 | 0.61 | 0.58 | 0.62 | 0.63 | 0.59 | 0.60 | |
| TwoLead | 0.70 | 0.86 | 0.69 | 0.77 | 0.71 | 0.66 | 0.66 | 0.68 | 0.79 | |
| No. | Dataset | Pattern Length | Target Disease | Sub-Pattern as Criteria | Shallow Domain Knowledge | |
|---|---|---|---|---|---|---|
| Start Index | End Index | |||||
| 1 | AF-D1 | 640 | Atrial Fibrillation | QRS | 70 | 300 |
| 2 | AF-D2 | 640 | Atrial Fibrillation | QRS | 70 | 300 |
| 3 | ECG200 | 96 | ischemia | T | 10 | 55 |
| 4 | ECG5000 | 140 | Congestive Heart Failure | P | 100 | 135 |
| 5 | TwoLead | 82 | Non-disease | Differences btw. Patterns | 0 | 70 |
| Dataset | Shallow Domain Knowledge | Accuracy (%) | Precision | Sensitivity | Specificity | F1-Score | ||
|---|---|---|---|---|---|---|---|---|
| Length | Start Index | End Index | ||||||
| AF-D1 | 0 | 0 | 0 | 20.00 | 0.17 | 0.20 | 0.60 | 0.18 |
| Optimized | 70 | 300 | 46.67 | 0.47 | 0.47 | 0.73 | 0.46 | |
| Max Pattern | 0 | 640 | 40.00 | 0.45 | 0.40 | 0.70 | 0.29 | |
| AF-D2 | 0 | 0 | 0 | 40.00 | 0.28 | 0.40 | 0.70 | 0.32 |
| Optimized | 70 | 300 | 60.00 | 0.70 | 0.60 | 0.80 | 0.60 | |
| Max Pattern | 0 | 640 | 46.67 | 0.52 | 0.47 | 0.73 | 0.48 | |
| ECG200 | 0 | 0 | 0 | 91.00 | 0.91 | 0.89 | 0.89 | 0.90 |
| Optimized | 10 | 55 | 95.00 | 0.95 | 0.95 | 0.95 | 0.95 | |
| Max Pattern | 0 | 96 | 92.00 | 0.93 | 0.90 | 0.90 | 0.91 | |
| ECG5000 | 0 | 0 | 0 | 94.02 | 0.68 | 0.55 | 0.98 | 0.58 |
| Optimized | 100 | 135 | 94.38 | 0.75 | 0.58 | 0.98 | 0.63 | |
| Max Pattern | 0 | 140 | 94.11 | 0.66 | 0.55 | 0.98 | 0.58 | |
| TwoLead | 0 | 0 | 0 | 56.19 | 0.72 | 0.56 | 0.56 | 0.47 |
| Optimized | 0 | 70 | 85.25 | 0.86 | 0.86 | 0.86 | 0.86 | |
| Max Pattern | 0 | 82 | 85.25 | 0.86 | 0.85 | 0.85 | 0.85 | |
| Dataset | Shallow Domain Knowledge | Accuracy (%) | Precision | Sensitivity | Specificity | F1-Score | |
|---|---|---|---|---|---|---|---|
| Start Index | End Index | ||||||
| TwoLead | 0 | 0 | 56.19 | 0.72 | 0.56 | 0.56 | 0.47 |
| 0 | 10 | 73.22 | 0.79 | 0.74 | 0.74 | 0.73 | |
| 0 | 20 | 69.62 | 0.81 | 0.70 | 0.70 | 0.67 | |
| 0 | 30 | 77.35 | 0.78 | 0.78 | 0.78 | 0.78 | |
| 0 | 40 | 64.71 | 0.66 | 0.65 | 0.65 | 0.65 | |
| 0 | 50 | 78.75 | 0.80 | 0.78 | 0.78 | 0.78 | |
| 0 | 60 | 72.52 | 0.73 | 0.73 | 0.73 | 0.73 | |
| 0 | 70 | 85.25 | 0.86 | 0.86 | 0.86 | 0.86 | |
| 0 | 82 | 85.25 | 0.86 | 0.85 | 0.85 | 0.85 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, S.; Lee, M. A Shallow Domain Knowledge Injection (SDK-Injection) Method for Improving CNN-Based ECG Pattern Classification. Appl. Sci. 2022, 12, 1307. https://doi.org/10.3390/app12031307
Oh S, Lee M. A Shallow Domain Knowledge Injection (SDK-Injection) Method for Improving CNN-Based ECG Pattern Classification. Applied Sciences. 2022; 12(3):1307. https://doi.org/10.3390/app12031307
Chicago/Turabian StyleOh, Soyeon, and Minsoo Lee. 2022. "A Shallow Domain Knowledge Injection (SDK-Injection) Method for Improving CNN-Based ECG Pattern Classification" Applied Sciences 12, no. 3: 1307. https://doi.org/10.3390/app12031307







