Classification of Sleep Stage with Biosignal Images Using Convolutional Neural Networks
Abstract
:1. Introduction
- (i)
- We propose a method for sleep staging classification, with only EEG and EOG signals, with good accuracy.
- (ii)
- A CNN framework is proposed for the classification. The EEG and EOG signals are converted into images of the time domain and frequency domain, and the images are fed to the CNN as an input.
- (iii)
- We demonstrate that the proposed method shows experimentally good performance on publicly available datasets:
2. Backgrounds
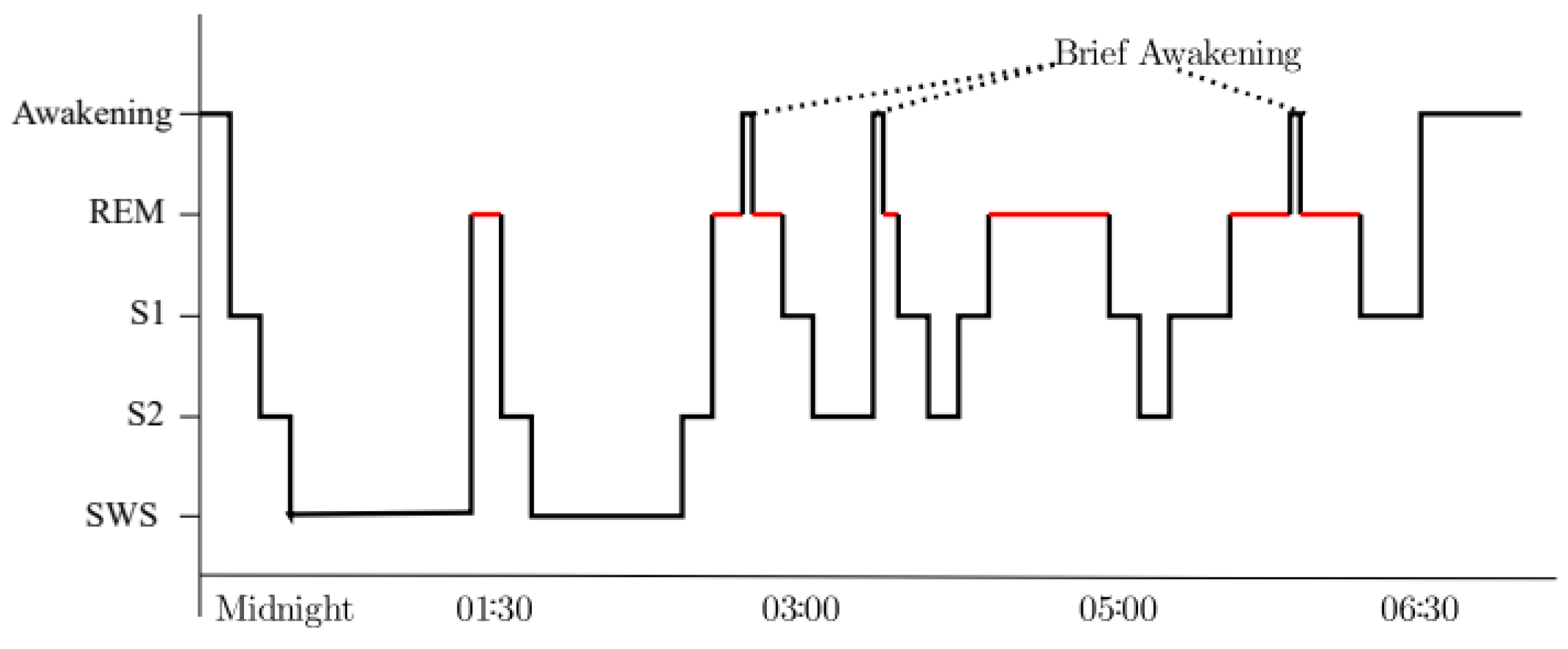
- The ambiguity of sleep scoring: There are no clear criteria to determine each stage. The stages are defined empirically by somnologists, and some sleep stages (SWS, REM) have identifiable characteristics, but the distinction between Wake, S1, and S2 is less clear. Therefore, many scoring disagreements exist in S1 (23–74% agreement for S1) [9].
- The cost of sleep scoring: Polysomnography takes a long time to analyze the biosignals recorded during sleep, since experts manually examine the results. Therefore, it is difficult to freely prescribe polysomnography to patients because it is an expensive medical treatment.
- Sleep-stage scoring can be reliable. The system will produce the same and stable results every time, while humans are error-prone.
- Somnologists can save an amount of time to label the recorded signals in polysomnography.
3. Related Works
- Temporal features: Temporal features explain the changes in brain signals over time. Each sleep stage is related to the brain signal and state. For example, an indicator of SWS is a change in peak amplitude or a change in frequencies.
- Spectral features: Frequency features are extracted from the signal using Fourier or wavelet transformation. Sleep stages are characterized by unique frequencies. Most often the brain wave is divided into delta (0.5–4 Hz), theta (4–8 Hz), alpha (8–12 Hz), sleep spindles (12–16 Hz), beta (12–40 Hz), and gamma (40–100 Hz) according to the frequency [5].
- Statistical features: Statistical features can describe the biosignals. The minimum, maximum, and average of the signal or the number of zero-crossing can be general features in the signals. Other statistical features are median, standard deviation, and skewness of the signals.
4. Sleep-Stage Classification Using CNN
4.1. Preprocessing and Input Data
4.2. Structure of CNN Model
5. Experimental Results
5.1. Sleep Dataset
5.2. Performance Measures
5.3. Experiments
- Exp1: Classification using EEG and EOG images in the time domains
- Exp2: Classification using EEG and EOG images in the frequency domains
- Exp3: Classification using EEG and EOG images in the time domain and frequency domain
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Definition of Sleep. Available online: https://medical-dictionary.thefreedictionary.com/sleep (accessed on 3 January 2022).
- Rasch, B.; Born, J. About Sleep’s Role in Memory. Physiol. Rev. 2013, 93, 681–766. [Google Scholar] [CrossRef] [PubMed]
- Carskadon, M.A.; Dement, W.C. Normal human sleep: An overview. In Principles and Practice of Sleep Medicine, 5th ed.; Kryger, M.H., Roth, T., Demen, W.C., Eds.; Elsevier Saunders: St. Louis, MO, USA, 2011; pp. 16–26. [Google Scholar]
- Jackson, C.L.; Redline, S.; Kawachi, I.; Hu, F.B. Association between sleep duration and diabetes in black and white adults. Diabetes Care 2013, 36, 3557–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associates Events: Rules, Terminology and Technical Specifications Version 2.6. Published Online 1 January 2021. Available online: https://aasm.org/clinical-resources/scoring-manual/ (accessed on 3 January 2022).
- Lee, Y.J.; Lee, J.Y.; Cho, J.H.; Choi, J.H. Inter-rater reliability of sleep stage scoring: A meta-analysis. J. Clin. Sleep Med. 2022, 18, 193–202. [Google Scholar] [CrossRef]
- Chriskos, P.; Frantzidis, C.A.; Nday, C.M.; Gkivogkli, P.T.; Bamidis, P.D.; Kourtidou-Papadeli, C. A review on current trends in automatic sleep staging through bio-signal recordings and future challenges. Sleep Med. Rev. 2021, 55, 101377. [Google Scholar] [CrossRef]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.C.; Oberye, J.J.L. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Rosenberg, R.S.; Hout, S.V. The American Academy of Sleep Medicine inter-scorer reliability program: Respiratory events. J. Clin. Sleep Med. 2014, 10, 447–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sen, B.; Peker, M.; Cavusoglu, A.; Celebi, F.V. A comparative study on classification of sleep stage based on EEG signals using feature selection and classification algorithms. J. Med. Syst. 2014, 38, 1–21. [Google Scholar] [CrossRef]
- Shea, K.O.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Dong, H.; Supratak, A.; Pan, W.; Wu, C.; Matthews, P.; Guo, Y. Mixed neural network approach for temporal sleep stage classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Zhang, X.; Wang, J.; Dang, X. The research of sleep staging based on single-lead electrocardiogram and deep neural network. Biomed. Eng. Lett. 2018, 8, 87–93. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic Sleep Stage Scoring with Single-Channel EEG Using Convolutional Neural Networks. arXiv 2016, arXiv:1610.01683. [Google Scholar]
- Mikkelsen, K.; de Vos, M. Personalizing deep learning models for automatic sleep staging. arXiv 2018, arXiv:1801.02645. [Google Scholar]
- Yildirim, O.; Baloglu, U.; Acharya, U. A Deep Learning Model for Automated Sleep Stages Classification Using PSG Signals. Int. J. Environ. Res. Public Health 2019, 16, 599. [Google Scholar] [CrossRef] [Green Version]
- Chambon, S.; Galtier, M.; Arnal, P.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreotti, F.; Phan, H.; Cooray, N.; Lo, C.; Hu, M.; De Vos, M. Multichannel Sleep Stage Classification and transfer Learning Using Convolutional Neural Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 171–174. [Google Scholar]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.; De Vos, M. Joint classification and prediction CNN framework for automatic sleep stage classification. Proc. IEEE Trans. Biomed. Eng. 2019, 66, 1285–1296. [Google Scholar] [CrossRef]
- Sors, A.; Bonnet, S.; Mirekc, S.; Vercueil, L.; Payen, J.-F. A convolutional neural network for sleep stage scoring from raw single-channel eeg. Biomed. Signal Process Control 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Andreotti, F.; Phan, H.; De Vos, M. Visualising Convolutional Neural Network Decisions in Automatic Sleep Scoring. In Proceedings Joint Workshop on Artificial Intelligence in Health (AIH); CEUR: Stockholm, Sweden, 2018; pp. 70–81. [Google Scholar]
- Vilamala, A.; Madsen, K.; Hansen, L. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. In Proceedings of the IEEE International Workshop on Machine Learning for Signal Processing (MLSP), Tokyo, Japan, 25–28 September 2017; pp. 1–6. [Google Scholar]
- Malafeev, A.; Laptev, D.; Bauer, S.; Omlin, X.; Wierzbicka, A.; Wichniak, A.; Jernajczyk, W.; Riener, R.; Buhmann, J.; Achermann, P. Automatic human sleep stage scoring using deep neural networks. Front Neurosci. 2018, 781. [Google Scholar] [CrossRef] [Green Version]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.; De Vos, M. Automatic sleep stage classification using single-channel EEG: Learning sequential features with attention-based recurrent neural networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 1452–1455. [Google Scholar]
- Patanaik, A.; Ong, J.; Gooley, J.; Ancoli-Israel, S.; Chee, M. An end-to-end framework for real-time automatic sleep stage classification. Sleep 2018, 41, zsy041. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y. Automatic sleep stage classification of single-channel EEG by using complex-valued convolutional neural network. Biomed. Eng./Biomed. Tech. 2017, 63, 177–190. [Google Scholar] [CrossRef]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A Model for Automatic Sleep Stage Scoring based on Raw Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [Green Version]
- Al-Saffar, A.A.M.; Tao, H.; Talab, M.A. Review of deep convolution neural network in image classification. In Proceedings of the International Conference on Radar, Antenna, Microwave, Electronics, and Telecommunications, Jakarta, Indonesia, 23–24 October 2017; pp. 26–31. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Gulli, A.; Pal, S. Deep Learning with Keras; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. Available online: tensorflow.org (accessed on 3 March 2021).
- Goldberger, A.; Amaral, L.; Glass, L.; Hausdorff, J.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rechtschaffen, K.; Kales, A. A Manual of Standardized Terminology Techniques and Scoring System for Sleep Stages of Human Subjects; Public Health Service, US Government Printing Office: Washington, DC, USA, 1968.
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; Vanderplas, J.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- O’Reilly, C.; Gosselin, N.; Carrier, J.; Nielsen, T. Montreal Archive of Sleep Studies: An open-access resource for instrument benchmarking and exploratory research. J. Sleep Res. 2014, 23, 628–635. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.; Redline, S. The National Sleep Research Resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, S.; Howard, B.; Iber, C.; Kiley, J.; Nieto, F.; O’Connor, G.; Rapoport, D.; Redline, S.; Robbins, J.; Samet, J.; et al. The Sleep Heart Health Study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar]








| Layer | Filter | Kernel | Output | Dropout | Activation |
|---|---|---|---|---|---|
| Input | (216,144) | ||||
| Conv2D | 32 | (3,3) | 0.25 | ReLU | |
| MaxPool | (2,2) | (108,72) | |||
| Conv2D | 64 | (3,3) | 0.25 | ReLU | |
| MaxPool | (2,2) | (54,36) | |||
| Flatten | (124,416) | ||||
| Dense | (256) | 0.5 | ReLU | ||
| Dense | (4) | Softmax |
| Stage | Precision | Recall | F1 Score | Weighted F1 Score | Accuracy |
|---|---|---|---|---|---|
| S1 | 0.77 | 0.48 | 0.59 | 0.84 | 0.85 |
| S2 | 0.84 | 0.96 | 0.90 | ||
| SWS | 0.99 | 1.00 | 1.00 | ||
| R | 0.78 | 0.70 | 0.74 |
| Stage | Precision | Recall | F1 Score | Weighted F1 Score | Accuracy |
|---|---|---|---|---|---|
| S1 | 0.78 | 0.48 | 0.60 | 0.86 | 0.86 |
| S2 | 0.85 | 0.95 | 0.90 | ||
| SWS | 0.96 | 0.97 | 0.97 | ||
| R | 0.83 | 0.84 | 0.84 |
| Stage | Precision | Recall | F1 Score | Weighted F1 Score | Accuracy |
|---|---|---|---|---|---|
| S1 | 0.78 | 0.91 | 0.84 | 0.94 | 0.94 |
| S2 | 1.00 | 1.00 | 1.00 | ||
| SWS | 1.00 | 1.00 | 1.00 | ||
| R | 0.91 | 0.78 | 0.84 |
| Time Domain | Freq. Domain | Time and Freq. Domain | |
|---|---|---|---|
| Accuracy | 86% (71.4–100) | 86% (72.1–95.2) | 94% (78.4–100) |
| Weighted F1 score | 85% (59–100) | 86% (60–97) | 94% (84–100) |
| Study | Dataset | Input | Method | Accuracy (%) |
|---|---|---|---|---|
| Tsinalis [14] | Sleep-EDF *, MASS ** | EEG | CNN | 74.8, 77.9 |
| Dong [12] | MASS | EOG | DNN | 81.4 |
| Supratak [27] | Sleep-EDFx, MASS | EEG | CNN + RNN | 82.0, 81.5 |
| Andreotti [18] | Sleep-EDF, MASS | EEG, EOG | CNN | 76.8, 79.4 |
| Mikkelsen [15] | Sleep-EDF | EEG, EOG | CNN | 84.0 |
| Chambon [17] | MASS | EEG, EOG, EMG | CNN | 79.9 |
| Sors [20] | SHHS1 *** | EEG | CNN | 87.0 |
| Phan [19] | Sleep-EDF | EEG, EOG | CNN | 82.3 |
| Yildirim [16] | Sleep-EDFx | EEG, EOG | CNN | 92.3 |
| This study | Sleep-EDFx | EEG, EOG | CNN | 94.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joe, M.-J.; Pyo, S.-C. Classification of Sleep Stage with Biosignal Images Using Convolutional Neural Networks. Appl. Sci. 2022, 12, 3028. https://doi.org/10.3390/app12063028
Joe M-J, Pyo S-C. Classification of Sleep Stage with Biosignal Images Using Convolutional Neural Networks. Applied Sciences. 2022; 12(6):3028. https://doi.org/10.3390/app12063028
Chicago/Turabian StyleJoe, Moon-Jeung, and Seung-Chan Pyo. 2022. "Classification of Sleep Stage with Biosignal Images Using Convolutional Neural Networks" Applied Sciences 12, no. 6: 3028. https://doi.org/10.3390/app12063028
APA StyleJoe, M.-J., & Pyo, S.-C. (2022). Classification of Sleep Stage with Biosignal Images Using Convolutional Neural Networks. Applied Sciences, 12(6), 3028. https://doi.org/10.3390/app12063028





