Survey of BERT-Base Models for Scientific Text Classification: COVID-19 Case Study
Abstract
:1. Introduction
1.1. Context
- First, to produce an appropriate dataset for training. Our research is oriented to multiclass classification rather than to multilabel classification. When we began our research in January 2020, we could not find any datasets on COVID-19 with short text that is classified according to categories. For this reason, we were required to build a new dataset on COVID-19 by using the PubMed search engine, and it was validated by experts from Community and Preventive Medicine at the Faculty of Medicine, Department of Epidemiology. The validation process took over 6 months. We advance a method to build the required dataset and address our needs.
- 2.
- Based on the literature, we considered concerns and issues that can now be addressed through natural language processing (NLP) based on different pre-trained language models including Glove [5], ELMo [6], OpenAI GPT model [7] and Bidirectional Encoder Representations from Transformers (abbreviated as BERT) [8]. Compared to the cited models, BERT provides better results for many use cases and without necessarily requiring a large amount of labelled data thanks to a “pre-training” phase without labels, allowing it to acquire a more detailed knowledge of the language. In addition, the BERT model uses a specific manner to handle several limits such as the reduced size of input text and the lack of vocabulary as was our case when we deal with the summary of scientific articles. Bearing in mind the several benefits of BERT, we propose the CovBERT model to help medical research epidemiologists fight against COVID-19. We have released the CovBERT model as a new resource to enhance the performance of NLP tasks in the scientific domain. Our proposed model is a pre-trained language model based on BERT and trained on our large corpus of scientific text, Cov-Dat-20. After training, we fine-tuned the CovBERT model with a specific subject related to COVID-19. Finally, we evaluated the CovBERT model on different numerously studied text classification datasets.
1.2. Traditional ML versus DL Models
- Feed an algorithm with data;
- Use this data to train a model;
- Test and deploy the model;
- Use the deployed model to perform an automated predictive task.
2. Related Works
3. Methodology
3.1. COV-Dat-20 Dataset Creation
- COVID-19: focus on the (1) COVID-19 treatment and (2) COVID-19 symptoms: (1) treatment, chloroquine, hydroxychloroquine, interferon beta-1a, remdesivir, lopinavir/ritonavir; (2) fever, headache, cough, chills, shortness of breath or difficulty breathing, muscle pain, repeated shaking with chills, new loss of taste or smell;
- Virology: genome sequencing, phylogenetic analysis, SARS-CoV-2, MERS-CoV, nomenclature, virus composition, virus layers;
- Public Health: COVID-19, interventions, awareness, behavior, behavioral change, coronavirus, pandemic, public health protection, public health measures;
- Mental Health: COVID-19, mental health disorders, SARS-CoV-2, neural taxonomies, personalized medicine, precision psychiatry, social connection, mental health, psychiatry.
3.2. Data Pre-Processing
- Lowercasing is a widespread approach to reduce all the text to lower case for simplicity.
- Tokenization: text pre-processing step, which involves splitting the text into tokens (words, sentences, etc.)
- Cleaning is a form of pre-processing to filter out useless data such as punctuation removal and stop-word removal (a stop word is a commonly used word in text and stop-word removal is a form pre-processing to filter out useless data).
- Lemmatization is an alternative approach to stemming for removing inflection.
3.3. Exploration of the BERT Model
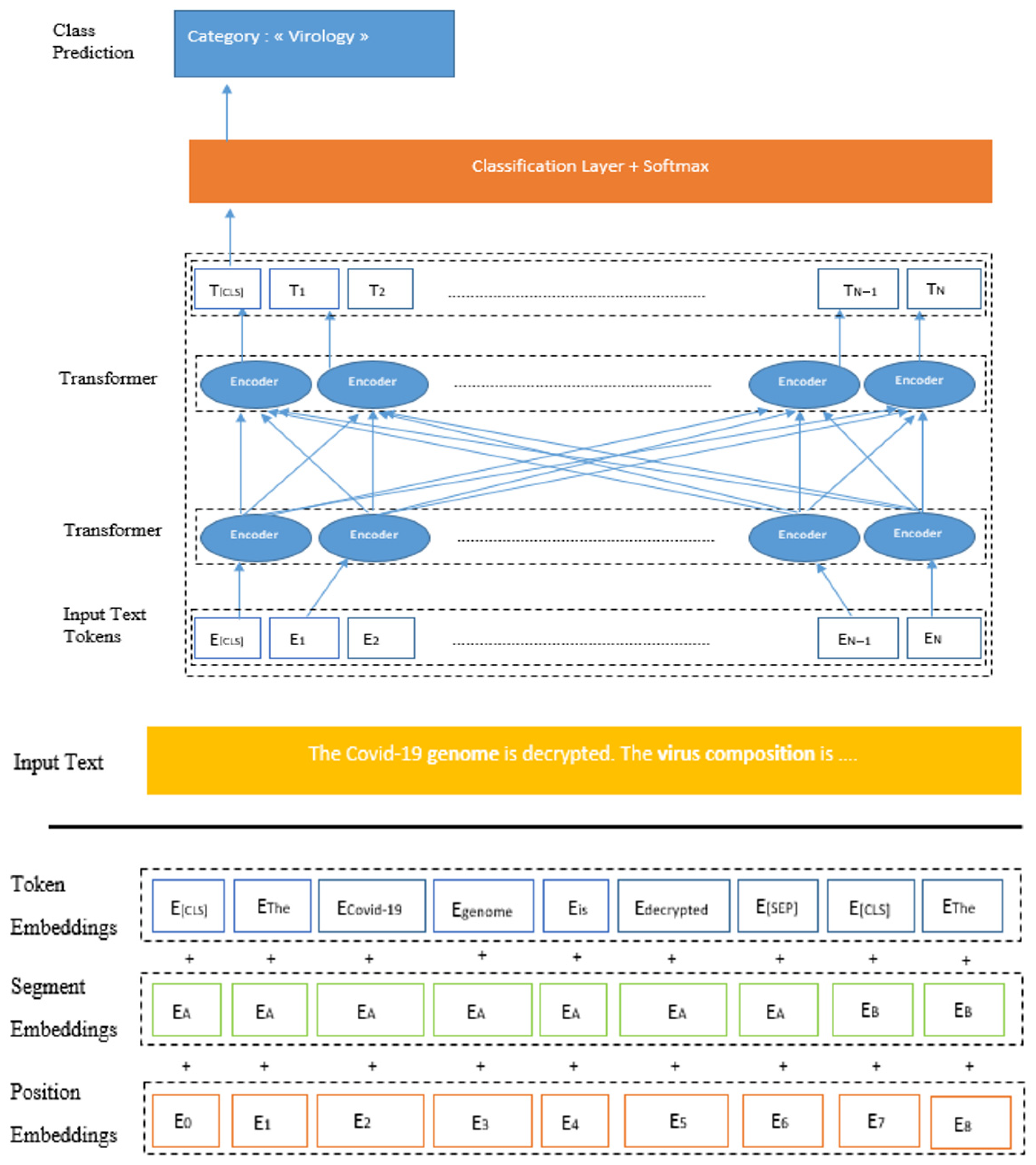
3.3.1. BERT-Base Characteristics
3.3.2. BERT-Base Operation
3.3.3. Contextual Embeddings in Biomedical and Scientific Domains
3.3.4. Self-Attention Mechanism
4. Pre-Training BERT-Base on COV-Dat-20
4.1. Importing Libraires
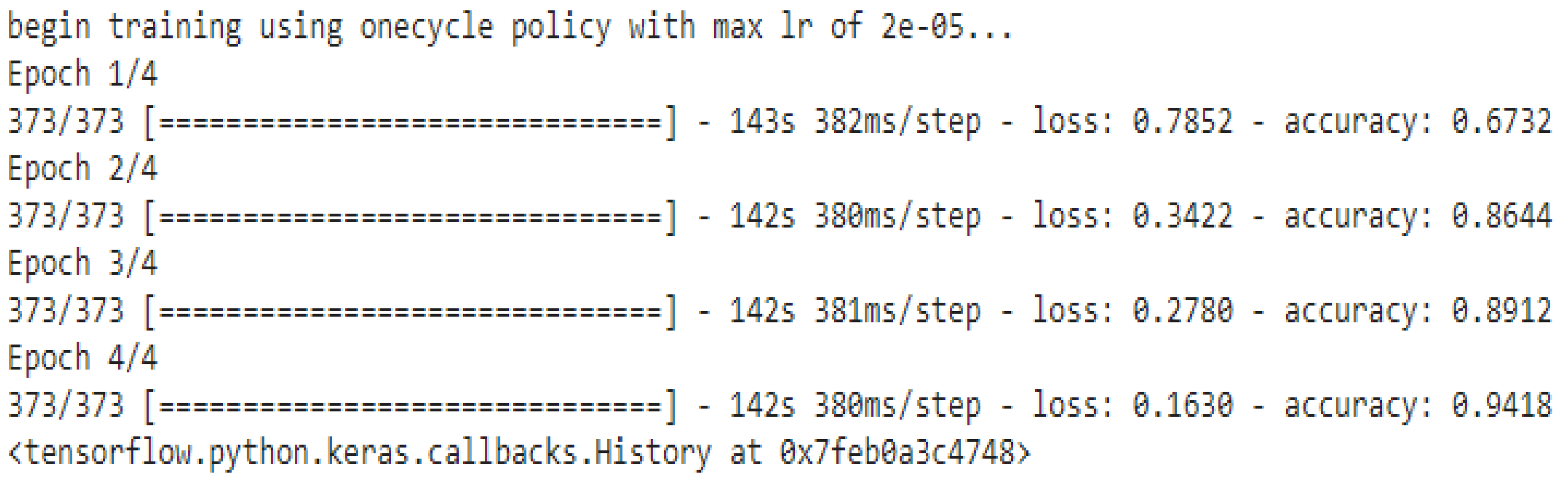
4.2. Needed Parameters for Training
- Max Length: 64;
- Batch size: 32;
- Learning rate (Adam): 2e-5;
- Number of epochs: 4;
- Seed val.: 42.
4.3. Model Characteristics
4.4. Evaluation Metrics
- TP: the positive class correctly predicted by the models;
- FP: the positive class incorrectly predicted by the models;
- TN: the false class correctly predicted by the models;
- FN: the false class incorrectly predicted by the models.
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zu, Z.; Jiang, M.; Xu, P.; Chen, W.; Ni, Q.; Lu, G.; Zhang, L. Coronavirus disease 2019 (COVID-19): A perspective from china. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worldometers for COVID-19. Available online: https://www.worldometers.info/ (accessed on 30 January 2020).
- Pan, S.; Yang, Q. A survey on transfer learning. Knowledge and Data Engineering. IEEE Trans. 2010, 1345–1359. [Google Scholar]
- Yousaf, A.; Asif, R.M.; Shakir, M.; Rehman, A.U.; Alassery, F.; Hamam, H.; Cheikhrouhou, O. A Novel Machine Learning-Based Price Forecasting for Energy Management Systems. Sustainability 2021, 13, 12693. [Google Scholar] [CrossRef]
- Pennington, J.; Socher, R.; Manning, C. Glove: Global vectors for word representation. In Proceedings of the Empiricial Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; Volume 12, pp. 1532–1543. [Google Scholar]
- Peters, M.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, New Orleans, LA, USA, 1–6 June 2018; pp. 2227–2237. [Google Scholar]
- Alt, C.; Hübner, M.; Hennig, L. Fine-tuning pre-trained transformer language models to distantly supervised relation extraction. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019; pp. 1388–1398. [Google Scholar] [CrossRef] [Green Version]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; pp. 4171–4186. [Google Scholar]
- Mirónczuk, M.; Protasiewicz, J. A recent overview of the state-of-the-art elements of text classification. Expert Syst. Appl. 2018, 106, 36–54. [Google Scholar] [CrossRef]
- Holzinger, A.; Kieseberg, P.; Weippl, E.; Tjoa, A.M. Current Advances, Trends and Challenges of Machine Learning and Knowledge Extraction: From Machine Learning to Explainable AI. In Proceedings of the International Cross-Domain Conference CD-MAKE 2018, Hamburg, Germany, 27–30 August 2018; pp. 1–8. [Google Scholar]
- Maas, A.; Daly, R.; Pham, P.; Huang, D.; Ng, A.; Potts, C. Learning word vectors for sentiment analysis. In Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies, Portland, OR, USA, 19–24 June 2011; pp. 142–150. [Google Scholar] [CrossRef]
- Zhang, D. Question classification using support vector machines. In Proceedings of the 26th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Toronto, ON, Canada, 28 July–1 August 2003; pp. 26–32. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Manning, C. Baselines and bigrams: Simple, good sentiment and topic classification. In Proceedings of the 50th Annual Meeting of the Association for Computational Linguistics: Short Papers, Jeju Island, Korea, 8–14 July 2012; Association for Computational Linguistics: Stroudsburg, PA, USA, 2012; pp. 90–94. [Google Scholar] [CrossRef]
- Wang, X.; Yin, S.; Shafiq, M.; Laghari, A.A.; Karim, S.; Cheikhrouhou, O.; Alhakami, W.; Hamam, H. A New V-Net Convolutional Neural Network Based on Four-Dimensional Hyperchaotic System for Medical Image Encryption. Secur. Commun. Netw. 2022, 2022, 4260804. [Google Scholar] [CrossRef]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modelling. IPS 2014 Workshop on Deep Learning. arXiv 2014, arXiv:1412.3555. [Google Scholar]
- Al-Shayea, Q. Artificial neural networks in medical diagnosis. J. Appl. Biomed. 2013, 11, 150–154. [Google Scholar] [CrossRef]
- Shi, M.; Wang, K.; Li, C. A C-LSTM with Word Embedding Model for News Text Classification. In Proceedings of the IEEE/ACIS 18th International Conference on Computer and Information Science (ICIS), Beijing, China, 17–19 June 2019; pp. 253–257. [Google Scholar]
- Xiao, Y.; Cho, K. Efficient character-level document classification by combining convolution and recurrent layers. arXiv 2016, arXiv:1602.00367. [Google Scholar]
- Wang, B. Disconnected recurrent neural networks for text categorization. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, Melbourne, Australia, 15–20 July 2018; pp. 2311–2320. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lu, H.; Wei, X.; Wei, G.; Behbahani, S.S.; Iseley, T.T. Application of artificial neural network in tunnel engineering: A systematic review. IEEE Access 2020, 8, 119527–119543. [Google Scholar] [CrossRef]
- Zheng, S.; Yang, M. A New Method of Improving BERT for Text Classification. In Proceedings of the Intelligence Science and Big Data Engineering. Big Data and Machine Learning, Nanjing, China, 17–20 October 2019; pp. 442–452. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Y.; Wei, B.; Li, Z.; Huang, X. Traditional chinese medicine clinical records classification using knowledge-powered document embedding. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016. [Google Scholar] [CrossRef]
- Figueroa, R.; Zeng-Treitler, Q.; Ngo, L.; Goryachev, S.; Wiechmann, E. Active learning for clinical text classification: Is it better than random sampling? J. Am. Med. Inform. Assoc.: JAMIA 2012, 19, 809–816. [Google Scholar] [CrossRef] [Green Version]
- Garla, V.; Brandt, C. Knowledge-based biomedical word sense disambiguation: An evaluation and application to clinical document classification. J. Am. Med. Inform. Assoc. 2012, 20, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Mao, C.; Luo, Y. Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. BMC Med. Inform. Decis. Mak. 2018, 19, 70–71. [Google Scholar] [CrossRef] [Green Version]
- Asgari-Chenaghlu, M. Word Vector Representation, Word2vec, Glove, and Many More Explained. Ph.D. Thesis, University of Tabriz, Tabriz, Iran, 2017. [Google Scholar] [CrossRef]
- Howard, J.; Ruder, S. Universal language model fine-tuning for text classification. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, Melbourne, Australia, 15–20 July 2018; pp. 328–339. [Google Scholar] [CrossRef] [Green Version]
- Radford, A.; Karthik, N.; Tim, S.; Ilya, S. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf (accessed on 1 October 2021).
- Salton, G.; Buckley, C. Term-weighting approaches in automatic text retrieval. Inform. Process. Man. 1988, 24, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Joulin, A.; Grave, E.; Bojanowski, P.; Mikolov, T. Bag of tricks for efficient text classification. In Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics, Valencia, Spain, 3–7 April 2017; pp. 427–431. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Bird, S. NLTK: The natural language toolkit. In Proceedings of the COLING/ACL on Interactive Presentation Sessions Association for Computational Linguistics 2006, Sydney, Australia, 17–18 July 2006; pp. 69–72. [Google Scholar]
- Yu, Y.; Si, X.; Hu, C.; Zhang, J. A review of recurrent neural networks: Lstm cells and network architectures. Neural Comput. 2019, 31, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Du, Q. A deep investigation into fasttext. In Proceedings of the IEEE 21st International Conference on High Performance Computing and Communications, Zhangjiajie, China, 10–12 August 2019; pp. 1714–1719. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So CKang, J. Biobert: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Altosaar, J.; Ranganath, R. Clinicalbert: Modeling clinical notes and predicting hospital readmission. arXiv 2019, arXiv:1904.05342. [Google Scholar]
- Beltagy, I.; Cohan, A.; Lo, K. Scibert: Pretrained contextualized embeddings for scientific text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) 2019, HongKong, China, 3–7 November 2019. [Google Scholar]
- Matthew, E.P.; Mark, N.; Robert, L.; Roy, S.; Vidur, J.; Sameer, S.; Noah, A.S. Knowledge Enhanced Contextual Word Representations. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), HongKong, China, 3–7 November 2019; pp. 43–54. [Google Scholar] [CrossRef] [Green Version]
- Ashish, V.; Noam, S.; Niki, P.; Jakob, U.; Llion, J.; Aidan, N.G.; Lukasz, K.; Illia, P. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 6000–6010. [Google Scholar]
- Khadhraoui, M.; Bellaaj, H.; Ben Ammar, M.; Hamam, H.; Jmaiel, M. Machine Learning Classification Models with SPD/ED Dataset: Comparative Study of Abstract Versus Full Article Approach. In Proceedings of the ICOST 2020, Hammamet, Tunisia, 24–26 June 2020; pp. 24–26. [Google Scholar] [CrossRef]




| Models | Summary | NLP Tasks | Datasets | Learning Type | Mono/Multi-Class | Accuracy Model/Bert | Domain |
|---|---|---|---|---|---|---|---|
| BoostingBERT | The model integrates multi-class boosting into the BERT model. The boosting technique is demonstrated to be able to be used to enhance the performance of BERT, instead of other techniques such as bagging or stacking. Based on the experimental results, BoostingBERT outperforms the bagging BERT constantly. Two approaches are compared, making use of the base transformer classifier in the BoostingBERT model: weights privacy vs. weights sharing, and the former one constantly outperforms the latter one. | Multiple NLP tasks | GLUE dataset | Ensemble learning | Multi-class | 82.93%/80.72% with CoLa dataset 93.35/92.55 with SST-2 dataset | Multi-domain |
| EduBERT | The use of pre-trained models has proven a great advance in learning analytics. They apply the BERT approach to the three LAK tasks previously explored on the MOOC forum data: detection of confusion, urgent intervention by teachers and classification of sentimentality. The experimental results have proven an improvement in performance beyond the state of the art. | Sentiment analysis (SA), named entity recognition (NER) and question answering (QA) | Stanford MOOCPosts dataset | Supervised/ unsupervised | Not mentioned | 89.78%/89.47% | Multi-domain |
| ALBERT | ALBERT introduces two optimizations to reduce model size: a factorization of the embedding layer and parameter sharing across the hidden layers of the network. The result of combining these two approaches results in a baseline model with only 12M parameters, compared to BERT’s 108M, with an accuracy of 80.1% on several NLP benchmarks compared with BERT’s 82.3% average. | Question answering | GLUE SQuAD RACE | Supervised/ unsupervised | Not mentioned | 88.7%/85.2% | Multi-domain |
| FinBERT | FinBERT is a language model based on BERT for financial NLP tasks. FinBERT is evaluated on two financial sentiment analysis datasets. The authors achieve the state of the art on FiQA sentiment scoring and Financial PhraseBank. They implement two other pre-trained language models, ULMFit and ELMo, for financial sentiment analysis to compare with FinBERT. Experiments are conducted to investigate the effects of further pre-training on the financial corpus, training strategies to prevent catastrophic forgetting and fine-tuning only a small subset of model layers for decreasing training time without a significant drop in performance. | Sentiment analysis and text classification | Financial sentiment analysis datasets | Supervised/unsupervised | Not Mentioned | 86% | Scientific domain |
| SciBERT | The authors release SCIBERT, a pre-trained language model based on BERT to address the scientific data. SCIBERT leverages unsupervised pre-training on a large multi-domain corpus of scientific publications to improve performance on downstream scientific NLP tasks. SCIBERT largely outperforms BERT and previous state-of-the-art models in a variety of biomedical text-mining tasks including sequence tagging, sentence classification and dependency parsing, with datasets from a variety of scientific domains. SCIBERT makes improvements over BERT. | Named entity recognition (NER), PICO extraction (PICO), text classification (CLS), relation classification (REL), dependency parsing (DEP) | Corpus of scientific text | Supervised/unsupervised | Not mentioned | 99.01%/88.85% | Scientific domain |
| KnowBert | KnowBert represents a general method to embed multiple knowledge bases (KBs) into large-scale models. The proposed model aims to enhance scientific data representations with structured, human-curated knowledge. For each KB, the retrieve of the relevant entity is based on an integrated entity linker; then, the contextual word representations are updated via a form of word-to-entity attention. After integrating WordNet and a subset of Wikipedia into BERT, KnowBert demonstrates improved perplexity, ability to recall facts as measured in a probing task and downstream performance on relationship extraction, entity typing and word sense disambiguation. KnowBert’s runtime is comparable to BERT’s, and it scales to large KBs. | Relation extraction, entity typing, word sense disambiguation | Wikipedia | Supervised/unsupervised | Not mentioned | 89.01%/89% | Scientific domain |
| ClinicalBert | The authors are exploring and releasing BERT models for clinical text: one for generic clinical text and another for discharge summaries specifically. The main approach demonstrates that using a domain-specific model enhances performance improvements on three common clinical NLP tasks compared with nonspecific embeddings. These domain-specific models are not as performant on two clinical de-identification tasks, and the authors argue that this is a natural consequence of the differences between de-identified source text and synthetically non-de-identified task text. | Readmission prediction, diagnosis predictions, mortality risk estimation | MIMIC-III dataset | Supervised/unsupervised | Not mentioned | 80.8%/77.6% | Biomedical domain |
| BlueBERT | The proposed approach is a BERT-base model pre-trained on PubMed abstracts and MIMIC-III clinical notes. Based on the BERT model, BlueBERT is specialized by an extra linear layer on top of the existing model to transform the output into 10 classes, one for each ICD-9 code. The authors implemented the BCEWithLogits loss for multi-class classification. Related to the BERT model’s architecture, BlueBERT introduces three small architectural variations: (1) adding three linear layers with ReLU non-linearity instead of just one linear layer, (2) freezing the BlueBERT weights from the first variant so that only the linear layer weights would be tuned and (3) adding a dropout layer after the BERT layer from the second variant. | Text classification | PubMed abstracts and MIMIC-III datasets | Supervised/unsupervised | Multi-class | 89.2%/86.9% | Biomedical domain |
| BioBERT | The BioBERT (Bidirectional Encoder Representations from Transformers for Biomedical Text Mining) model is a domain-specific language representation model pre-trained on large-scale biomedical corpora. Based on experimental results, BioBERT largely outperforms BERT and previous state-of-the-art models in a variety of biomedical text-mining tasks when pre-trained on biomedical corpora. BioBERT outperforms on the following three representative biomedical text-mining tasks: biomedical named entity recognition, biomedical relation extraction and biomedical question answering. The analysis results show that pre-training BERT on biomedical corpora helps it to understand complex biomedical texts. | NER Biomedical relation extraction Bio question answering | PubMed abstracts/4,5B PMC full papers/13,5B | Supervised/ unsupervised | Not mentioned | 89.04% /88.30% | Biomedical Domain |
| Category | Description |
|---|---|
| COVID-19 | It is interested in scientific papers of probable treatment and the different symptoms related to COVID-19. |
| Virology | It deals with scientific papers about the study of viruses, genome sequencing, etc. |
| Public Health | It focuses on scientific papers about the study, prevention, control, in particular through vaccination, and epidemiological data against COVID-19. |
| Mental Health | It spotlights several scientific papers about the impact of COVID-19 on mental health. |
| Model | NLP Tasks | Dataset /Size | Characteristics | Hyperparameters | Learning Period | Methods |
|---|---|---|---|---|---|---|
| BioBERT [35] | NER Biomedical relation extraction Bio question answering | PubMed Abstracts/4,5B PMC Full Papers/13,5B | Biomedical domain | Sentence length: 128–512 tokens | 23 days 8 NVIDIA V100 (32GB) GPUs | Word piece tokenization Pre-training BERT on biomedical corpora: Naver Smart ML Fine-tuning BioBERT |
| ClinicalBERT [36] | Readmission prediction Diagnosis predictions Mortality risk estimation | MIMIC-III | Clinical domain | Sequence length: 128–512 tokens | Amazon Web Services using a single K80 GPU | Subword embeddings Self-attention mechanism |
| SciBERT [37] | NER Text classification Relation classification Dependency parsing | Semantic Scolar/1.14M | Scientific domain | Sentence length: 128–512 tokens | 5 days + 2 days TPU v3 with 8 cores | Finetuning BERT: Frozen BERT embeddings Contextualize word embeddings |
| KnowBERT [38] | Relation extraction Entity typing Word sense disambiguation | Wikipedia | Knowledge domain | - | - | Mention-span representations Retrieval of relevant entity embeddings Recontextualization of entity span embeddings |
| Models | Domain | NLP Tasks |
|---|---|---|
| roberta-base | Multi-Domain | 1. Named Entity Recognition 2. Sequence Classification 3. Question Answering |
| albert-base-v1 | Multi-Domain | 1. Sequence Classification 2. Question Answering |
| allenai/scibert_scivocab_uncased | Scientific Domain | 1. Named Entity Recognition (NER) 2. PICO Extraction 3. Text Classification 4. Relation Classification (REL) 5. Dependency Parsing (DEP) |
| allenai/scibert_scivocab_cased | Scientific Domain | 1. Named Entity Recognition (NER) 2. PICO Extraction 3. Text Classification 4. Relation Classification (REL) 5. Dependency Parsing (DEP) |
| emilyalsentzer/Bio_ClinicalBERT | Biomedical Domain | 1 Biomedical Named Entity Recognition 2. Biomedical Relation Extraction 3. Biomedical Question Answering |
| dmis-lab/biobert-base-cased-v1.1 | Biomedical Domain | 1. Biomedical Named Entity Recognition 2. Biomedical Relation Extraction 3. Biomedical Question Answering |
| monologg/biobert_v1.1_pubmed | Biomedical Domain | 1. Biomedical Named Entity Recognition 2. Biomedical Relation Extraction 3. Biomedical Question Answering |
| dmis-lab/biobert-v1.1 | Biomedical Domain | 1. Biomedical Named Entity Recognition 2. Biomedical Relation Extraction 3. Biomedical Question Answering |
| gsarti/biobert-nli | Biomedical Domain | 1. Biomedical Named Entity Recognition 2. Biomedical Relation Extraction 3. Biomedical Question Answering |
| CovBERT | Biomedical and Scientific Domains | 1. Text Classification |
| Models | Accuracy | Average Loss | Recall | Precision | F1 Metric |
|---|---|---|---|---|---|
| roberta-base | 83% | 29% | 51% | 68% | 57% |
| albert-base-v1 | 84% | 39% | 41% | 56% | 47% |
| allenai/scibert_scivocab_uncased | 84% | 33% | 70% | 71% | 69% |
| allenai/scibert_scivocab_cased | 84% | 30% | 74% | 74% | 73% |
| emilyalsentzer/Bio_ClinicalBERT | 87% | 25% | 83% | 82% | 82% |
| dmis-lab/biobert-base-cased-v1.1 | 87% | 14% | 68% | 68% | 66% |
| monologg/biobert_v1.1_pubmed | 87% | 17% | 77% | 79% | 76% |
| dmis-lab/biobert-v1.1 | 88% | 19% | 68% | 68% | 66% |
| gsarti/biobert-nli | 89% | 19% | 66% | 71% | 65% |
| CovBERT | 94% | 18% | 88% | 86% | 86% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadhraoui, M.; Bellaaj, H.; Ammar, M.B.; Hamam, H.; Jmaiel, M. Survey of BERT-Base Models for Scientific Text Classification: COVID-19 Case Study. Appl. Sci. 2022, 12, 2891. https://doi.org/10.3390/app12062891
Khadhraoui M, Bellaaj H, Ammar MB, Hamam H, Jmaiel M. Survey of BERT-Base Models for Scientific Text Classification: COVID-19 Case Study. Applied Sciences. 2022; 12(6):2891. https://doi.org/10.3390/app12062891
Chicago/Turabian StyleKhadhraoui, Mayara, Hatem Bellaaj, Mehdi Ben Ammar, Habib Hamam, and Mohamed Jmaiel. 2022. "Survey of BERT-Base Models for Scientific Text Classification: COVID-19 Case Study" Applied Sciences 12, no. 6: 2891. https://doi.org/10.3390/app12062891
APA StyleKhadhraoui, M., Bellaaj, H., Ammar, M. B., Hamam, H., & Jmaiel, M. (2022). Survey of BERT-Base Models for Scientific Text Classification: COVID-19 Case Study. Applied Sciences, 12(6), 2891. https://doi.org/10.3390/app12062891








