Quenching Efficiency of Quantum Dots Conjugated to Lipid Bilayers on Graphene Oxide Evaluated by Fluorescence Single Particle Tracking
Abstract
:1. Introduction
2. Materials and Methods
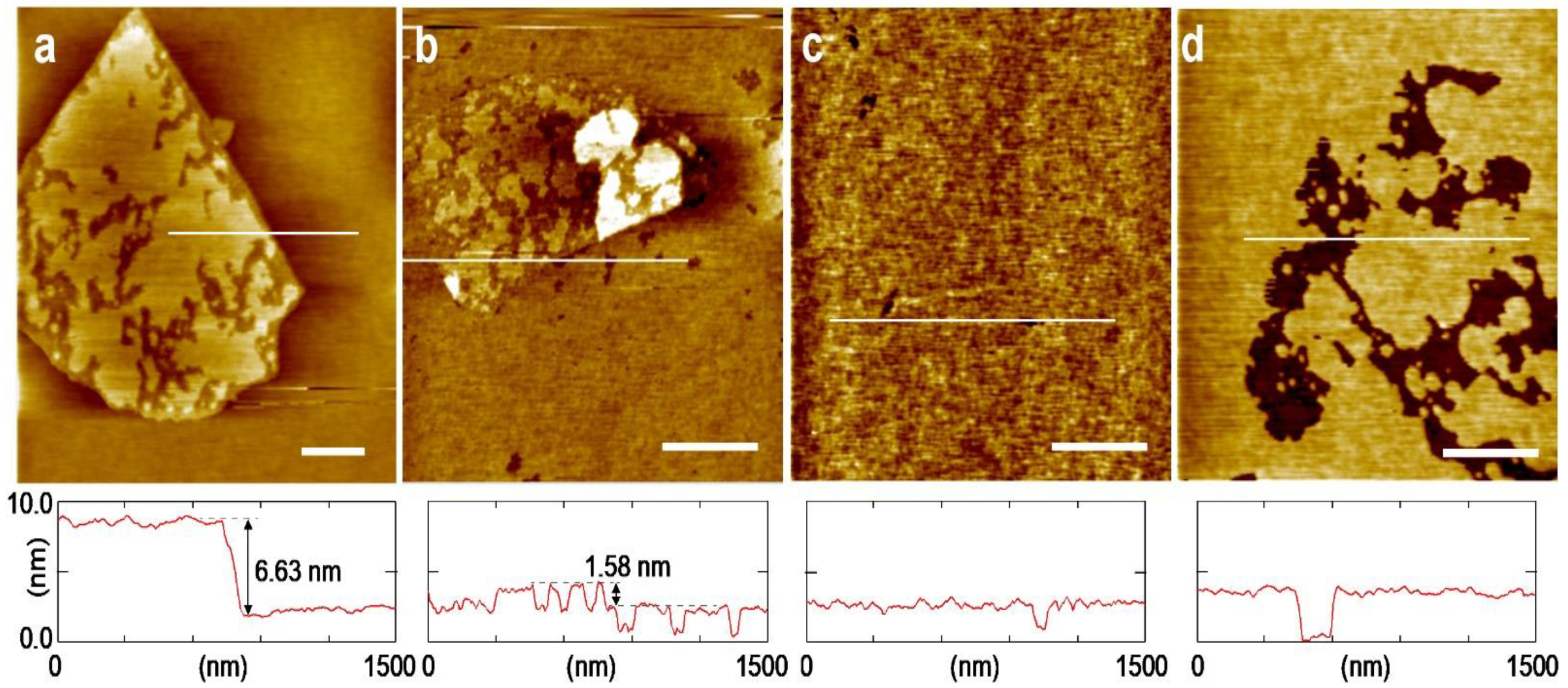
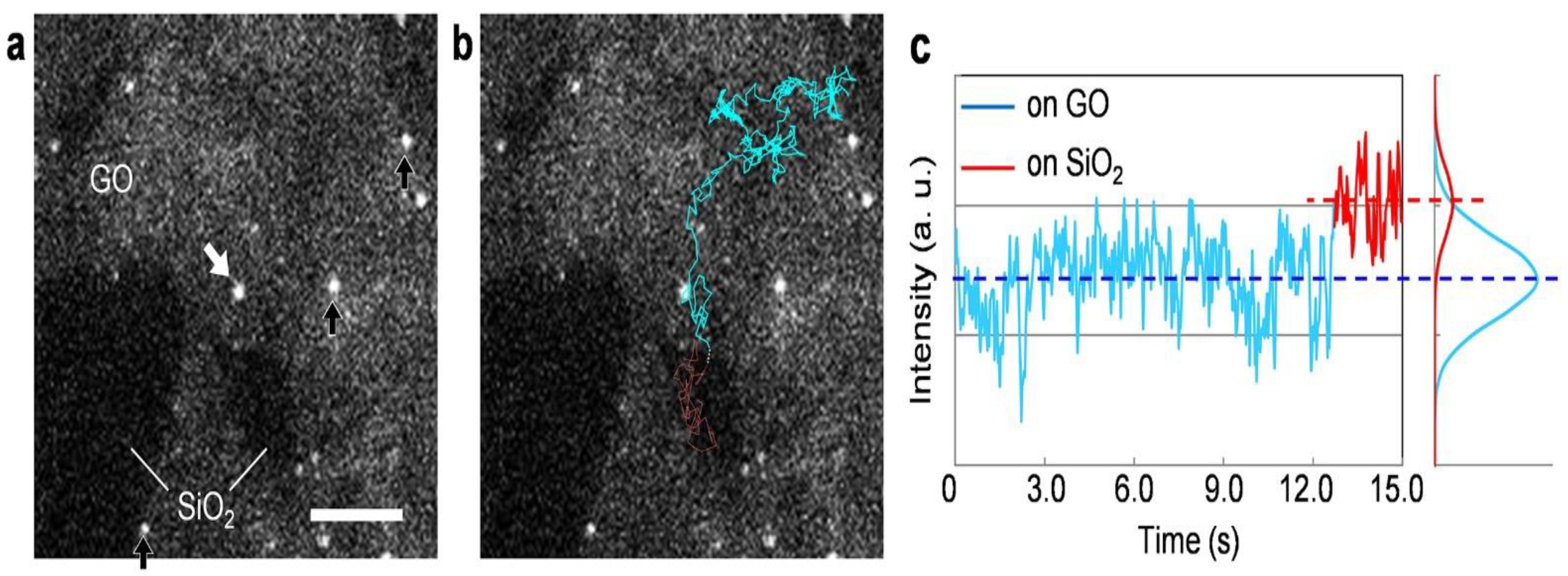
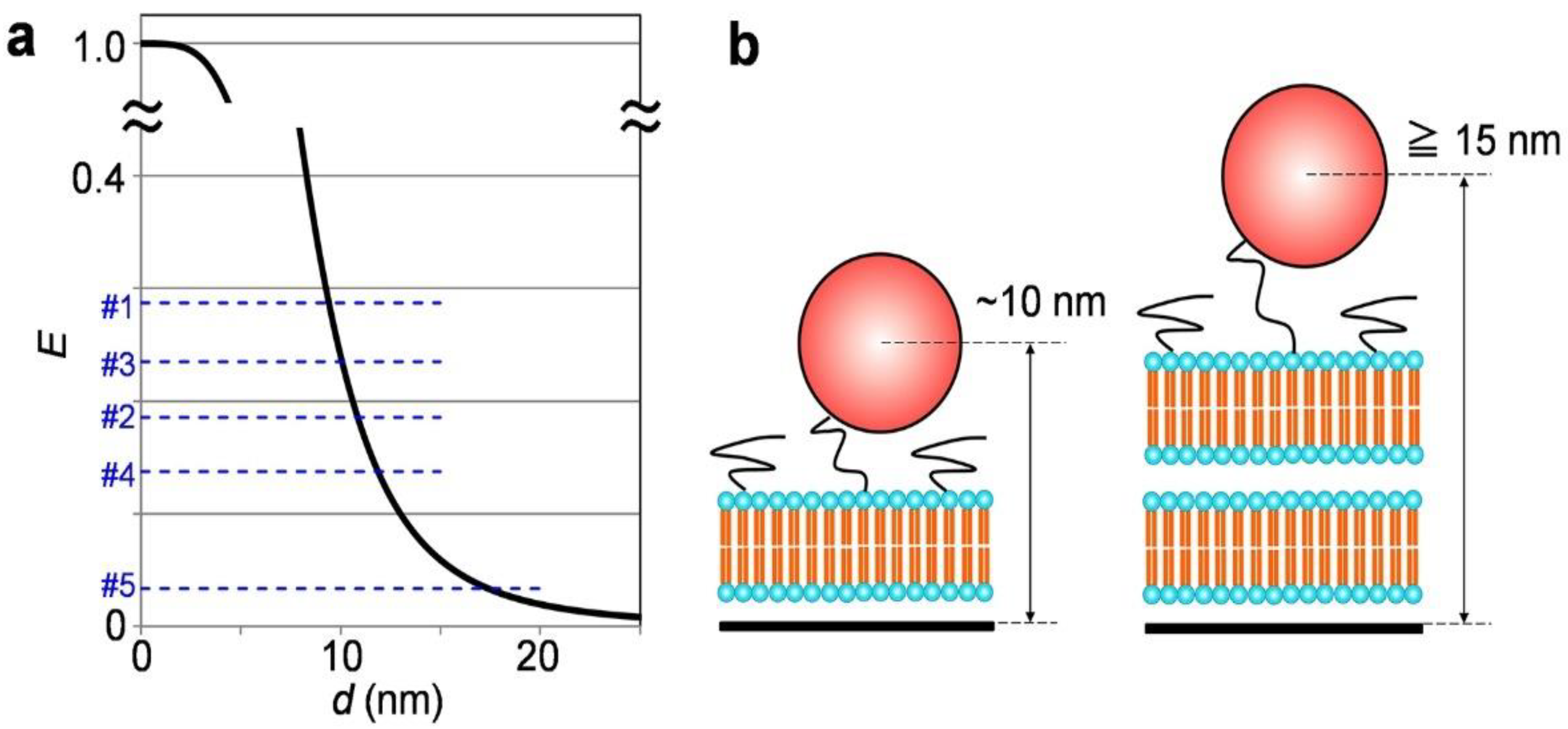
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rothman, J.E. The Principle of Membrane Fusion in the Cell (Nobel Lecture). Angew. Chem. Int. Ed. 2014, 53, 12676–12694. [Google Scholar] [CrossRef] [PubMed]
- Tillman, T.S.; Cascio, M. Effects of membrane lipids on ion channel structure and function. Cell Biochem. Biophys. 2003, 38, 161–190. [Google Scholar] [CrossRef]
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghal, M.M.R.; Hossain, F.; Yamazaki, M. Action of antimicrobial peptides and cell-penetrating peptides on membrane potential revealed by the single GUV method. Biophys. Rev. 2020, 12, 339–348. [Google Scholar] [CrossRef]
- Komiya, M.; Kato, M.; Tadaki, D.; Ma, T.; Yamamoto, H.; Tero, R.; Tozawa, Y.; Niwano, M.; Hirano-Iwata, A. Advances in Artificial Cell Membrane Systems as a Platform for Reconstituting Ion Channels. Chem. Rec. 2020, 20, 730–742. [Google Scholar] [CrossRef]
- Morigaki, K.; Tanimoto, Y. Evolution and development of model membranes for physicochemical and functional studies of the membrane lateral heterogeneity. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2012–2017. [Google Scholar] [CrossRef]
- Khorshid, M.; Losada-Pérez, P.; Wackers, G.; Yongabi, D.; Renner, F.U.; Thoelen, R.; Wagner, P. Real-time monitoring of interactions between Ebola fusion peptide and solid-supported phospholipid membranes: Effect of peptide concentration and layer geometry. Phys. Med. 2017, 4, 1–7. [Google Scholar] [CrossRef]
- Uchihashi, T.; Scheuring, S. Applications of high-speed atomic force microscopy to real-time visualization of dynamic biomolecular processes. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 229–240. [Google Scholar] [CrossRef]
- Castellana, E.T.; Cremer, P.S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. [Google Scholar] [CrossRef]
- Tanaka, M.; Sackmann, E. Polymer-supported membranes as models of the cell surface. Nature 2005, 437, 656–663. [Google Scholar] [CrossRef]
- Jackman, J.; Knoll, W.; Cho, N.-J. Biotechnology Applications of Tethered Lipid Bilayer Membranes. Materials 2012, 5, 2637–2657. [Google Scholar] [CrossRef] [Green Version]
- Tero, R. Substrate Effects on the Formation Process, Structure and Physicochemical Properties of Supported Lipid Bilayers. Materials 2012, 5, 2658–2680. [Google Scholar] [CrossRef]
- Cote, L.J.; Kim, J.; Tung, V.C.; Luo, J.; Kim, F.; Huang, J. Graphene oxide as surfactant sheets. Pure Appl. Chem. 2011, 83, 95–110. [Google Scholar] [CrossRef]
- Chen, D.; Feng, H.; Li, J. Graphene Oxide: Preparation, Functionalization, and Electrochemical Applications. Chem. Rev. 2012, 112, 6027–6053. [Google Scholar] [CrossRef]
- Lu, C.-H.; Yang, H.-H.; Zhu, C.-L.; Chen, X.; Chen, G.-N. A graphene platform for sensing biomolecules. Angew. Chem. Int. Ed. Engl. 2009, 48, 4785–4787. [Google Scholar] [CrossRef]
- Ueno, Y.; Furukawa, K.; Matsuo, K.; Inoue, S.; Hayashi, K.; Hibino, H. Molecular design for enhanced sensitivity of a FRET aptasensor built on the graphene oxide surface. Chem. Commun. 2013, 49, 10346–10348. [Google Scholar] [CrossRef] [Green Version]
- Loh, K.P.; Bao, Q.; Eda, G.; Chhowalla, M. Graphene oxide as a chemically tunable platform for optical applications. Nat. Chem. 2010, 2, 1015–1024. [Google Scholar] [CrossRef]
- Bonanni, A.; Loo, A.H.; Pumera, M. Graphene for impedimetric biosensing. TrAC Trends Anal. Chem. 2012, 37, 12–21. [Google Scholar] [CrossRef]
- Li, J.-L.; Bao, H.-C.; Hou, X.-L.; Sun, L.; Wang, X.-G.; Gu, M. Graphene Oxide Nanoparticles as a Nonbleaching Optical Probe for Two-Photon Luminescence Imaging and Cell Therapy. Angew. Chemie Int. Ed. 2012, 51, 1830–1834. [Google Scholar] [CrossRef]
- Goto, Y.; Yoshida, N.; Umeyama, Y.; Yamada, T.; Tero, R.; Hiraishi, A. Enhancement of Electricity Production by Graphene Oxide in Soil Microbial Fuel Cells and Plant Microbial Fuel Cells. Front. Bioeng. Biotechnol. 2015, 3, 42. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, S.; Das, P.; Maity, P.P.; Mondal, S.; Ghosh, S.; Dhara, S.; Das, N.C. Green Reduced Graphene Oxide Toughened Semi-IPN Monolith Hydrogel as Dual Responsive Drug Release System: Rheological, Physicomechanical, and Electrical Evaluations. J. Phys. Chem. B 2018, 122, 7201–7218. [Google Scholar] [CrossRef]
- Ghosh, S.; Ganguly, S.; Das, P.; Das, T.K.; Bose, M.; Singha, N.K.; Das, A.K.; Das, N.C. Fabrication of Reduced Graphene Oxide/Silver Nanoparticles Decorated Conductive Cotton Fabric for High Performing Electromagnetic Interference Shielding and Antibacterial Application. Fibers Polym. 2019, 20, 1161–1171. [Google Scholar] [CrossRef]
- Kim, J.; Kim, F.; Huang, J. Seeing graphene-based sheets. Mater. Today 2010, 13, 28–38. [Google Scholar] [CrossRef]
- Lu, C.; Li, J.; Zhang, X.; Zheng, A.; Yang, H.; Chen, X.; Chen, G.-N. General approach for monitoring peptide-protein interactions based on graphene-peptide complex. Anal. Chem. 2011, 83, 7276–7282. [Google Scholar] [CrossRef]
- Huang, A.; Li, W.; Shi, S.; Yao, T. Quantitative Fluorescence Quenching on Antibody-conjugated Graphene Oxide as a Platform for Protein Sensing. Sci. Rep. 2017, 7, 40772. [Google Scholar] [CrossRef] [Green Version]
- Swathi, R.S.; Sebastian, K.L. Resonance energy transfer from a dye molecule to graphene. J. Chem. Phys. 2008, 129, 054703. [Google Scholar] [CrossRef]
- Swathi, R.S.; Sebastian, K.L. Long range resonance energy transfer from a dye molecule to graphene has (distance)(-4) dependence. J. Chem. Phys. 2009, 130, 086101. [Google Scholar] [CrossRef]
- Huang, P.-J.J.; Liu, J. DNA-Length-Dependent Fluorescence Signaling on Graphene Oxide Surface. Small 2012, 8, 977–983. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, Y.; Tsuzuki, K.; Iwasa, S.; Ishikawa, R.; Sandhu, A.; Tero, R. Fabrication of Supported Lipid Bilayer on Graphene Oxide. J. Phys. Conf. Ser. 2012, 352, 012017. [Google Scholar] [CrossRef]
- Okamoto, Y.; Motegi, T.; Iwasa, S.; Sandhu, A.; Tero, R. Fluidity evaluation of cell membrane model formed on graphene oxide with single particle tracking using quantum dot. Jpn. J. Appl. Phys. 2015, 54, 04DL09. [Google Scholar] [CrossRef]
- Hummers, W.S., Jr.; Offeman, R.E. Preparation of graphitic oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- Tung, V.C.; Allen, M.J.; Yang, Y.; Kaner, R.B. High-throughput solution processing of large-scale graphene. Nat. Nanotechnol. 2009, 4, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, K.; Okamoto, Y.; Iwasa, S.; Ishikawa, R.; Sandhu, A.; Tero, R. Reduced Graphene Oxide as the Support for Lipid Bilayer Membrane. J. Phys. Conf. Ser. 2012, 352, 012016. [Google Scholar] [CrossRef] [Green Version]
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of solid-supported lipid bilayers: An integrated view. Langmuir 2006, 22, 3497–3505. [Google Scholar] [CrossRef]
- Murcia, M.J.; Minner, D.E.; Mustata, G.-M.; Ritchie, K.; Naumann, C.A. Design of quantum dot-conjugated lipids for long-term, high-speed tracking experiments on cell surfaces. J. Am. Chem. Soc. 2008, 130, 15054–15062. [Google Scholar] [CrossRef] [Green Version]
- Tero, R.; Sazaki, G.; Ujihara, T.; Urisu, T. Anomalous Diffusion in Supported Lipid Bilayers Induced by Oxide Surface Nanostructures. Langmuir 2011, 27, 9662–9665. [Google Scholar] [CrossRef]
- Okamoto, Y.; Motegi, T.; Morita, K.; Takagi, T.; Amii, H.; Kanamori, T.; Sonoyama, M.; Tero, R. Lateral Diffusion and Molecular Interaction in a Bilayer Membrane Consisting of Partially Fluorinated Phospholipids. Langmuir 2016, 32, 10712–10718. [Google Scholar] [CrossRef]
- Motegi, T.; Yamazaki, K.; Ogino, T.; Tero, R. Substrate-Induced Structure and Molecular Dynamics in a Lipid Bilayer Membrane. Langmuir 2017, 33, 14748–14755. [Google Scholar] [CrossRef]
- Sbalzarini, I.F.; Koumoutsakos, P. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 2005, 151, 182–195. [Google Scholar] [CrossRef]
- Maekawa, T.; Chin, H.; Nyu, T.; Sut, T.N.; Ferhan, A.R.; Hayashi, T.; Cho, N.J. Molecular diffusion and nano-mechanical properties of multi-phase supported lipid bilayers. Phys. Chem. Chem. Phys. 2019, 21, 16686–16693. [Google Scholar] [CrossRef]
- Kakimoto, Y.; Tachihara, Y.; Okamoto, Y.; Miyazawa, K.; Fukuma, T.; Tero, R. Morphology and Physical Properties of Hydrophilic-Polymer-Modified Lipids in Supported Lipid Bilayers. Langmuir 2018, 34, 7201–7209. [Google Scholar] [CrossRef]
- Kaufmann, S.; Borisov, O.; Textor, M.; Reimhult, E. Mechanical properties of mushroom and brush poly(ethylene glycol)-phospholipid membranes. Soft Matter 2011, 7, 9267–9275. [Google Scholar] [CrossRef]
- Tero, R.; Watanabe, H.; Urisu, T. Supported phospholipid bilayer formation on hydrophilicity-controlled silicon dioxide surfaces. Phys. Chem. Chem. Phys. 2006, 8, 3885–3894. [Google Scholar] [CrossRef]
- Hayashi, T.; Tanaka, Y.; Koide, Y.; Tanaka, M.; Hara, M. Mechanism underlying bioinertness of self-assembled monolayers of oligo(ethyleneglycol)-terminated alkanethiols on gold: Protein adsorption, platelet adhesion, and surface forces. Phys. Chem. Chem. Phys. 2012, 14, 10196–10206. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, S.; Qin, H.; Zhou, J.; Peng, X. Oxygen Stabilizes Photoluminescence of CdSe/CdS Core/Shell Quantum Dots via Deionization. J. Am. Chem. Soc. 2020, 142, 4254–4264. [Google Scholar] [CrossRef]
- Watson, A.; Wu, X.; Bruchez, M. Lighting Up Cells with Quantum Dots. Biotechniques 2003, 34, 296–303. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Berti, L.; Medintz, I.L. Materials for Fluorescence Resonance Energy Transfer Analysis: Beyond Traditional Donor–Acceptor Combinations. Angew. Chemie Int. Ed. 2006, 45, 4562–4589. [Google Scholar] [CrossRef]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Medintz, I.L.; Mattoussi, H. Quantum dot-based resonance energy transfer and its growing application in biology. Phys. Chem. Chem. Phys. 2009, 11, 17–45. [Google Scholar] [CrossRef]
- Wegner, K.D.; Hildebrandt, N. Quantum dots: Bright and versatile in vitro and in vivo fluorescence imaging biosensors. Chem. Soc. Rev. 2015, 44, 4792–4834. [Google Scholar] [CrossRef] [Green Version]
- Clapp, A.R.; Medintz, I.L.; Mattoussi, H. Förster resonance energy transfer investigations using quantum-dot fluorophores. Chemphyschem 2006, 7, 47–57. [Google Scholar] [CrossRef]
- Medintz, I.L.; Clapp, A.R.; Mattoussi, H.; Goldman, E.R.; Fisher, B.; Mauro, J.M. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nat. Mater. 2003, 2, 630–638. [Google Scholar] [CrossRef]
- Gaudreau, L.; Tielrooij, K.J.; Prawiroatmodjo, G.E.D.K.; Osmond, J.; de Abajo, F.J.G.; Koppens, F.H.L. Universal Distance-Scaling of Nonradiative Energy Transfer to Graphene. Nano Lett. 2013, 13, 2030–2035. [Google Scholar] [CrossRef] [Green Version]
- Morales-Narváez, E.; Pérez-López, B.; Pires, L.B.; Merkoçi, A. Simple Förster resonance energy transfer evidence for the ultrahigh quantum dot quenching efficiency by graphene oxide compared to other carbon structures. Carbon N. Y. 2012, 50, 2987–2993. [Google Scholar] [CrossRef]
- Morales-Narváez, E.; Merkoçi, A. Graphene Oxide as an Optical Biosensing Platform: A Progress Report. Adv. Mater. 2019, 31, 1805043. [Google Scholar] [CrossRef] [Green Version]

| Number | IGO (a.u.) | ISiO2 (a.u.) | E | d (nm) |
|---|---|---|---|---|
| 1 a | 14.4 | 20.2 | 0.29 | 9.4 |
| 2 b | 14.5 | 17.8 | 0.19 | 10.9 |
| 3 b | 22.5 | 29.4 | 0.23 | 10.1 |
| 4 b | 15.1 | 17.5 | 0.13 | 11.9 |
| 5 b | 20.2 | 20.9 | 0.03 | 17.4 |
| 6 b | 13.3 | 12.9 | −0.03 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okamoto, Y.; Iwasa, S.; Tero, R. Quenching Efficiency of Quantum Dots Conjugated to Lipid Bilayers on Graphene Oxide Evaluated by Fluorescence Single Particle Tracking. Appl. Sci. 2022, 12, 3733. https://doi.org/10.3390/app12083733
Okamoto Y, Iwasa S, Tero R. Quenching Efficiency of Quantum Dots Conjugated to Lipid Bilayers on Graphene Oxide Evaluated by Fluorescence Single Particle Tracking. Applied Sciences. 2022; 12(8):3733. https://doi.org/10.3390/app12083733
Chicago/Turabian StyleOkamoto, Yoshiaki, Seiji Iwasa, and Ryugo Tero. 2022. "Quenching Efficiency of Quantum Dots Conjugated to Lipid Bilayers on Graphene Oxide Evaluated by Fluorescence Single Particle Tracking" Applied Sciences 12, no. 8: 3733. https://doi.org/10.3390/app12083733
APA StyleOkamoto, Y., Iwasa, S., & Tero, R. (2022). Quenching Efficiency of Quantum Dots Conjugated to Lipid Bilayers on Graphene Oxide Evaluated by Fluorescence Single Particle Tracking. Applied Sciences, 12(8), 3733. https://doi.org/10.3390/app12083733






