Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare
Abstract
:1. Introduction
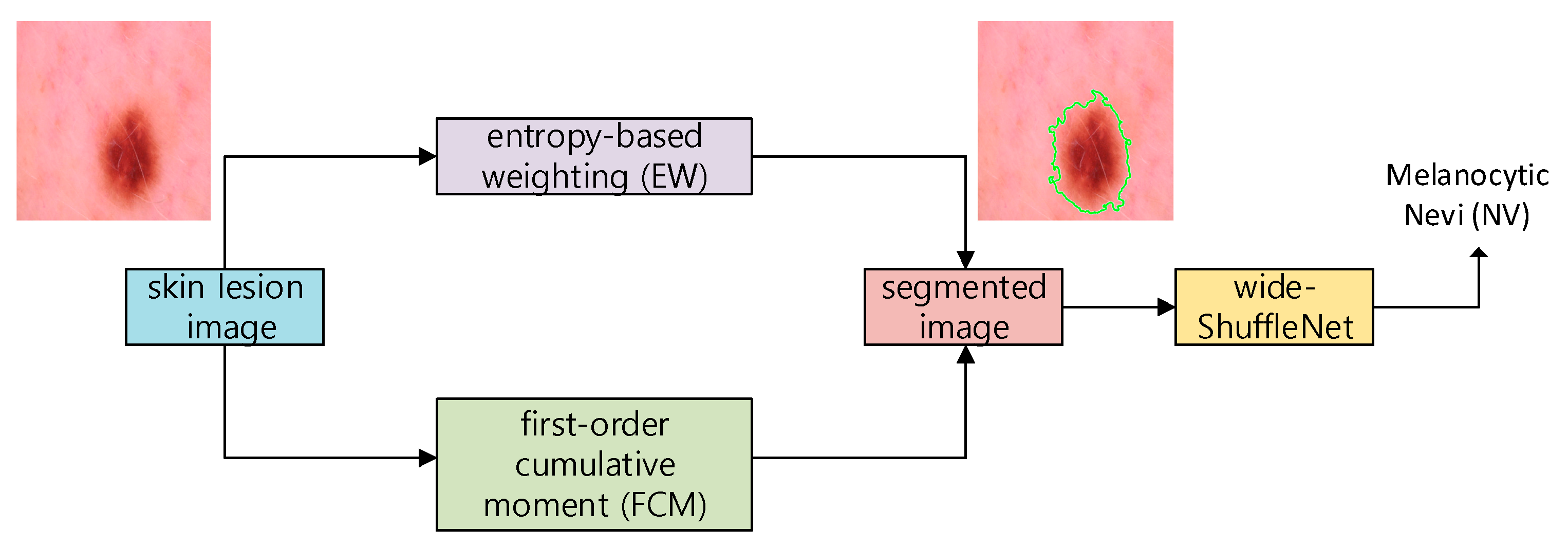
- We propose a novel method to segment the skin image using the entropy-based weighting (EW) and first-order cumulative moment (FCM) of the skin image.
- A two-dimensional wide-ShuffleNet network is applied to classify the segmented image after applying EW-FCM. To the best of our knowledge, EW-FCM and wide-ShuffleNet are novel approaches.
- Based on our numerical results on HAM10000 and ISIC2019 datasets, the proposed framework is more efficient and accurate than state-of-the-art methods.
2. Related Works
2.1. ML Approaches
2.2. DL Approaches
2.2.1. Non-Segmentation DL Approaches
2.2.2. Segmentation DL Approaches
3. Methodology
3.1. EW-FCM Segmentation Technique
3.2. Wide-ShuffleNet
4. Experiment
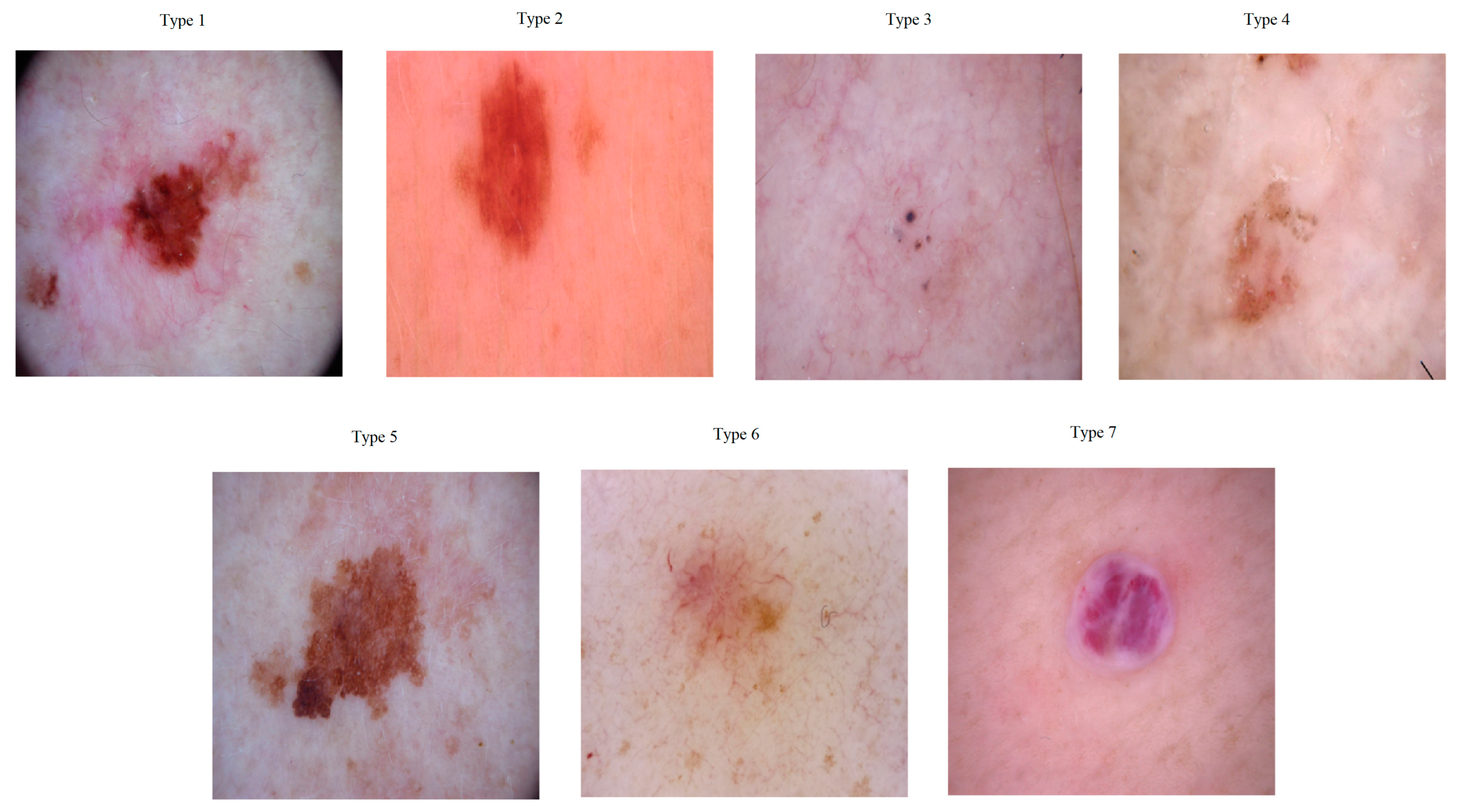
4.1. Datasets
4.2. Evaluation
4.3. Implementation Details
4.4. Comparison of the HAM10000 and ISIC 2019 Datasets
4.4.1. Comparison with Non-Segmentation Methods
4.4.2. Comparison with Segmentation Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AKIEC | Actinic Keratoses and Intraepithelial Carcinoma |
| BCC | Basal Cell Carcinoma |
| BKL | Benign Keratosis-like Lesions |
| BN | Batch Normalization |
| CNN | Convolutional Neural Network |
| DF | Dermatofibroma |
| DL | Deep Learning |
| DWConv | Depthwise Separable Convolution |
| EW | Entropy-based Weighting |
| EWS | Entropy-based Weighting Scheme (including Otsu) |
| FCM | First-order Cumulative Moment |
| GConv | Group Convolution |
| GPU | Graphics Processing Unit |
| KNN | K-Nearest Neighbor |
| MEL | Melanoma |
| ML | Machine Learning |
| NV | Melanocytic Nevi |
| SCC | Squamous Cell Carcinoma |
| SVM | Support Vector Machine |
| VASC | Vascular Lesions |
| WHO | World Health Organization |
References
- Rey-Barroso, L.; Peña-Gutiérrez, S.; Yáñez, C.; Burgos-Fernández, F.J.; Vilaseca, M.; Royo, S. Optical technologies for the improvement of skin cancer diagnosis: A review. Sensors 2021, 21, 252. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions using transfer learning and augmentation with Alex-net. PLoS ONE 2019, 14, e0217293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zicari, R.V.; Ahmed, S.; Amann, J.; Braun, S.A.; Brodersen, J.; Bruneault, F.; Wurth, R. Co-Design of a trustworthy AI System in healthcare: Deep learning based skin lesion classifier. Front. Hum. Dyn. 2021, 3, 40. [Google Scholar] [CrossRef]
- Mishra, N.; Celebi, M. An overview of melanoma detection in dermoscopy images using image processing and machine learning. arXiv 2016, arXiv:1601.07843. [Google Scholar]
- World Health Organization. Radiation: Ultraviolet (UV) Radiation and Skin Cancer. Available online: https://www.who.int/news-room/questions-and-answers/item/radiation-ultraviolet-(uv)-radiation-and-skin-cancer#:~:text=Currently%2C%20between%202%20and%203,skin%20cancer%20in%20their%20lifetime (accessed on 19 October 2021).
- Jerant, A.F.; Johnson, J.T.; Sheridan, C.D.; Caffrey, T.J. Early detection and treatment of skin cancer. Am. Fam. Physician 2000, 62, 357–368. [Google Scholar]
- Trufant, J.; Jones, E. Skin cancer for primary care. In Common Dermatologic Conditions in Primary Care; John, J.R., Edward, F.R., Jr., Eds.; Springer: Cham, Switzerland, 2019; pp. 171–208. [Google Scholar]
- Barata, C.; Celebi, M.E.; Marques, J.S. A survey of feature extraction in dermoscopy image analysis of skin cancer. IEEE J. Biomed. Health Inform. 2019, 23, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Celebi, M.E.; Kingravi, H.A.; Uddin, B.; Iyatomi, H.; Aslandogan, Y.A.; Stoecker, W.V.; Moss, R.H. A methodological approach to the classification of dermoscopy images. Comput. Med. Imaging Graph. 2007, 31, 362–373. [Google Scholar] [CrossRef] [Green Version]
- Tommasi, T.; La Torre, E.; Caputo, B. Melanoma recognition using representative and discriminative kernel classifiers. In Proceedings of the International Workshop on Computer Vision Approaches to Medical Image Analysis (CVAMIA), Graz, Austria, 12 May 2006; pp. 1–12. [Google Scholar] [CrossRef]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P.C. A methodological approach to classify typical and atypical pigment network patterns for melanoma diagnosis. Biomed. Signal Process. Control 2018, 44, 25–37. [Google Scholar] [CrossRef]
- Taner, A.; Öztekin, Y.B.; Duran, H. Performance analysis of deep learning CNN models for variety classification in hazelnut. Sustainability 2021, 13, 6527. [Google Scholar] [CrossRef]
- Wang, W.; Siau, K. Artificial intelligence, machine learning, automation, robotics, future of work and future of humanity: A review and research agenda. J. Database Manag. 2019, 30, 61–79. [Google Scholar] [CrossRef]
- Samuel, A.L. Some studies in machine learning using the game of checkers. II—Recent progress. In Computer Games I; Springer: Berlin/Heidelberg, Germany, 1988; pp. 366–400. [Google Scholar]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, J.; Zhao, Y.; Zhu, S.; He, Y.; Zhang, C. Variety identification of single rice seed using hyperspectral imaging combined with convolutional neural network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef] [Green Version]
- Acquarelli, J.; Van, L.T.; Gerretzen, J.; Tran, T.N.; Buydens, L.M.; Marchiori, E. Convolutional neural networks for vibrational spectroscopic data analysis. Anal. Chim. Acta 2017, 954, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lin, T.; Xu, J.; Luo, X.; Ying, Y. DeepSpectra: An end-to-end deep learning approach for quantitative spectral analysis. Anal. Chim. Acta 2019, 1058, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, Y.; Li, X.; Lau, R.Y.; Zhang, X.; Huang, X. Hyperspectral image classification with deep learning models. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Yu, X.; Tang, L.; Wu, X.; Lu, H. Nondestructive freshness discriminating of shrimp using visible/near-infrared hyperspectral imaging technique and deep learning algorithm. Food Anal. Methods 2018, 11, 768–780. [Google Scholar] [CrossRef]
- Yue, J.; Mao, S.; Li, M. A deep learning framework for hyperspectral image classification using spatial pyramid pooling. Remote Sens. Lett. 2016, 7, 875–884. [Google Scholar] [CrossRef]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep learning meets hyperspectral image analysis: A multidisciplinary review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef] [Green Version]
- Thurnhofer-Hemsi, K.; López-Rubio, E.; Domínguez, E.; Elizondo, D.A. Skin lesion classification by ensembles of deep convolutional networks and regularly spaced shifting. IEEE Access 2021, 9, 112193–112205. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, C.; Thurnhofer-Hemsi, K.; Soroushmehr, R.; Mishra, A.; Gryak, J.; Dominguez, E.; Najarian, K.; Lopez-Rubio, E. Diabetic wound segmentation using convolutional neural networks. In Proceedings of the 41th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 1002–1005. [Google Scholar]
- Thurnhofer-Hemsi, K.; Domínguez, E. Analyzing digital image by deep learning for melanoma diagnosis. In Advances in Computational Intelligence; Rojas, I., Joya, G., Catala, A., Eds.; Springer: Cham, Switzerland, 2019; pp. 270–279. [Google Scholar]
- Thurnhofer-Hemsi, K.; Domínguez, E. A convolutional neural network framework for accurate skin cancer detection. Neural Process. Lett. 2021, 53, 3073–3093. [Google Scholar] [CrossRef]
- Codella, N.C.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar] [CrossRef] [Green Version]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Jiang, N.; Hu, H.; Ji, Z.; Zhang, W. Image classification based on quantum K-nearest-neighbor algorithm. Quantum Inf. Process. 2018, 17, 239. [Google Scholar] [CrossRef]
- Sumithra, R.; Suhil, M.; Guru, D.S. Segmentation and classification of skin lesions for disease diagnosis. Procedia Comput. Sci. 2015, 45, 76–85. [Google Scholar] [CrossRef] [Green Version]
- Sajid, P.M.; Rajesh, D.A. Performance evaluation of classifiers for automatic early detection of skin cancer. J. Adv. Res. Dyn. Control. Syst. 2018, 10, 454–461. [Google Scholar]
- Zhang, S.; Wu, Y.; Chang, J. Survey of image recognition algorithms. In Proceedings of the 2020 IEEE 4th Information Technology, Networking, Electronic and Automation Control Conference (ITNEC), Chongqing, China, 12–14 June 2020; pp. 542–548. [Google Scholar] [CrossRef]
- Alam, M.; Munia, T.T.K.; Tavakolian, K.; Vasefi, F.; MacKinnon, N.; Fazel-Rezai, R. Automatic detection and severity measurement of eczema using image processing. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1365–1368. [Google Scholar] [CrossRef]
- Immagulate, I.; Vijaya, M.S. Categorization of non-melanoma skin lesion diseases using support vector machine and its variants. Int. J. Med. Imaging 2015, 3, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, P.K.; Chandra, S. An improved bag of dense features for skin lesion recognition. J. King Saud Univ. Comput. Inf. Sci. 2019; in press. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support vector machines for classification. In Efficient Learning Machines; Apress: Berkeley, CA, USA, 2015; pp. 39–66. [Google Scholar]
- Hsu, W. Bayesian classification. In Encyclopedia of Database Systems, 2nd ed.; Liu, L., Özsu, M.T., Eds.; Springer: New York, NY, USA, 2018; pp. 3854–3857. [Google Scholar]
- Tahmassebi, A.; Gandomi, A.; Schulte, M.; Goudriaan, A.; Foo, S.; Meyer-Base, A. Optimized naive-bayes and decision tree approaches for fMRI smoking cessation classification. Complexity 2018, 2018, 2740817. [Google Scholar] [CrossRef]
- Seixas, J.L.; Mantovani, R.G. Decision trees for the detection of skin lesion patterns in lower limbs ulcers. In Proceedings of the 2016 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 15–17 December 2018; pp. 677–681. [Google Scholar] [CrossRef]
- Arasi, M.A.; El-Horbaty, E.S.M.; El-Sayed, A. Classification of dermoscopy images using naive bayesian and decision tree techniques. In Proceedings of the 2018 1st Annual International Conference on Information and Sciences (AiCIS), Fallujah, Iraq, 20–21 November 2018; pp. 7–12. [Google Scholar] [CrossRef]
- Hamad, M.A.; Zeki, A.M. Accuracy vs. cost in decision trees: A survey. In Proceedings of the 2018 International Conference on Innovation and Intelligence for Informatics, Computing, and Technologies (3ICT), Sakhier, Bahrain, 18–20 November 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Serte, S.; Demirel, H. Gabor wavelet-based deep learning for skin lesion classification. Comput. Biol. Med. 2019, 113, 103423. [Google Scholar] [CrossRef]
- Menegola, A.; Tavares, J.; Fornaciali, M.; Li, L.T.; Avila, S.; Valle, E. RECOD Titans at ISIC Challenge 2017. 2017. Available online: https://arxiv.org/abs/1703.04819 (accessed on 19 October 2021).
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, Y.; Otomo, Y.; Ogata, Y.; Nakamura, Y.; Fujita, R.; Ishitsuka, Y.; Fujimoto, M. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses boardcertified dermatologists in skin tumour diagnosis. Br. J. Dermatol. 2019, 180, 373–381. [Google Scholar] [CrossRef]
- Mahbod, A.; Ecker, R.; Ellinger, I. Skin lesion classification using hybrid deep neural networks. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1229–1233. [Google Scholar]
- Harangi, B. Skin lesion classification with ensembles of deep convolutional neural networks. Biomed. Inf. 2018, 86, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Nyíri, T.; Kiss, A. Novel ensembling methods for dermatological image classification. In Proceedings of the International Conference on Theory and Practice of Natural Computing, Dublin, Ireland, 12–14 December 2018; pp. 438–448. [Google Scholar]
- Matsunaga, K.; Hamada, A.; Minagawa, A.; Koga, H. Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. arXiv 2017, arXiv:1703.03108. [Google Scholar]
- Li, Y.; Shen, L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef] [Green Version]
- Díaz, I.G. Incorporating the knowledge of dermatologists to convolutional neural networks for the diagnosis of skin lesions. arXiv 2017, arXiv:1703.01976v1. [Google Scholar]
- Son, H.M.; Jeon, W.; Kim, J.; Heo, C.Y.; Yoon, H.J.; Park, J.U.; Chung, T.M. AI-based localization and classification of skin disease with erythema. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Al-Masni, M.A.; Kim, D.H.; Kim, T.S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef]
- Truong, M.T.N.; Kim, S. Automatic image thresholding using Otsu’s method and entropy weighting scheme for surface defect detection. Soft Comput. 2018, 22, 4197–4203. [Google Scholar] [CrossRef]
- Zhan, Y.; Zhang, G. An improved OTSU algorithm using histogram accumulation moment for ore segmentation. Symmetry 2019, 11, 431. [Google Scholar] [CrossRef] [Green Version]
- Zade, S. Medical-Image-Segmentation. Available online: https://github.com/mathworks/Medical-Image-Segmentation/releases/tag/v1.0 (accessed on 19 October 2021).
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems 28 (NIPS 2015), Montreal, QC, Canada, 7–12 December 2015; pp. 91–99. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. arXiv 2017, arXiv:1707.01083. [Google Scholar]
- He, K.; Sun, J. Convolutional neural networks at constrained time cost. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 5353–5360. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 8–10 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016. [Google Scholar] [CrossRef] [Green Version]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. Squeezenet: Alexnet-level accuracy with 50× fewer parameters and 0.5 mb model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. arXiv 2016, arXiv:1611.05431. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Zhang, T.; Qi, G.; Xiao, B.; Wang, J. Interleaved group convolutions for deep neural networks. arXiv 2017, arXiv:1707.02725. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. arXiv 2016, arXiv:1610.02357. [Google Scholar]
- Zagoruyko, S.; Komodaki, N. Wide residual networks. arXiv 2016, arXiv:1605.07146. [Google Scholar]
- Hoang, H.H.; Trinh, H.H. Improvement for convolutional neural networks in image classification using long skip connection. Appl. Sci. 2021, 11, 2092. [Google Scholar] [CrossRef]
- Yahya, A.A.; Tan, J.; Hu, M. A novel handwritten digit classification system based on convolutional neural network approach. Sensors 2021, 21, 6273. [Google Scholar] [CrossRef]
- Xu, B.; Wang, N.; Chen, T.; Li, M. Empirical evaluation of rectified activations in convolutional network. arXiv 2015, arXiv:1505.00853. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Zanddizari, H.; Nguyen, N.; Zeinali, B.; Chang, J.M. A new preprocessing approach to improve the performance of CNN-based skin lesion classification. Med. Biol. Eng. Comput. 2021, 59, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Milton, M.A.A. Automated skin lesion classification using ensemble of deep neural networks in ISIC 2018: Skin lesion analysis towards melanoma detection challenge. arXiv 2019, arXiv:1901.10802. [Google Scholar]
- Ray, S. Disease classification within dermascopic images using features extracted by ResNet50 and classification through deep forest. arXiv 2018, arXiv:1807.05711. [Google Scholar]
- Perez, F.; Avila, S.; Valle, E. Solo or ensemble? Choosing a CNN architecture for melanoma classification. arXiv 2019, arXiv:1904.12724. [Google Scholar]
- Gessert, N.; Sentker, T.; Madesta, F.; Schmitz, R.; Kniep, H.; Baltruschat, I.; Werner, R.; Schlaefer, A. Skin lesion diagnosis using ensembles, unscaled multi-crop evaluation and loss weighting. arXiv 2018, arXiv:1808.01694. [Google Scholar]
- Mobiny, A.; Singh, A.; Van Nguyen, H. Risk-aware machine learning classifier for skin lesion diagnosis. J. Clin. Med. 2019, 8, 1241. [Google Scholar] [CrossRef] [Green Version]
- Naga, S.P.; Rao, T.; Balas, V. A systematic approach for identification of tumor regions in the human brain through HARIS algorithm. In Deep Learning Techniques for Biomedical and Health Informatics; Academic Press: Cambridge, MA, USA, 2020; pp. 97–118. [Google Scholar] [CrossRef]
- Cetinic, E.; Lipic, T.; Grgic, S. Fine-tuning convolutional neural networks for fine art classification. Expert Syst. Appl. 2018, 114, 107–118. [Google Scholar] [CrossRef]
- Rathod, J.; Waghmode, V.; Sodha, A.; Bhavathankar, P. Diagnosis of skin diseases using convolutional neural networks. In Proceedings of the 2018 Second International Conference on Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 29–31 March 2018; pp. 1048–1051. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Hartanto, C.A.; Wibowo, A. Development of mobile skin cancer detection using faster R-CNN and MobileNet V2 model. In Proceedings of the 2020 7th International Conference on Information Technology, Computer, and Electrical Engineering (ICITACEE), Semarang, Indonesia, 24–25 September 2020; pp. 58–63. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]






| Methods | Dataset | Number of Images | Number of Classes |
|---|---|---|---|
| Menegola et al. [44] | ISIC 2017 | 2000 | 2 |
| Han et al. [45] | Asan | 17,125 | 12 |
| Esteva et al. [46] | DERMOFIT | 1300 | 10 |
| Fujisawa et al. [47] | University of Tsukuba Hospital | 6009 | 21 |
| Mahbod et al. [48] | ISIC 2017 | 2000 | 2 |
| Harangi [49] | ISIC 2017 | 2000 | 2 |
| Nyiri and Kiss [50] | ISIC 2017 | 2000 | 2 |
| Matsunaga et al. [51] | ISIC 2017 | 2000 | 2 |
| Li and Shen [52] | ISIC 2017 | 2000 | 2 |
| Gonzalez-Diaz [53] | ISIC 2017 | 2000 | 2 |
| Unet [54] | DERMNET | 15,851 | 18 |
| Frcn [55] | HAM10000 | 10,015 | 7 |
| Class Name | HAM10000 Number of Images | ISIC2019 Number of Images |
|---|---|---|
| AKIEC | 327 | 867 |
| BCC | 514 | 3323 |
| BKL | 1099 | 2624 |
| DF | 115 | 239 |
| MEL | 1113 | 4522 |
| NV | 6705 | 12,875 |
| SCC | - | 628 |
| VASC | 142 | 253 |
| Total | 10,015 | 25,331 |
| Experiment | Method | ACC |
|---|---|---|
| Experiment 1 HAM10000 (80% training, 20% testing) | PNASNet [77] | 76.00% |
| ResNet-50 + gcForest [78] | 80.04% | |
| VGG-16 + GoogLeNet Ensemble [79] | 81.50% | |
| Densenet-121 with SVM [80] | 82.70% | |
| DenseNet-169 [80] | 81.35% | |
| Bayesian DenseNet-169 [81] | 83.59% | |
| Shifted MobileNetV2 [23] | 81.90% | |
| Shifted GoogLeNet [23] | 80.50% | |
| Shifted 2-Nets [23] | 83.20% | |
| The proposed method | 84.80% | |
| Experiment 2 HAM 10000 (90% training, 10% testing) | HARTS [82] | 77.00% |
| FTNN [83] | 79.00% | |
| CNN [84] | 80.00% | |
| VGG19 [85] | 81.00% | |
| MobileNet V1 [68] | 82.00% | |
| MobileNet V2 [86] | 84.00% | |
| MobileNet V2-LSTM [29] | 85.34% | |
| The proposed method | 86.33% | |
| Experiment 3 ISIC 2019 (90% training, 10% testing) | VGG19 [85] | 80.17% |
| ResNet-152 [64] | 84.15% | |
| Efficient-B0 [87] | 81.75% | |
| Efficient-B7 [87] | 84.87% | |
| The proposed method | 82.56% |
| Methods | ||||
|---|---|---|---|---|
| Shifted MobileNetV2 | Shifted GoogLeNet | Shifted 2-Nets | The Proposed Method | |
| Specificity | 95.20% | 94.70% | 95.30% | 96.03% |
| Sensitivity | 65.90% | 58.10% | 64.40% | 70.71% |
| Precision | 71.40% | 68.50% | 76.10% | 75.15% |
| F1 score | 67.00% | 60.80% | 67.80% | 72.61% |
| Accuracy | 81.90% | 80.50% | 83.20% | 84.80% |
| Parameters | 3.4 M | 7 M | 10.4 M | 1.8 M |
| Method | Accuracy | Specificity | Sensitivity | Parameters |
|---|---|---|---|---|
| HARTS [82] | 77.00% | 83.00% | 78.21% | - |
| FTNN [83] | 79.00% | 84.00% | 79.54% | - |
| VGG19 [85] | 81.00% | 87.00% | 82.46% | 143 M |
| MobileNet V1 [68] | 82.00% | 89.00% | 84.04% | 4.2 M |
| MobileNet V2 [86] | 84.00% | 90.00% | 86.41% | 3.4 M |
| MobileNet V2-LSTM [29] | 85.34% | 92.00% | 88.24% | 3.4 M |
| The proposed method | 86.33% | 97.72% | 86.33% | 1.8 M |
| Method | Accuracy | Specificity | Sensitivity | Parameters |
|---|---|---|---|---|
| VGG19 [85] | 80.17% | - | - | 143 M |
| ResNet-152 [64] | 84.15% | - | - | 50 M |
| Efficient-B0 [87] | 81.75% | - | - | 5 M |
| Efficient-B7 [87] | 84.87% | 66 M | ||
| The proposed method | 82.56% | 97.51% | 82.56% | 1.8 M |
| Experiment | Method | ACC | Sensitivity | Specificity |
|---|---|---|---|---|
| Experiment 1 HAM10000 (80% training, 20% testing) | Original Otsu + wide-ShuffleNet | 79.62% | 79.62% | 96.60% |
| Otsu Momentum [57] + wide-ShuffleNet | 81.51% | 81.51% | 96.92% | |
| Image Entropy [56] + wide-ShuffleNet | 83.91% | 83.91% | 97.32% | |
| U-Net + wide-ShuffleNet | 85.10% | 85.10% | 97.52% | |
| U-Net + EfficientNet-B0 [54] | 85.65% | 85.65% | 97.61% | |
| FrCN + Inception-ResNet-v2 [55] | 87.74% | - | - | |
| FrCN + Inception-v3 [55] | 88.05% | - | - | |
| FrCN + DenseNet-201 [55] | 88.70% | - | - | |
| EW-FCM + wide-ShuffleNet | 84.80% | 84.80% | 97.48% | |
| Experiment 2 HAM 10000 (90% training, 10% testing) | Original Otsu + wide-ShuffleNet | 80.14% | 80.14% | 96.69% |
| Otsu Momentum [57] + wide-ShuffleNet | 82.54% | 82.54% | 97.09% | |
| Image Entropy [56] + wide-ShuffleNet | 84.83% | 84.83% | 97.47% | |
| EW-FCM + wide-ShuffleNet | 86.33% | 86.33% | 97.72% | |
| Experiment 3 ISIC 2019 (90% training, 10% testing) | Original Otsu + wide-ShuffleNet | 78.55% | 78.55% | 96.94% |
| Otsu Momentum [57] + wide-ShuffleNet | 80.34% | 80.34% | 97.19% | |
| Image Entropy [56] + wide-ShuffleNet | 81.20% | 81.20% | 97.31% | |
| EW-FCM + wide-ShuffleNet | 82.56% | 82.56% | 97.51% |
| Experiment | Method | ACC | Sensitivity | Specificity |
|---|---|---|---|---|
| Experiment 1 HAM10000 (80% training, 20% testing) | Original Image + ShuffleNet | 76.83% | 76.83% | 96.14% |
| Original Image + wide-ShuffleNet | 77.88% | 77.88% | 96.31% | |
| EW-FCM + ShuffleNet | 83.66% | 83.66% | 97.28% | |
| EW-FCM + wide-ShuffleNet | 84.80% | 84.80% | 97.48% | |
| EW-FCM + EfficientNet-B0 | 85.50% | 85.50% | 97.58% | |
| Experiment 2 HAM 10000 (90% training, 10% testing) | Original Image + ShuffleNet | 77.25% | 77.25% | 96.21% |
| Original Image + wide-ShuffleNet | 78.64% | 78.64% | 96.44% | |
| EW-FCM + ShuffleNet | 84.43% | 84.43% | 97.41% | |
| EW-FCM + wide-ShuffleNet | 86.33% | 86.33% | 97.72% | |
| EW-FCM + EfficientNet-B0 | 87.23% | 87.23% | 97.87% | |
| Experiment 3 ISIC 2019 (90% training, 10% testing) | Original Image + ShuffleNet | 74.23% | 74.23% | 96.32% |
| Original Image + wide-ShuffleNet | 75.73% | 75.73% | 96.53% | |
| EW-FCM + ShuffleNet | 81.62% | 81.62% | 97.38% | |
| EW-FCM + wide-ShuffleNet | 82.56% | 82.56% | 97.51% | |
| EW-FCM + EfficientNet-B0 | 84.66% | 84.66% | 97.81% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoang, L.; Lee, S.-H.; Lee, E.-J.; Kwon, K.-R. Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare. Appl. Sci. 2022, 12, 2677. https://doi.org/10.3390/app12052677
Hoang L, Lee S-H, Lee E-J, Kwon K-R. Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare. Applied Sciences. 2022; 12(5):2677. https://doi.org/10.3390/app12052677
Chicago/Turabian StyleHoang, Long, Suk-Hwan Lee, Eung-Joo Lee, and Ki-Ryong Kwon. 2022. "Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare" Applied Sciences 12, no. 5: 2677. https://doi.org/10.3390/app12052677
APA StyleHoang, L., Lee, S.-H., Lee, E.-J., & Kwon, K.-R. (2022). Multiclass Skin Lesion Classification Using a Novel Lightweight Deep Learning Framework for Smart Healthcare. Applied Sciences, 12(5), 2677. https://doi.org/10.3390/app12052677






