Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds
Abstract
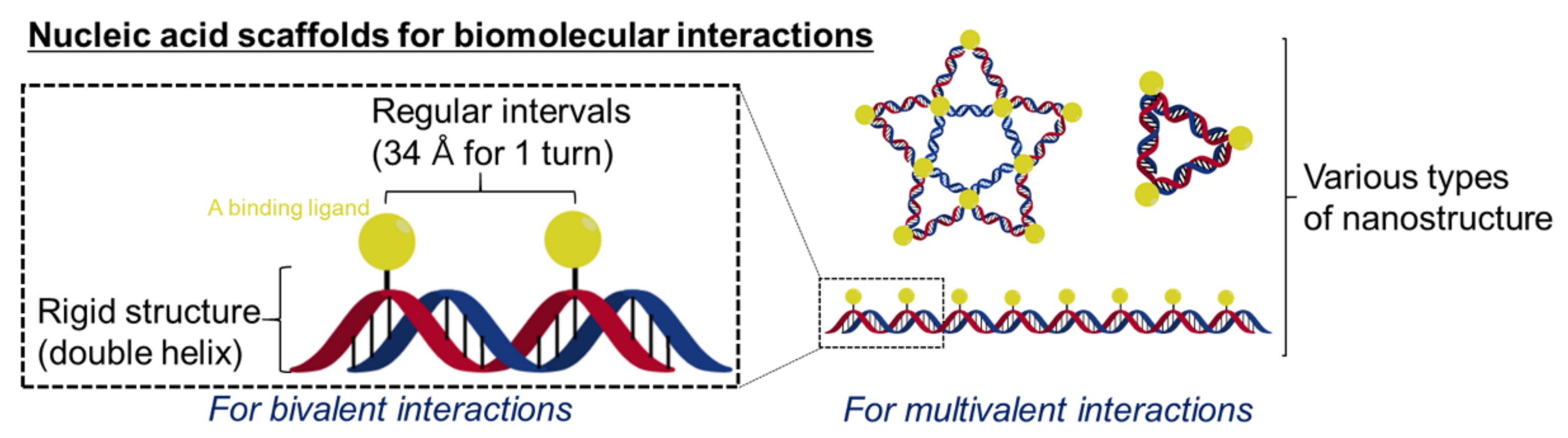
:1. Introduction
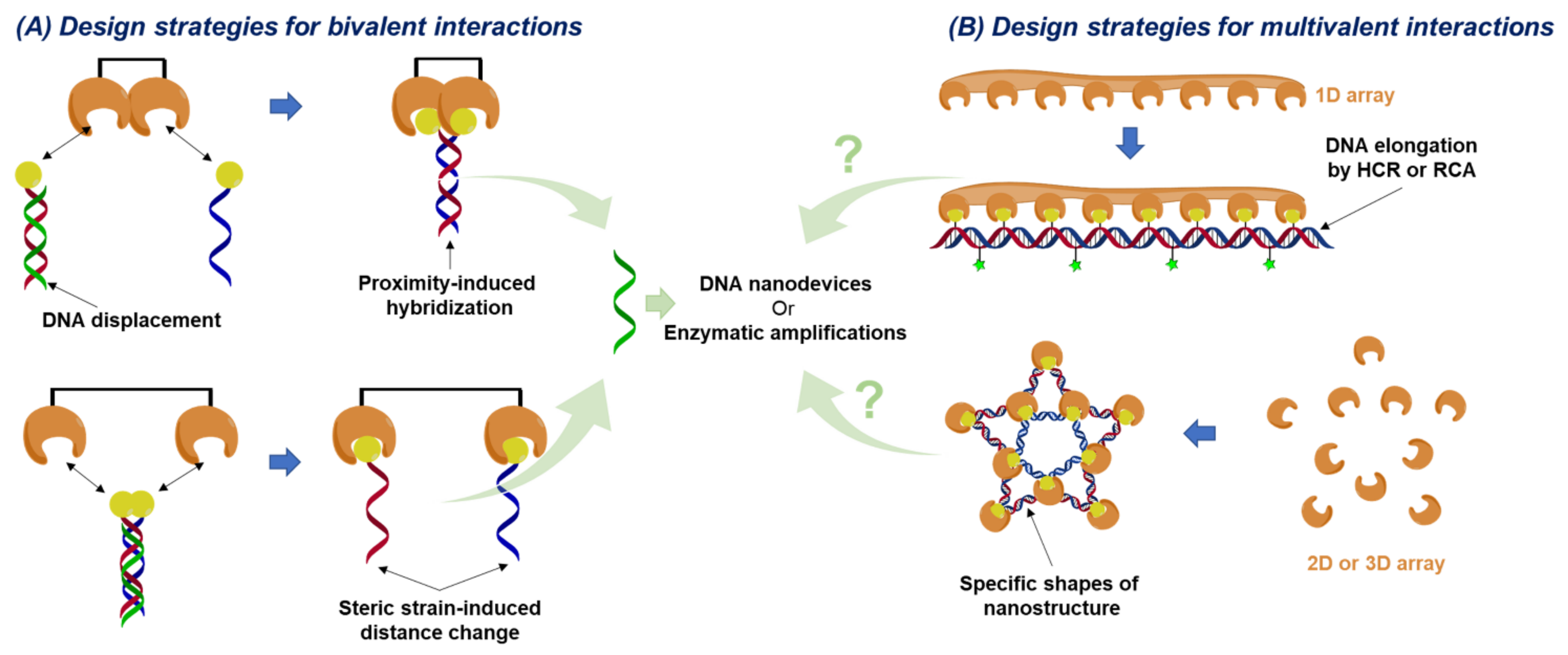
2. Biosensors Based on Bivalent Binding Modes
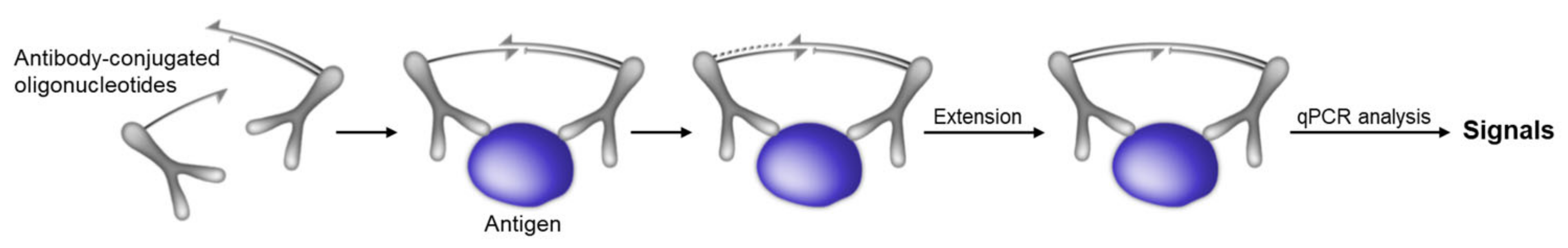
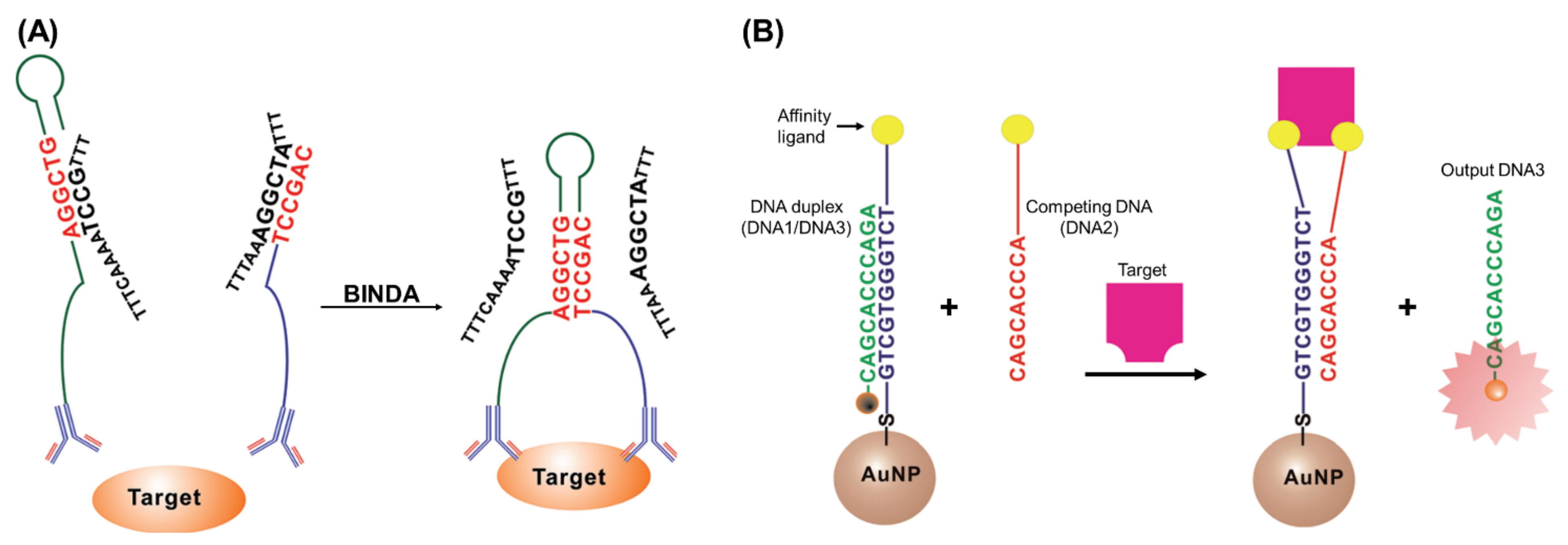
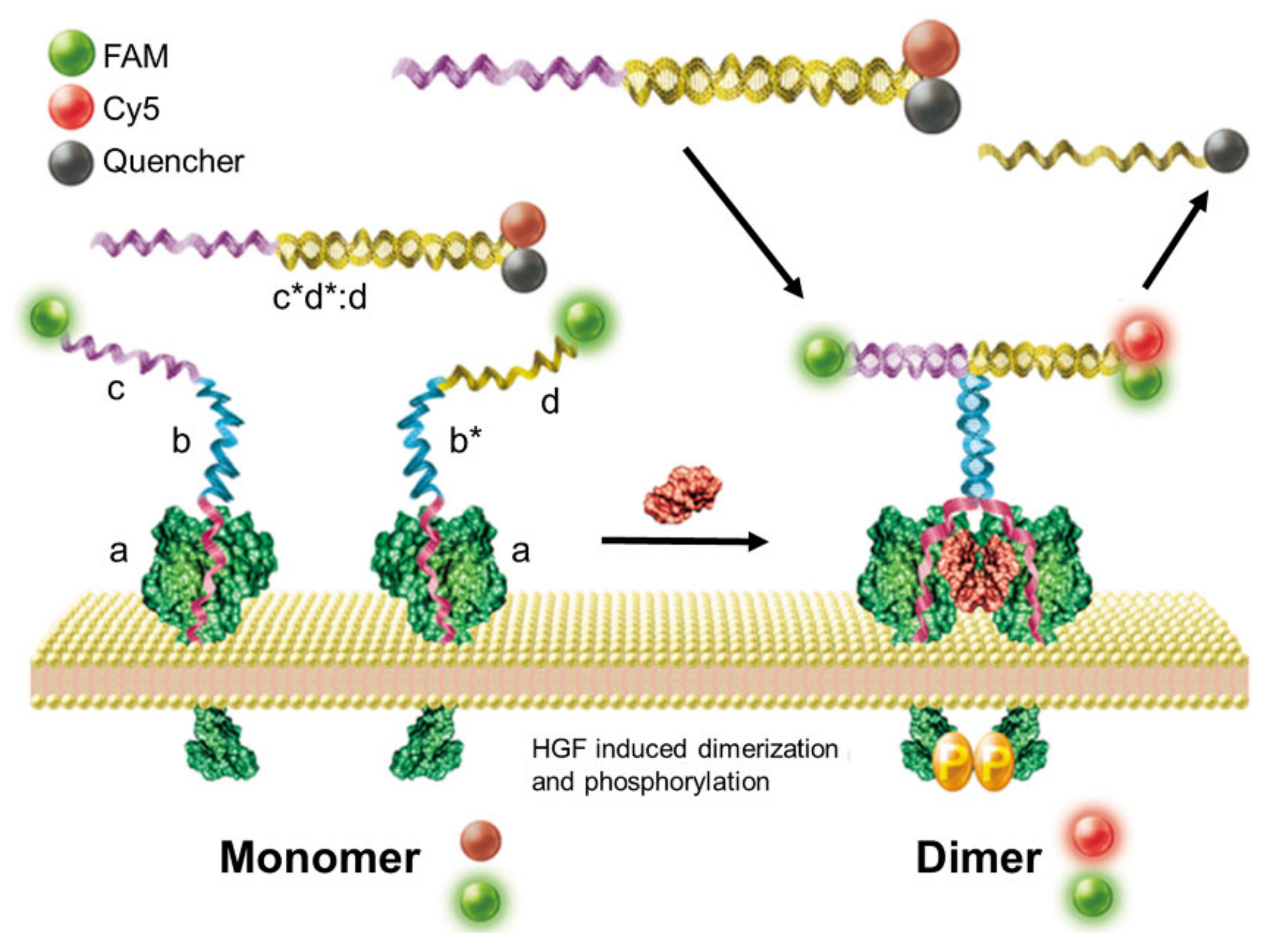
2.1. Biosensors Based on Proximity-Induced Hybridization
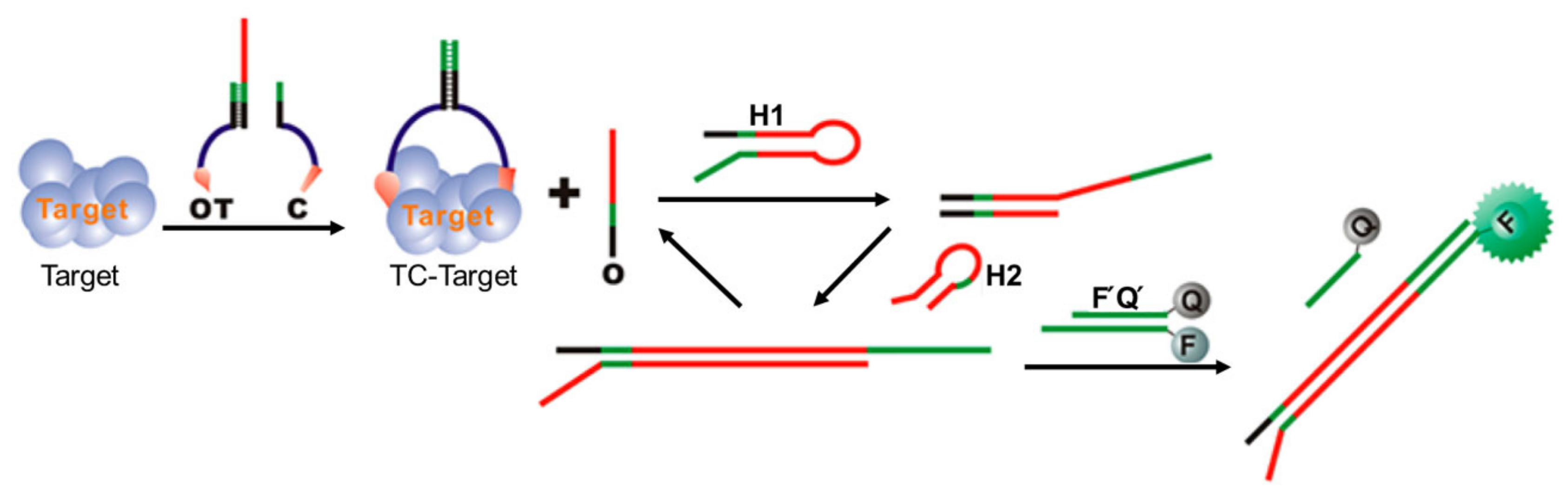
2.2. Biosensors Based on DNA Displacement
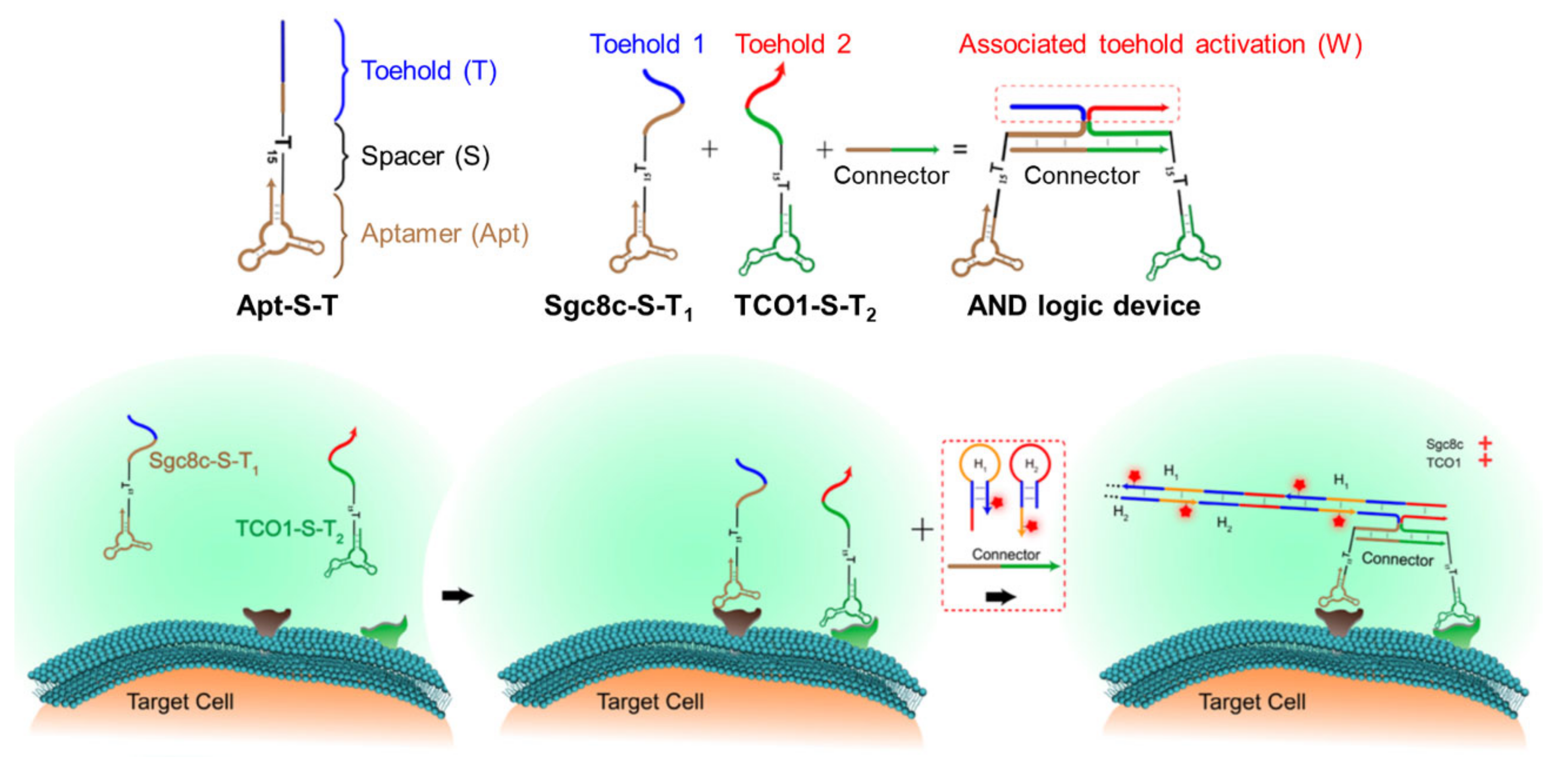
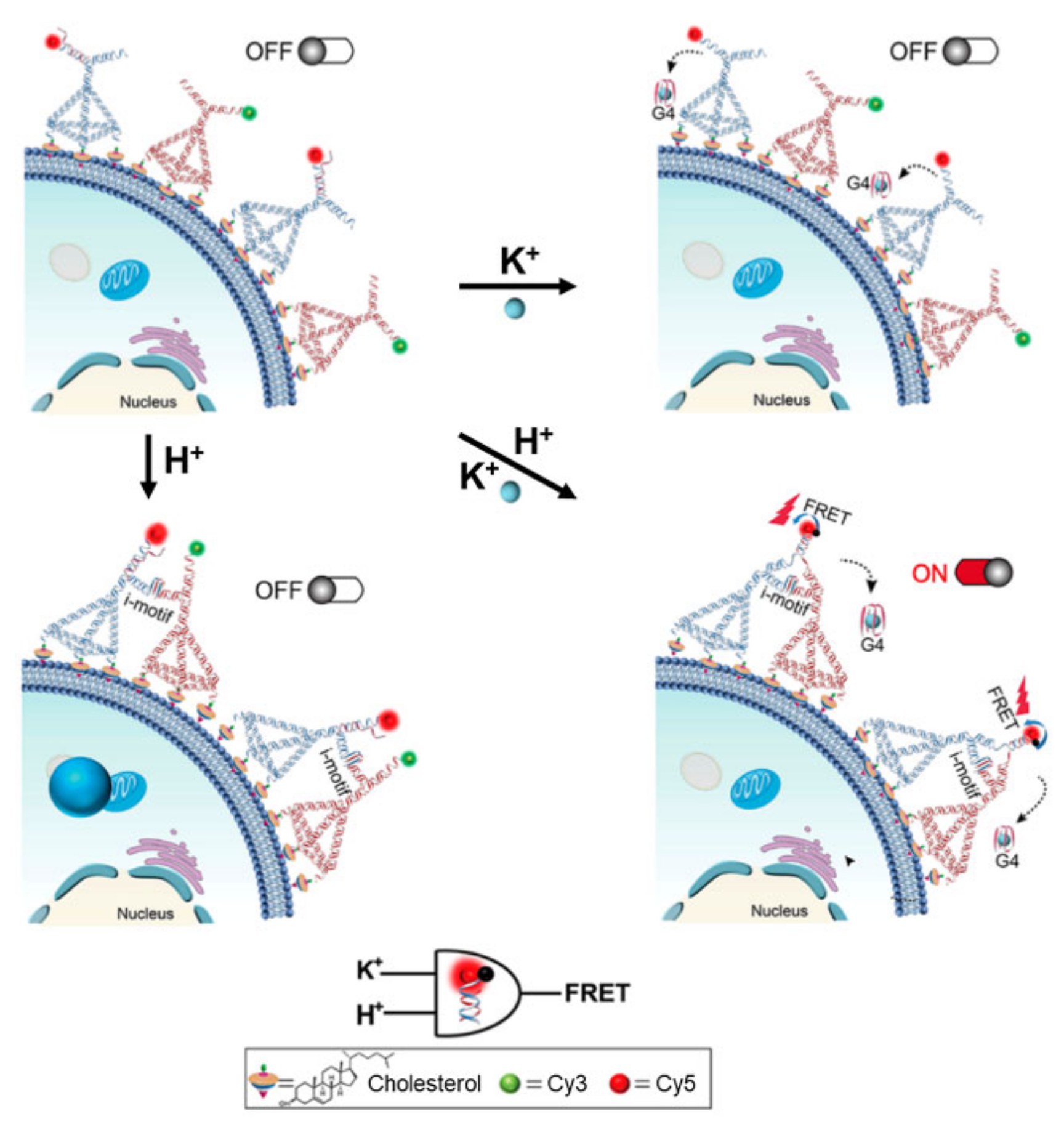
2.3. Biosensors Based on DNA Nanodevices
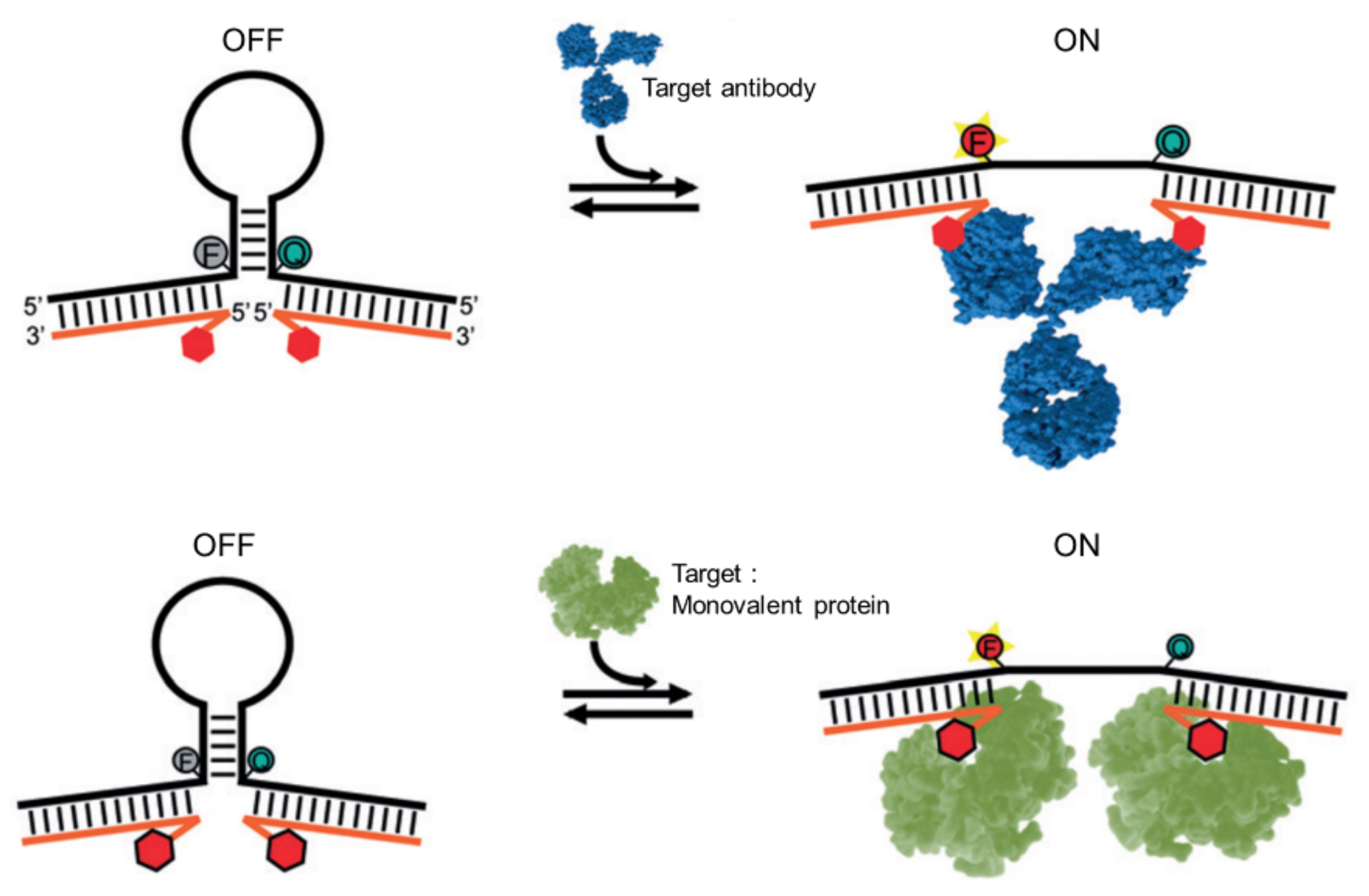
2.4. Biosensors Based on Steric Strain-Induced Distance Change
3. Biosensors Based on Multivalent Binding Modes
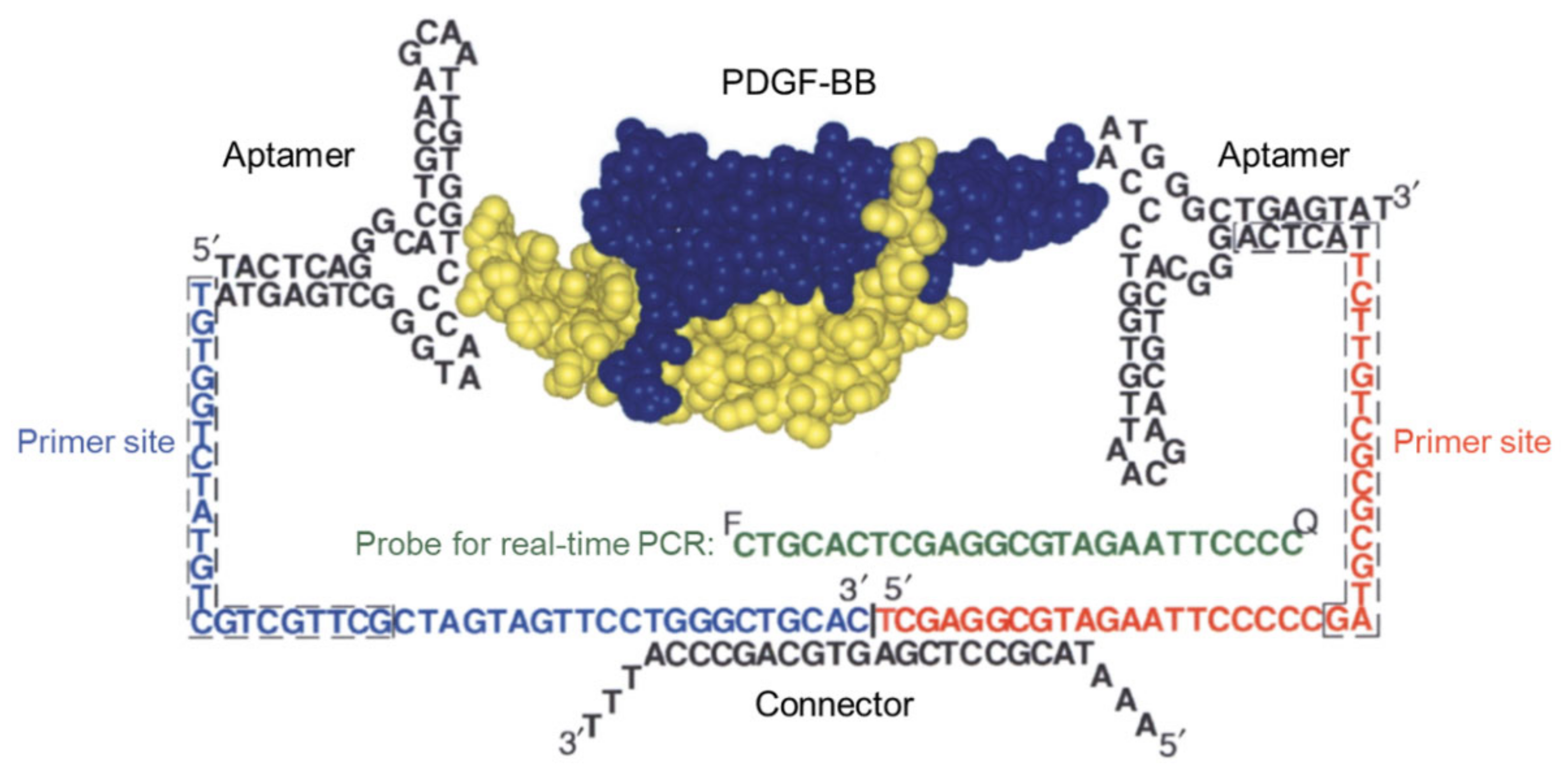
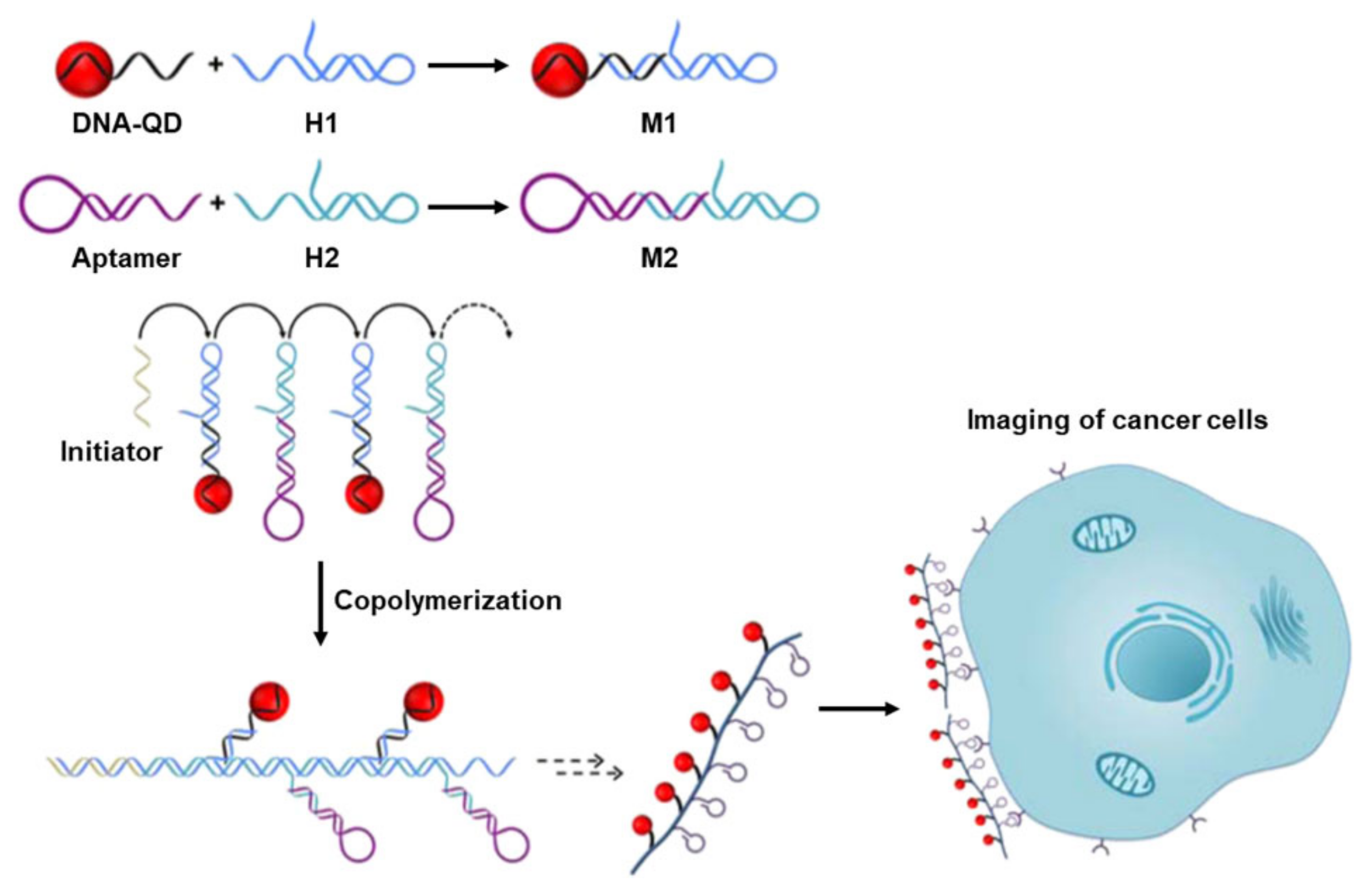
3.1. Biosensors Based on DNA Chain Elongation
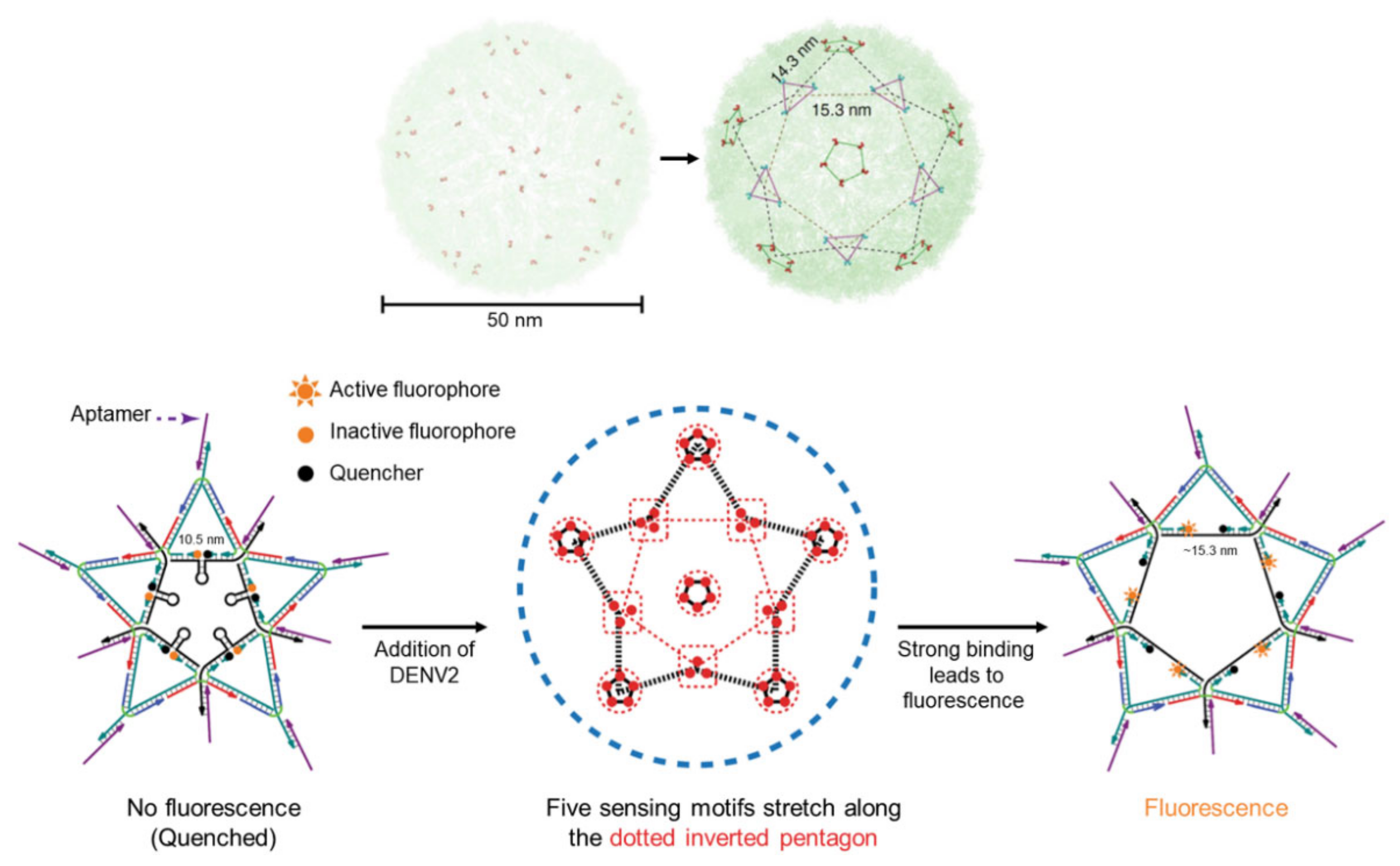
3.2. Biosensors Based on DNA Nanostructures
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.W.; Haag, R. Multivalency as a Chemical Organization and Action Principle. Angew. Chem. Int. Ed. 2012, 51, 10472–10498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, S.C. Virology. Looking inside adenovirus. Science 2010, 329, 1026–1027. [Google Scholar] [CrossRef]
- Bandlow, V.; Liese, S.; Lauster, D.; Ludwig, K.; Netz, R.R.; Herrmann, A.; Seitz, O. Spatial Screening of Hemagglutinin on Influenza A Virus Particles: Sialyl-LacNAc Displays on DNA and PEG Scaffolds Reveal the Requirements for Bivalency Enhanced Interactions with Weak Monovalent Binders. J. Am. Chem. Soc. 2017, 139, 16389–16397. [Google Scholar] [CrossRef]
- Kiessling, L.L.; Gestwicki, J.E.; Strong, L.E. Synthetic multivalent ligands as probes of signal transduction. Angew. Chem. Int. Ed. 2006, 45, 2348–2368. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Jung, Y. Applying Multivalent Biomolecular Interactions for Biosensors. Chem-Eur. J. 2018, 24, 19103–19109. [Google Scholar] [CrossRef]
- Diezmann, F.; Seitz, O. DNA-guided display of proteins and protein ligands for the interrogation of biology. Chem. Soc. Rev. 2011, 40, 5789–5801. [Google Scholar] [CrossRef] [PubMed]
- Yeldell, S.B.; Seitz, O. Nucleic acid constructs for the interrogation of multivalent protein interactions. Chem. Soc. Rev. 2020, 49, 6848–6865. [Google Scholar] [CrossRef]
- Englund, E.A.; Wang, D.Y.; Fujigaki, H.; Sakai, H.; Micklitsch, C.M.; Ghirlando, R.; Martin-Manso, G.; Pendrak, M.L.; Roberts, D.D.; Durell, S.R.; et al. Programmable multivalent display of receptor ligands using peptide nucleic acid nanoscaffolds. Nat. Commun. 2012, 3, 614. [Google Scholar] [CrossRef]
- Abendroth, F.; Bujotzek, A.; Shan, M.; Haag, R.; Weber, M.; Seitz, O. DNA-Controlled Bivalent Presentation of Ligands for the Estrogen Receptor. Angew. Chem. Int. Ed. 2011, 50, 8592–8596. [Google Scholar] [CrossRef]
- Dubel, N.; Liese, S.; Scherz, F.; Seitz, O. Exploring the Limits of Bivalency by DNA-Based Spatial Screening. Angew. Chem. Int. Ed. 2019, 58, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, H.; Diezmann, F.; Seitz, O. DNA as a Molecular Ruler: Interrogation of a Tandem SH2 Domain with Self-Assembled, Bivalent DNA-Peptide Complexes. Angew. Chem. Int. Ed. 2011, 50, 4146–4150. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.; Murat, P.; Defrancq, E. Recent developments in oligonucleotide conjugation. Chem. Soc. Rev. 2010, 39, 2054–2070. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef] [PubMed]
- Wilner, O.I.; Willner, I. Functionalized DNA Nanostructures. Chem. Rev. 2012, 112, 2528–2556. [Google Scholar] [CrossRef]
- Meng, H.M.; Liu, H.; Kuai, H.L.; Peng, R.Z.; Mo, L.T.; Zhang, X.B. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 2016, 45, 2583–2602. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Chen, T.S.; Zhang, Q.Q.; Zhou, M.R.; Zhu, X.L. Application of DNA nanodevices for biosensing. Analyst 2020, 145, 3481–3489. [Google Scholar] [CrossRef]
- Bhatia, D.; Wunder, C.; Johannes, L. Self-assembled, Programmable DNA Nanodevices for Biological and Biomedical Applications. ChemBioChem 2021, 22, 763–778. [Google Scholar] [CrossRef]
- Engelen, W.; Janssen, B.M.G.; Merkx, M. DNA-based control of protein activity. Chem. Commun. 2016, 52, 3598–3610. [Google Scholar] [CrossRef] [Green Version]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gustafsdottir, S.M.; Ostman, A.; Landegren, U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef]
- Robinson, P.V.; Tsai, C.T.; de Groot, A.E.; McKechnie, J.L.; Bertozzi, C.R. Glyco-seek: Ultrasensitive Detection of Protein-Specific Glycosylation by Proximity Ligation Polymerase Chain Reaction. J. Am. Chem. Soc. 2016, 138, 10722–10725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.Q.; Liu, C.L.; Wang, X.Q.; Xu, B.; Liu, X.M.; Li, Y.; Xia, J.; Li, Y.; Zhang, C.; Li, D.N.; et al. O-GlcNAcylation Quantification of Certain Protein by the Proximity Ligation Assay and Clostridium perfringen OGA(D298N)(CpOGA(D298N)). ACS Chem. Biol. 2021, 16, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.Q.; Tian, T.; Lu, Y.Z.; Liu, D.; Huang, M.J.; Zhu, L.; Zhu, Z.; Song, Y.L.; Yang, C.Y. Tracing Tumor-Derived Exosomal PD-L1 by Dual-Aptamer Activated Proximity-Induced Droplet Digital PCR. Angew. Chem. Int. Ed. 2021, 60, 7582–7586. [Google Scholar] [CrossRef]
- Abasiyanik, M.F.; Wolfe, K.; Van Phan, H.; Lin, J.; Laxman, B.; White, S.R.; Verhoef, P.A.; Mutlu, G.M.; Patel, B.; Tay, S. Ultrasensitive digital quantification of cytokines and bacteria predicts septic shock outcomes. Nat. Commun. 2020, 11, 2607. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, M.; Gustafsdottir, S.M.; Schallmeiner, E.; Jarvius, J.; Bjarnegard, M.; Betsholtz, C.; Landegren, U.; Fredriksson, S. Cytokine detection by antibody-based proximity ligation. Proc. Natl. Acad. Sci. USA 2004, 101, 8420–8424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Hu, J.M.; Sollie, R.S.; Easley, C.J. Improvement of Sensitivity and Dynamic Range in Proximity Ligation Assays by Asymmetric Connector Hybridization. Anal. Chem. 2010, 82, 6976–6982. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, S.; Dixon, W.; Ji, H.; Koong, A.C.; Mindrinos, M.; Davis, R.W. Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods 2007, 4, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Gustafsdottir, S.M.; Schlingemann, J.; Rada-Iglesias, A.; Schallmeiner, E.; Kamali-Moghaddam, M.; Wadelius, C.; Landegren, U. In vitro analysis of DNA-protein interactions by proximity ligation. Proc. Natl. Acad. Sci. USA 2007, 104, 3067–3072. [Google Scholar] [CrossRef] [Green Version]
- Heyduk, T. Practical biophysics: Sensors for rapid detection of biological targets utilizing target-induced oligonucleotide annealing. Biophys. Chem. 2010, 151, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, M.; Eriksson, A.; Tran, B.; Assarsson, E.; Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011, 39, e102. [Google Scholar] [CrossRef]
- Li, G.; Moellering, R.E. A Concise, Modular Antibody-Oligonucleotide Conjugation Strategy Based on Disuccinimidyl Ester Activation Chemistry. ChemBioChem 2019, 20, 1599–1605. [Google Scholar] [CrossRef]
- Dovgan, I.; Koniev, O.; Kolodych, S.; Wagner, A. Antibody-Oligonucleotide Conjugates as Therapeutic, Imaging, and Detection Agents. Bioconjug. Chem. 2019, 30, 2483–2501. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; He, L.; Hu, Y.S.; Luo, Z.F.; Zhang, J.J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020, 11, 12157–12164. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.B.; Deng, B.; Wu, P.; Li, F.; Li, X.F.; Le, X.C.; Zhang, H.Q.; Hou, X.D. Amplified binding-induced homogeneous assay through catalytic cycling of analyte for ultrasensitive protein detection. Chem. Commun. 2016, 52, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Genshaft, A.S.; Li, S.; Gallant, C.J.; Darmanis, S.; Prakadan, S.M.; Ziegler, C.G.K.; Lundberg, M.; Fredriksson, S.; Hong, J.; Regev, A.; et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 2016, 17, 188. [Google Scholar] [CrossRef] [PubMed]
- Larssen, P.; Wik, L.; Czarnewski, P.; Eldh, M.; Lof, L.; Ronquist, K.G.; Dubois, L.; Freyhult, E.; Gallant, C.J.; Oelrich, J.; et al. Tracing Cellular Origin of Human Exosomes Using Multiplex Proximity Extension Assays. Mol. Cell. Proteomics 2017, 16, 502–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gidlof, O.; Evander, M.; Rezeli, M.; Marko-Varga, G.; Laurell, T.; Erlinge, D. Proteomic profiling of extracellular vesicles reveals additional diagnostic biomarkers for myocardial infarction compared to plasma alone. Sci. Rep. 2019, 9, 8991. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.J.; Ahmad, K.Z.; Li, S.J.; Warden, A.R.; Su, J.; Zhang, Y.; Yu, Y.Y.; Zhi, X.; Ding, X.T. Pre-coated interface proximity extension reaction assay enables trace protein detection with single-digit accuracy. Biosens. Bioelectron. 2021, 183, 113211. [Google Scholar] [CrossRef] [PubMed]
- Berggrund, M.; Enroth, S.; Lundberg, M.; Assarsson, E.; Stalberg, K.; Lindquist, D.; Hallmans, G.; Grankvist, K.; Olovsson, M.; Gyllensten, U. Identification of Candidate Plasma Protein Biomarkers for Cervical Cancer Using the Multiplex Proximity Extension Assay. Mol. Cell. Proteomics 2019, 18, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Amin, R.A.; Gallant, C.J.; Muthelo, P.M.; Landegren, U. Sensitive Measurement of Drug-Target Engagement by a Cellular Thermal Shift Assay with Multiplex Proximity Extension Readout. Anal. Chem. 2021, 93, 10999–11009. [Google Scholar] [CrossRef]
- Rodriguez, L.; Pekkarinen, P.T.; Lakshmikanth, T.; Tan, Z.Y.; Consiglio, C.R.; Pou, C.; Chen, Y.; Mugabo, C.H.; Nguyen, N.A.; Nowlan, K.; et al. Systems-Level Immunomonitoring from Acute to Recovery Phase of Severe COVID-19. Cell Rep. Med. 2020, 1, 100078. [Google Scholar] [CrossRef] [PubMed]
- Schueder, F.; Lara-Gutierrez, J.; Haas, D.; Beckwith, K.S.; Yin, P.; Ellenberg, J.; Jungmann, R. Super-Resolution Spatial Proximity Detection with Proximity-PAINT. Angew. Chem. Int. Ed. 2021, 60, 716–720. [Google Scholar] [CrossRef]
- Du, M.Y.; Zheng, J.; Tian, S.B.; Liu, Y.C.; Zheng, Z.H.; Wang, H.Z.; Xia, J.B.; Ji, X.H.; He, Z.K. DNAzyme Walker for Homogeneous Detection of Enterovirus EV71 and CVB3. Anal. Chem. 2021, 93, 5606–5611. [Google Scholar] [CrossRef]
- Bertucci, A.; Porchetta, A.; Ricci, F. Antibody-Templated Assembly of an RNA Mimic of Green Fluorescent Protein. Anal. Chem. 2018, 90, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.M.; Wang, T.Y.; Kim, J.; Shannon, C.; Easley, C.J. Quantitation of Femtomolar Protein Levels via Direct Readout with the Electrochemical Proximity Assay. J. Am. Chem. Soc. 2012, 134, 7066–7072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, B.; Chen, J.B.; Zhang, H.Q. Assembly of Multiple DNA Components through Target Binding toward Homogeneous, Isothermally Amplified, and Specific Detection of Proteins. Anal. Chem. 2014, 86, 7009–7016. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.W.; Wu, J.; Zhang, Y.; Yan, F.; Ju, H.X. Proximity Hybridization Regulated DNA Biogate for Sensitive Electrochemical Immunoassay. Anal. Chem. 2014, 86, 7494–7499. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, P.; Hou, X.; Hou, X.D.; Chen, J.B. Accelerating DNA nanomotor by branched DNAzyme for ultrasensitive optical detection of thrombin. Microchem. J. 2018, 139, 260–267. [Google Scholar] [CrossRef]
- Shen, H.W.; Liu, L.Y.; Yuan, Z.W.; Liu, Q.; Li, B.Y.; Zhang, M.; Tang, H.J.; Zhang, J.; Zhao, S.Q. Novel cytosensor for accurate detection of circulating tumor cells based on a dual-recognition strategy and BSA@Ag@Ir metallic-organic nanoclusters. Biosens. Bioelectron. 2021, 179, 113102. [Google Scholar] [CrossRef]
- Zhou, H.; Yu, Q.; Wang, H.Y.; Zhu, W.J.; Liu, J.; Wang, Z.H. A general scattering proximity immunoassay with the formation of dimer of gold nanoparticle. Talanta 2021, 233, 122515. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Mansour, H.; Watson, C.J.F.; Tang, Y.N.; MacNeil, A.J.; Li, F. Amplified detection of nucleic acids and proteins using an isothermal proximity CRISPR Cas12a assay. Chem. Sci. 2021, 12, 2133–2137. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Wang, H.P.; Dong, R.L.; Li, S.Y.; Wang, Z.G.; Liu, S.L.; Pang, D.W. Proximity-induced exponential amplification reaction triggered by proteins and small molecules. Chem. Commun. 2021, 57, 4714–4717. [Google Scholar] [CrossRef]
- Pellejero, L.B.; Mahdifar, M.; Ercolani, G.; Watson, J.; Brown, T.; Ricci, F. Using antibodies to control DNA-templated chemical reactions. Nat. Commun. 2020, 11, 6242. [Google Scholar] [CrossRef]
- Zhang, H.Q.; Li, X.F.; Le, X.C. Binding-Induced DNA Assembly and Its Application to Yoctomole Detection of Proteins. Anal. Chem. 2012, 84, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, H.Q.; Lai, C.; Li, X.F.; Le, X.C. A Molecular Translator that Acts by Binding-Induced DNA Strand Displacement for a Homogeneous Protein Assay. Angew. Chem. Int. Ed. 2012, 51, 9317–9320. [Google Scholar] [CrossRef]
- Liang, H.; Chen, S.; Li, P.P.; Wang, L.P.; Li, J.Y.; Li, J.; Yang, H.H.; Tan, W.H. Nongenetic Approach for Imaging Protein Dimerization by Aptamer Recognition and Proximity-Induced DNA Assembly. J. Am. Chem. Soc. 2018, 140, 4186–4190. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.L.; Yang, P.; Du, L.J.; Xia, L.Y.; Chen, J.B.; Hou, X.D. A signal conversion system using binding-induced strand displacement for disease biomarker assay. Luminescence 2021, 36, 1483–1490. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Lin, R.Y.; Li, T.; Liu, F.; Li, N. The fast detection of streptavidin based on the initial reaction rate of the binding-induced DNA strand-displacement reaction. Anal. Methods 2016, 8, 6701–6704. [Google Scholar] [CrossRef]
- Yang, J.M.; Dou, B.T.; Yuan, R.; Xiang, Y. Aptamer/Protein Proximity Binding-Triggered Molecular Machine for Amplified Electrochemical Sensing of Thrombin. Anal. Chem. 2017, 89, 5138–5143. [Google Scholar] [CrossRef]
- Yang, J.M.; Dou, B.T.; Yuan, R.; Xiang, Y. Proximity Binding and Metal Ion-Dependent DNAzyme Cyclic Amplification-Integrated Aptasensor for Label-Free and Sensitive Electrochemical Detection of Thrombin. Anal. Chem. 2016, 88, 8218–8223. [Google Scholar] [CrossRef]
- Man, Y.; Liu, J.B.; Wu, J.; Yin, L.; Pei, H.; Wu, Q.; Xia, Q.F.; Ju, H.X. An anchored monopodial DNA walker triggered by proximity hybridization for amplified amperometric biosensing of nucleic acid and protein. Anal. Chim. Acta 2020, 1107, 48–54. [Google Scholar] [CrossRef]
- Yun, W.; You, L.F.; Li, F.K.; Wu, H.; Chen, L.; Yang, L.Z. Proximity ligation assay induced and DNAzyme powered DNA motor for fluorescent detection of thrombin. Spectrochim. Acta A 2019, 207, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Gan, H.Y.; Wu, J.; Ju, H.X. Molecular Machine Powered Surface Programmatic Chain Reaction for Highly Sensitive Electrochemical Detection of Protein. Anal. Chem. 2018, 90, 5503–5508. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, K.; Jin, J.J.; Zhang, X.F. Binding-induced output of catalyst DNA for efficient payload of DNAzyme on magnetic beads by catalyzed hairpin assembly. Microchem. J. 2021, 168, 106490. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, K.X.; Liu, G.C.; Liu, M.J.; Liu, Y.; Li, J.H. Label-Free Nanopore Proximity Bioassay for Platelet-Derived Growth Factor Detection. Anal. Chem. 2015, 87, 5677–5682. [Google Scholar] [CrossRef]
- Fan, W.; Chen, J.B.; Du, H.; Hu, C.J.; Yang, P.; Hou, X.D. Activation of catalytic DNAzyme by binding-induced DNA displacement for homogeneous assay. Luminescence 2021, 36, 1498–1506. [Google Scholar] [CrossRef]
- Tang, Y.A.; Wang, Z.X.; Yang, X.L.; Chen, J.B.; Liu, L.N.; Zhao, W.A.; Le, X.C.; Li, F. Constructing real-time, wash-free, and reiterative sensors for cell surface proteins using binding-induced dynamic DNA assembly. Chem. Sci. 2015, 6, 5729–5733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, R.Z.; Zheng, X.F.; Lyu, Y.F.; Xu, L.J.; Zhang, X.B.; Ke, G.L.; Liu, Q.L.; You, C.J.; Huan, S.Y.; Tan, W.H. Engineering a 3D DNA-Logic Gate Nanomachine for Bispecific Recognition and Computing on Target Cell Surfaces. J. Am. Chem. Soc. 2018, 140, 9793–9796. [Google Scholar] [CrossRef]
- Li, F.; Lin, Y.W.; Le, X.C. Binding-Induced Formation of DNA Three-Way Junctions and Its Application to Protein Detection and DNA Strand Displacement. Anal. Chem. 2013, 85, 10835–10841. [Google Scholar] [CrossRef]
- Ren, K.W.; Wu, J.; Yan, F.; Zhang, Y.; Ju, H.X. Immunoreaction-triggered DNA assembly for one-step sensitive ratiometric electrochemical biosensing of protein biomarker. Biosens. Bioelectron. 2015, 66, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, P.; Hou, X.; Zhou, R.X.; Hou, X.D.; Chen, J.B. Expanding DNA nanomachine functionality through binding-induced DNA output for application in clinical diagnosis. Chem. Commun. 2019, 55, 3610–3613. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Q.; Wang, C.; Hao, L.J.; Gao, R.; Li, F.; Liu, S.F. Proximity recognition and polymerase-powered DNA walker for one-step and amplified electrochemical protein analysis. Biosens. Bioelectron. 2019, 128, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Bracaglia, S.; Ranallo, S.; Plaxco, K.W.; Ricci, F. Programmable, Multiplexed DNA Circuits Supporting Clinically Relevant, Electrochemical Antibody Detection. ACS Sens. 2021, 6, 2442–2448. [Google Scholar] [CrossRef]
- Mocenigo, M.; Porchetta, A.; Rossetti, M.; Brass, E.; Tonini, L.; Puzzi, L.; Tagliabue, E.; Triulzi, T.; Marini, B.; Ricci, F.; et al. Rapid, Cost-Effective Peptide/Nucleic Acid-Based Platform for Therapeutic Antibody Monitoring in Clinical Samples. ACS Sens. 2020, 5, 3109–3115. [Google Scholar] [CrossRef]
- Porchetta, A.; Ippodrino, R.; Marini, B.; Caruso, A.; Caccuri, F.; Ricci, F. Programmable Nucleic Acid Nanoswitches for the Rapid, Single-Step Detection of Antibodies in Bodily Fluids. J. Am. Chem. Soc. 2018, 140, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Shi, S.W.; Yang, X.L.; Wang, Y.; Qi, H.L.; Gao, Q.; Zhang, C.X. Separation-Free Electrogenerated Chemiluminescence Immunoassay Incorporating Target Assistant Proximity Hybridization and Dynamically Competitive Hybridization of a DNA Signal Probe. Anal. Chem. 2020, 92, 884–891. [Google Scholar] [CrossRef]
- Yang, X.L.; Liu, W.H.; Chan, D.C.H.; Ahmed, S.U.; Wang, H.; Wang, Z.; Nemr, C.R.; Kelley, S.O. Fluorescent Droplet Cytometry for On-Cell Phenotype Tracking. J. Am. Chem. Soc. 2020, 142, 14805–14809. [Google Scholar] [CrossRef] [PubMed]
- Li, N.X.; Du, M.Y.; Liu, Y.C.; Ji, X.H.; He, Z.K. Multipedal DNA Walker Biosensors Based on Catalyzed Hairpin Assembly and Isothermal Strand-Displacement Polymerase Reaction for the Chemiluminescent Detection of Proteins. ACS Sens. 2018, 3, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.S.; Xin, C.; Liu, S.; Liu, Y.; Liu, S.F. Affinity Binding-Induced Hg2+ Release and Quantum Dot Doping for General, Label-Free, and Homogenous Fluorescence Protein Assay. ACS Sens. 2018, 3, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lin, Y.W.; Lau, A.; Tang, Y.N.; Chen, J.B.; Le, X.C. Binding-Induced Molecular Amplifier as a Universal Detection Platform for Biomolecules and Biomolecular Interaction. Anal. Chem. 2018, 90, 8651–8657. [Google Scholar] [CrossRef]
- Gan, H.Y.; Wu, J.; Ju, H.X. Proximity hybridization-induced on particle DNA walker for ultrasensitive protein detection. Anal. Chim. Acta 2019, 1074, 142–149. [Google Scholar] [CrossRef]
- Qing, M.; Sun, Z.; Wang, L.; Du, S.Z.; Zhou, J.; Tang, Q.; Luo, H.Q.; Li, N.B. CRISPR/Cas12a-regulated homogeneous electrochemical aptasensor for amplified detection of protein. Sens. Actuators B Chem. 2021, 348, 130713. [Google Scholar] [CrossRef]
- Kong, G.Z.; Zhang, M.; Xiong, M.Y.; Fu, X.Y.; Ke, G.L.; Zhang, X.B. DNA nanostructure-based fluorescent probes for cellular sensing. Anal. Methods 2020, 12, 1415–1429. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.Q.; Wang, Z.X.; Li, X.K.; Li, X.F.; Le, X.C. Dynamic DNA Assemblies Mediated by Binding-Induced DNA Strand Displacement. J. Am. Chem. Soc. 2013, 135, 2443–2446. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.H.; Zhen, D.S.; Jiang, L.; Zhou, X.M. Binding-Induced 3D-Bipedal DNA Walker for Cascade Signal Amplification Detection of Thrombin Combined with Catalytic Hairpin Assembly Strategy. Anal. Chem. 2019, 91, 15317–15324. [Google Scholar] [CrossRef]
- Li, X.L.; Wu, Y.Y.; Niu, J.J.; Jiang, D.G.; Xiao, D.; Zhou, C.S. One-step sensitive thrombin detection based on a nanofibrous sensing platform. J. Mater. Chem. B 2019, 7, 5161–5169. [Google Scholar] [CrossRef]
- Hua, X.Y.; Zheng, T.; Zhao, J.M.; Xu, W.J. Recycling of Proximity Binding-Based DNA Architecture Driven by Hairpin Strand Displacement for Amplified Electrochemical Aptasensor. J. Electrochem. Soc. 2019, 166, B1689–B1694. [Google Scholar] [CrossRef]
- Tang, Y.N.; Lin, Y.W.; Yang, X.L.; Wang, Z.X.; Le, X.C.; Li, F. Universal Strategy To Engineer Catalytic DNA Hairpin Assemblies for Protein Analysis. Anal. Chem. 2015, 87, 8063–8066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelen, W.; Meijer, L.H.H.; Somers, B.; de Greef, T.F.A.; Merkx, M. Antibody-controlled actuation of DNA-based molecular circuits. Nat. Commun. 2017, 8, 14473. [Google Scholar] [CrossRef]
- Liu, J.; Lai, T.; Mu, K.J.; Zhou, Z. Strip biosensor for amplified detection of nerve growth factor-beta based on a molecular translator and catalytic DNA circuit. Analyst 2014, 139, 4874–4878. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Zhang, C.; Lv, C.; Sun, Y.; Zhang, M.Z.; Zhao, Y.M.; Yang, L.L.; Han, D.; Tan, W.H. Construction of a Multiple-Aptamer-Based DNA Logic Device on Live Cell Membranes via Associative Toehold Activation for Accurate Cancer Cell Identification. J. Am. Chem. Soc. 2019, 141, 12738–12743. [Google Scholar] [CrossRef]
- Bi, S.; Yue, S.Z.; Zhang, S.S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Gan, X.Y.; Jiang, B.Y.; Yuan, R.; Xiang, Y. Electrode immobilization-free and sensitive electrochemical sensing of thrombin via magnetic nanoparticle-decorated DNA polymers. Sens. Actuators B Chem. 2021, 331, 129395. [Google Scholar] [CrossRef]
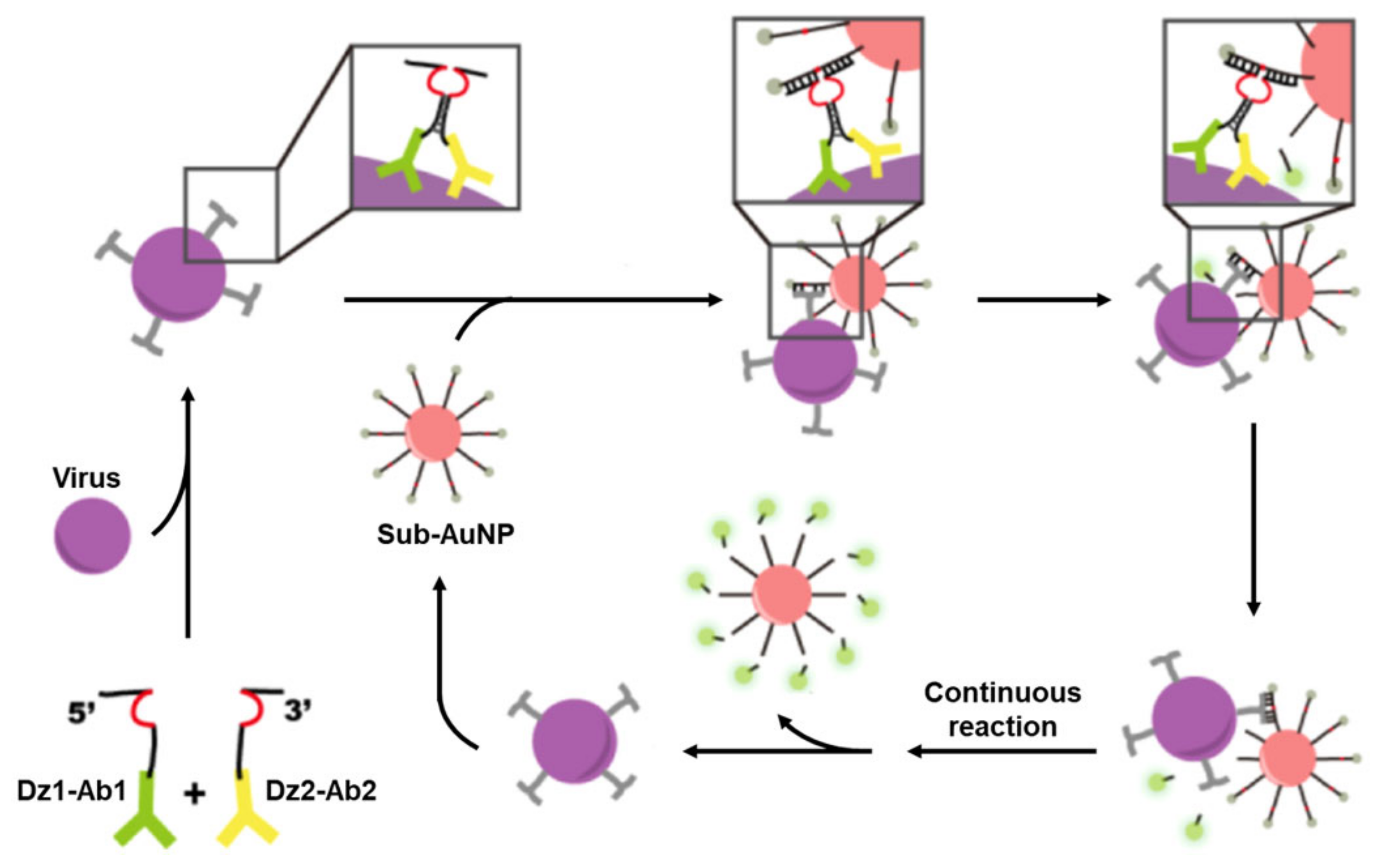
- Du, M.Y.; Mao, G.B.; Tian, S.B.; Liu, Y.C.; Zheng, J.; Ke, X.L.; Zheng, Z.H.; Wang, H.Z.; Ji, X.H.; He, Z.K. Target-Induced Cascade Amplification for Homogeneous Virus Detection. Anal. Chem. 2019, 91, 15099–15106. [Google Scholar] [CrossRef]
- Li, N.; Liu, L.; Xiang, M.H.; Liu, J.W.; Yu, R.Q.; Jiang, J.H. Proximity- induced hybridization chain assembly with small- molecule linked DNA for single- step amplified detection of antibodies. Chem. Commun. 2019, 55, 4387–4390. [Google Scholar] [CrossRef] [PubMed]
- Ranallo, S.; Rossetti, M.; Plaxco, K.W.; Vallee-Belisle, A.; Ricci, F. A Modular, DNA-Based Beacon for Single-Step Fluorescence Detection of Antibodies and Other Proteins. Angew. Chem. Int. Ed. 2015, 54, 13214–13218. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, M.; Trofymchuk, K.; Ranallo, S.; Ricci, F.; Steiner, F.; Cole, F.; Glembockyte, V.; Tinnefeld, P. Single antibody detection in a DNA origami nanoantenna. iScience 2021, 24, 103072. [Google Scholar] [CrossRef]
- Ranallo, S.; Prevost-Tremblay, C.; Idili, A.; Vallee-Belisle, A.; Ricci, F. Antibody-powered nucleic acid release using a DNA-based nanomachine. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.S.; Zhang, S.S.; Wang, L.; Liu, S.F. A Modular Nanoswitch for Mix-and-Detect Protein Assay Based on Binding-Induced Cascade Dissociation of Kissing Complex. ChemBioChem 2018, 19, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Vorobyeva, M.; Vorobjev, P.; Venyaminova, A. Multivalent Aptamers: Versatile Tools for Diagnostic and Therapeutic Applications. Molecules 2016, 21, 1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; He, X.W.; Luo, X.C.; Wang, L.; Ma, N. DNA-Programmed Quantum Dot Polymerization for Ultrasensitive Molecular Imaging of Cancer Cells. Anal. Chem. 2016, 88, 9355–9358. [Google Scholar] [CrossRef]
- Li, Z.; Wang, G.L.; Shen, Y.; Guo, N.N.; Ma, N. DNA-Templated Magnetic Nanoparticle-Quantum Dot Polymers for Ultrasensitive Capture and Detection of Circulating Tumor Cells. Adv. Funct. Mater. 2018, 28, 1707152. [Google Scholar] [CrossRef]
- Ding, C.P.; Zhang, C.L.; Yin, X.Y.; Cao, X.Y.; Cai, M.F.; Xian, Y.Z. Near-Infrared Fluorescent Ag2S Nanodot-Based Signal Amplification for Efficient Detection of Circulating Tumor Cells. Anal. Chem. 2018, 90, 6702–6709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Ali, M.M.; Eckert, M.A.; Kang, D.K.; Chen, Y.Y.; Sender, L.S.; Fruman, D.A.; Zhao, W.A. A polyvalent aptamer system for targeted drug delivery. Biomaterials 2013, 34, 9728–9735. [Google Scholar] [CrossRef]
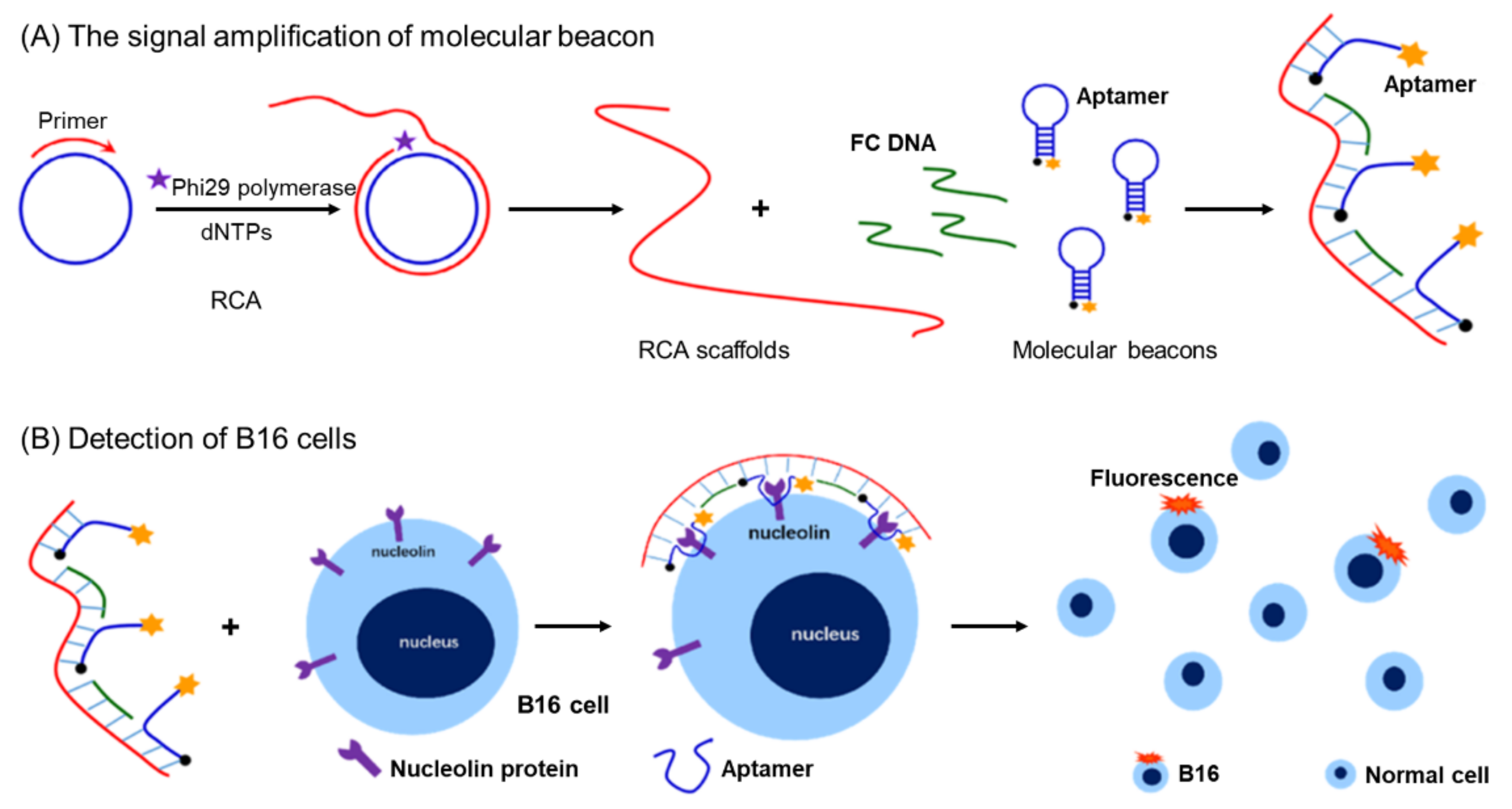
- Zhang, Z.Q.; Wang, S.S.; Ma, J.; Zhou, T.; Wang, F.; Wang, X.F.; Zhang, G.D. Rolling Circle Amplification-Based Polyvalent Molecular Beacon Probe-Assisted Signal Amplification Strategies for Sensitive Detection of B16 Cells. ACS Biomater. Sci. Eng. 2020, 6, 3114–3121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.X.; Chen, F.; Li, Q.; Wang, L.H.; Fan, C.H. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, S.B.; Yu, X.; Hu, S.Y.; Lu, Y.; Wu, Z.S. Periodically Ordered, Nuclease-Resistant DNA Nanowires Decorated with Cell-Specific Aptamers as Selective Theranostic Agents. Angew. Chem. Int. Ed. 2020, 59, 17540–17547. [Google Scholar] [CrossRef] [PubMed]
- Li, W.S.; Yang, X.H.; He, L.L.; Wang, K.M.; Wang, Q.; Huang, J.; Liu, J.B.; Wu, B.; Xu, C.C. Self-Assembled DNA Nanocentipede as Multivalent Drug Carrier for Targeted Delivery. ACS Appl. Mater. Inter. 2016, 8, 25733–25740. [Google Scholar] [CrossRef]
- Krissanaprasit, A.; Key, C.M.; Pontula, S.; LaBean, T.H. Self-Assembling Nucleic Acid Nanostructures Functionalized with Aptamers. Chem. Rev. 2021, 121, 13797–13868. [Google Scholar] [CrossRef]
- Liu, X.W.; Yan, H.; Liu, Y.; Chang, Y. Targeted Cell-Cell Interactions by DNA Nanoscaffold-Templated Multivalent Bispecific Aptamers. Small 2011, 7, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Zhang, J.; Wan, H.; Yan, C.Y.; Xia, F. Rationally Designed Multivalent Aptamers Targeting Cell Surface for Biomedical Applications. ACS Appl. Mater. Interfaces 2021, 13, 9369–9389. [Google Scholar] [CrossRef]
- Machida, T.; Novoa, A.; Gillon, E.; Zheng, S.S.; Claudinon, J.; Eierhoff, T.; Imberty, A.; Romer, W.; Winssinger, N. Dynamic Cooperative Glycan Assembly Blocks the Binding of Bacterial Lectins to Epithelial Cells. Angew. Chem. Int. Ed. 2017, 56, 6762–6766. [Google Scholar] [CrossRef] [Green Version]
- Chidchob, P.; Offenbartl-Stiegert, D.; McCarthy, D.; Luo, X.; Li, J.N.; Howorka, S.; Sleiman, H.F. Spatial Presentation of Cholesterol Units on a DNA Cube as a Determinant of Membrane Protein-Mimicking Functions. J. Am. Chem. Soc. 2019, 141, 1100–1108. [Google Scholar] [CrossRef]
- Huang, D.; Patel, K.; Perez-Garrido, S.; Marshall, J.F.; Palma, M. DNA Origami Nanoarrays for Multivalent Investigations of Cancer Cell Spreading with Nanoscale Spatial Resolution and Single-Molecule Control. ACS Nano 2019, 13, 728–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissanaprasit, A.; Key, C.M.; Froehlich, K.; Pontula, S.; Mihalko, E.; Dupont, D.M.; Andersen, E.S.; Kjems, J.; Brown, A.C.; LaBean, T.H. Multivalent Aptamer-Functionalized Single-Strand RNA Origami as Effective, Target-Specific Anticoagulants with Corresponding Reversal Agents. Adv. Healthc. Mater. 2021, 10, 2001826. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Sun, P.C.; Su, J.J.; Zhang, N.; Gu, H.Z.; Zhao, Y.X. DNA nanotechnology-facilitated ligand manipulation for targeted therapeutics and diagnostics. J. Control. Release 2021, 340, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Rinker, S.; Ke, Y.G.; Liu, Y.; Chhabra, R.; Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 2008, 3, 418–422. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Li, Q.; Wang, L.H.; Zhang, G.J.; Fan, C.H. Framework-Nucleic-Acid-Enabled Biosensor Development. ACS Sens. 2018, 3, 903–919. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.; Lundin, V.; Petrova, E.; Fordos, F.; Benson, E.; Al-Amin, A.; Herland, A.; Blokzijl, A.; Hogberg, B.; Teixeira, A.I. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 2014, 11, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xun, K.Y.; Zheng, L.Y.; Peng, X.Y.; Qiu, L.P.; Tan, W.H. DNA-Based Dynamic Mimicry of Membrane Proteins for Programming Adaptive Cellular Interactions. J. Am. Chem. Soc. 2021, 143, 4585–4592. [Google Scholar] [CrossRef] [PubMed]
- Xun, K.Y.; Pei, K.; Liu, X.J.; Peng, X.Y.; Du, Y.L.; Qiu, L.P.; Tan, W.H. Cell-Membrane-Anchored DNA Nanoplatform for Programming Cellular Interactions. J. Am. Chem. Soc. 2019, 141, 18013–18020. [Google Scholar]
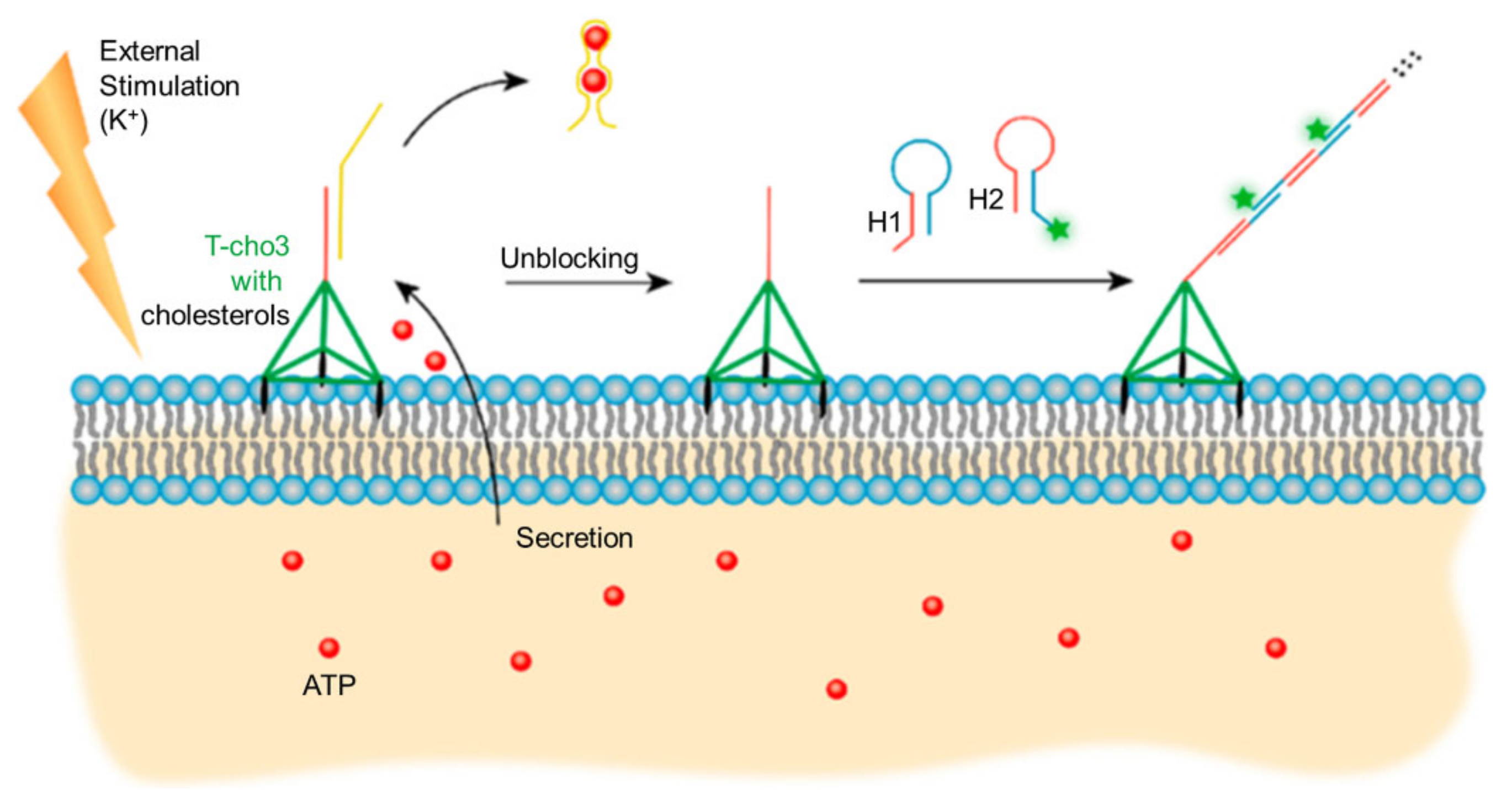
- Peng, P.; Wang, Q.W.; Du, Y.; Wang, H.H.; Shi, L.L.; Li, T. Extracellular Ion-Responsive Logic Sensors Utilizing DNA Dimeric Nanoassemblies on Cell Surface and Application to Boosting AS1411 Internalization. Anal. Chem. 2020, 92, 9273–9280. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.W.; Sun, Y.H.; Li, J.; Li, Q.; Lv, M.; Zhu, B.; Tian, T.; Cheng, D.F.; Xia, J.Y.; Zhang, L.; et al. Multiple-Armed Tetrahedral DNA Nanostructures for Tumor-Targeting, Dual-Modality in Vivo Imaging. ACS Appl. Mater. Interfaces 2016, 8, 4378–4384. [Google Scholar] [CrossRef] [PubMed]
- Song, L.L.; Wang, Z.R.; Liu, J.B.; Wang, T.; Jiang, Q.; Ding, B.Q. Tumor-Targeted DNA Bipyramid for in Vivo Dual -Modality Imaging. ACS Appl. Bio Mater. 2020, 3, 2854–2860. [Google Scholar] [CrossRef]
- Li, M.; Ding, H.M.; Lin, M.H.; Yin, F.F.; Song, L.; Mao, X.H.; Li, F.; Ge, Z.L.; Wang, L.H.; Zuo, X.L.; et al. DNA Framework-Programmed Cell Capture via Topology-Engineered Receptor-Ligand Interactions. J. Am. Chem. Soc. 2019, 141, 18910–18915. [Google Scholar] [CrossRef]
- Gao, Y.; Li, Q.; Zhang, J.; Wu, C.; Shen, Z.; Xue, C.; Chang, H.T.; Wu, Z.S. Bead-String-Shaped DNA Nanowires with Intrinsic Structural Advantages and Their Potential for Biomedical Applications. ACS Appl. Mater. Inter. 2020, 12, 3341–3353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Liu, Q.J.; Tan, J.Z.; Zhan, X.X.; Liu, T.; Wang, Y.T.; Lu, G.; Wu, M.H.; Zhang, Y.Q. Coating with flexible DNA network enhanced T-cell activation and tumor killing for adoptive cell therapy. Acta Pharm. Sin. B. 2021, 11, 1965–1977. [Google Scholar] [CrossRef]
- Liu, J.X.; Li, W.W.; Li, R.S.; Yin, X.Z.; He, S.L.; Hu, J.Q.; Ruan, S.C. Programmable DNA Framework Sensors for In Situ Cell-Surface pH Analysis. Anal. Chem. 2021, 93, 12170–12174. [Google Scholar] [CrossRef]
- Liu, C.; Huang, Z.K.; Jiang, W.; Liu, X.J.; Li, J.; Han, X.Y.; Tu, H.J.; Qiu, L.P.; Tan, W.H. Programmable pH-Responsive DNA Nanosensors for Imaging Exocytosis and Retrieval of Synaptic Vesicles. Anal. Chem. 2020, 92, 3620–3626. [Google Scholar] [CrossRef] [PubMed]
- Kwon, P.S.; Ren, S.; Kwon, S.J.; Kizer, M.E.; Kuo, L.; Xie, M.; Zhu, D.; Zhou, F.; Zhang, F.M.; Kim, D.; et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2020, 12, 26–35. [Google Scholar] [CrossRef]
- Bandlow, V.; Lauster, D.; Ludwig, K.; Hilsch, M.; Reiter-Scherer, V.; Rabe, J.P.; Bottcher, C.; Herrmann, A.; Seitz, O. Sialyl-LacNAc-PNA DNA Concatamers by Rolling-Circle Amplification as Multivalent Inhibitors of Influenza A Virus Particles. ChemBioChem 2019, 20, 159–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachem, G.; Wamhoff, E.C.; Silberreis, K.; Kim, D.; Baukmann, H.; Fuchsberger, F.; Dernedde, J.; Rademacher, C.; Seitz, O. Rational Design of a DNA-Scaffolded High-Affinity Binder for Langerin. Angew. Chem. Int. Ed. 2020, 59, 21016–21022. [Google Scholar] [CrossRef]
- Matsuura, K.; Hibino, M.; Yamada, Y.; Kobayashi, K. Construction of glyco-clusters by self-organization of site-specifically glycosylated oligonucleotides and their cooperative amplification of lectin-recognition. J. Am. Chem. Soc. 2001, 123, 357–358. [Google Scholar] [CrossRef] [PubMed]
- Gorska, K.; Huang, K.T.; Chaloin, O.; Winssinger, N. DNA-Templated Homo- and Heterodimerization of Peptide Nucleic Acid Encoded Oligosaccharides that Mimick the Carbohydrate Epitope of HIV. Angew. Chem. Int. Ed. 2009, 48, 7695–7700. [Google Scholar] [CrossRef]
- Dix, A.V.; Conroy, J.L.; George Rosenker, K.M.; Sibley, D.R.; Appella, D.H. PNA-Based Multivalent Scaffolds Activate the Dopamine D2 Receptor. ACS Med. Chem. Lett. 2015, 6, 425–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.J.; Kim, J.E.; Kim, D.K.; Park, J.H.; Lee, K.B.; Na, D.H. Glucosamine-conjugated Anionic Poly(amidoamine) Dendrimers Inhibit Interleukin-8 Production by Helicobacter pylori in Gastric Epithelial Cells. Bull. Korean Chem. Soc. 2016, 37, 596–599. [Google Scholar] [CrossRef]
- Rosenzweig, B.A.; Ross, N.T.; Tagore, D.M.; Jayawickramarajah, J.; Saraogi, I.; Hamilton, A.D. Multivalent Protein Binding and Precipitation by Self-Assembling Molecules on a DNA Pentaplex Scaffold. J. Am. Chem. Soc. 2009, 131, 5020–5021. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Choi, H.; Heo, Y.; Kim, C.; Kim, M.; Kim, K.T. Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds. Appl. Sci. 2022, 12, 1717. https://doi.org/10.3390/app12031717
Kim H, Choi H, Heo Y, Kim C, Kim M, Kim KT. Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds. Applied Sciences. 2022; 12(3):1717. https://doi.org/10.3390/app12031717
Chicago/Turabian StyleKim, Hokyung, Hayeon Choi, Yoonji Heo, Cheoljae Kim, Min Kim, and Ki Tae Kim. 2022. "Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds" Applied Sciences 12, no. 3: 1717. https://doi.org/10.3390/app12031717
APA StyleKim, H., Choi, H., Heo, Y., Kim, C., Kim, M., & Kim, K. T. (2022). Biosensors Based on Bivalent and Multivalent Recognition by Nucleic Acid Scaffolds. Applied Sciences, 12(3), 1717. https://doi.org/10.3390/app12031717







