A Neural Network-Based Method for Respiratory Sound Analysis and Lung Disease Detection
Abstract
1. Introduction
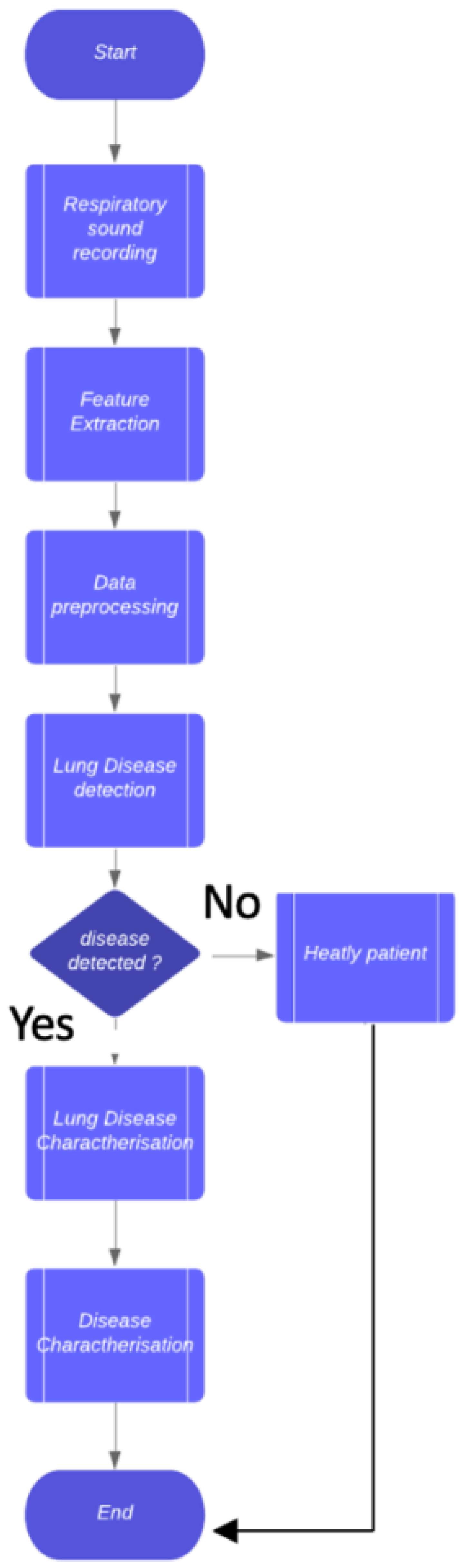
- a two-step method composed of a classifier is proposed: the first one aims to discriminate between healthy patients and patients affected by a generic lung disease, while the second model is devoted to detecting the specific lung disease;
- we exploit a feature vector directly obtained from respiratory sounds, which, to the best of the authors’ knowledge, has never been previously considered;
- in the experimental analysis, we use two datasets, obtained from real-world patients, composed of respiratory sounds, collected and labelled from two different institutions (the first one in Portugal and the second one in Greece);
- for conclusion validity, we analyse the effectiveness of the considered feature vector with different supervised machine learning techniques, by showing that machine learning can be helpful in the automatic detection of lung diseases;
- we obtain an F-Measure of 0.983 in lung disease detection;
- we obtain an F-Measure equal to 0.923 in lung disease characterisation, i.e., in the discrimination between asthma, bronchiectasis, bronchiolitis, chronic obstructive pulmonary disease, pneumonia, and lower or upper respiratory tract infection.
2. Materials and Methods
2.1. Materials
2.2. Methods
- Chromagram (CR): this feature is related to a chromagram representation automatically gathered from a waveform ( feature);
- Root Mean Square (RMS): this feature (i.e., RMS) is related the value of the mean square as the root that is obtained for each audio frame that is gathered from the sound sample under analysis ( feature);
- Spectral Centroid (SC): this feature is symptomatic of the “centre of mass” for a sound sample and is obtained as the mean related to the frequencies of the audio ( feature);
- Bandwidth: it is related the bandwidth of the spectrum ( feature);
- Spectral Roll-Off (SR): it is expressed as the frequency related to a certain percentage of the total spectral of the energy ( feature);
- Tonnetz (T): it is computed from the the tonal centroid ( feature).
- Mel-Frequency Cepstral Coefficient: this feature (i.e., MEL), whose acronym is related to a feature vector (ranging from 10 to 20 different numerical features 10–20), is devoted to representing the shape of a spectral envelope ( feature);
- Zero Crossing Rate (ZCR): this value is related to the rate of an audio time series ( feature);
- Poly (P): it is computed as the fitting coefficient related to an nth-order polynomial ( feature).
2.3. Study Design
3. Study Evaluation
3.1. Experiment Settings
- generation of set for the training, i.e., T⊂D;
- generation of an evaluation set T;
- execution of the model training T;
- application of the model previously generated to each element of the set.
3.2. Descriptive Statistics
3.3. Classification Performance
3.4. Model Analysis
4. Related Work
5. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tálamo, C.; de Oca, M.M.; Halbert, R.; Perez-Padilla, R.; Jardim, J.R.B.; Muino, A.; Lopez, M.V.; Valdivia, G.; Pertuzé, J.; Moreno, D.; et al. Diagnostic labeling of COPD in five Latin American cities. Chest 2007, 131, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Bohadana, A.; Izbicki, G.; Kraman, S.S. Fundamentals of lung auscultation. N. Engl. J. Med. 2014, 370, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Proctor, J.; Rickards, E. How to perform chest auscultation and interpret the findings. Nurs. Times 2020, 116, 23–26. [Google Scholar]
- Bahoura, M.; Pelletier, C. New parameters for respiratory sound classification. In Proceedings of the CCECE 2003-Canadian Conference on Electrical and Computer Engineering. Toward a Caring and Humane Technology (Cat. No. 03CH37436), Montreal, QC, Canada, 4–7 May 2003; Volume 3, pp. 1457–1460. [Google Scholar]
- Pasterkamp, H.; Kraman, S.S.; Wodicka, G.R. Respiratory sounds: Advances beyond the stethoscope. Am. J. Respir. Crit. Care Med. 1997, 156, 974–987. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, R.; Sundaraj, K.; Ahamed, N.U.; Arjunan, A.; Sundaraj, S. Computer-based respiratory sound analysis: A systematic review. IETE Tech. Rev. 2013, 30, 248–256. [Google Scholar] [CrossRef]
- Rocha, B.; Filos, D.; Mendes, L.; Vogiatzis, I.; Perantoni, E.; Kaimakamis, E.; Natsiavas, P.; Oliveira, A.; Jácome, C.; Marques, A.; et al. A respiratory sound database for the development of automated classification. In Precision Medicine Powered by pHealth and Connected Health; Springer: Berlin/Heidelberg, Germany, 2018; pp. 33–37. [Google Scholar]
- Guntupalli, K.K.; Alapat, P.M.; Bandi, V.D.; Kushnir, I. Validation of automatic wheeze detection in patients with obstructed airways and in healthy subjects. J. Asthma 2008, 45, 903–907. [Google Scholar] [CrossRef] [PubMed]
- de Lima Hedayioglu, F.; Coimbra, M.T.; da Silva Mattos, S. A Survey of Audio Processing Algorithms for Digital Stethoscopes. In Proceedings of the HEALTHINF, Porto, Portugal, 14–17 January 2009; pp. 425–429. [Google Scholar]
- Leng, S.; San Tan, R.; Chai, K.T.C.; Wang, C.; Ghista, D.; Zhong, L. The electronic stethoscope. Biomed. Eng. Online 2015, 14, 66. [Google Scholar] [CrossRef]
- McKinney, M.; Breebaart, J. Features for audio and music classification. In Proceedings of the ISMIR (International Conference on Music Information Retrieval), Baltimore, MD, USA, 27–30 October 2003. [Google Scholar]
- Breebaart, J.; McKinney, M.F. Features for audio classification. In Algorithms in Ambient Intelligence; Springer: Berlin/Heidelberg, Germany, 2004; pp. 113–129. [Google Scholar]
- Müller, M.; Kurth, F.; Clausen, M. Audio Matching via Chroma-Based Statistical Features. In Proceedings of the ISMIR (International Conference on Music Information Retrieval), London, UK, 11–15 September 2005; Volume 2005, p. 6. [Google Scholar]
- Valero, X.; Alias, F. Gammatone cepstral coefficients: Biologically inspired features for non-speech audio classification. IEEE Trans. Multimed. 2012, 14, 1684–1689. [Google Scholar] [CrossRef]
- Alías, F.; Socoró, J.C.; Sevillano, X. A review of physical and perceptual feature extraction techniques for speech, music and environmental sounds. Appl. Sci. 2016, 6, 143. [Google Scholar] [CrossRef]
- Chiţu, A.G.; Rothkrantz, L.J.; Wiggers, P.; Wojdel, J.C. Comparison between different feature extraction techniques for audio-visual speech recognition. J. Multimodal User Interfaces 2007, 1, 7–20. [Google Scholar] [CrossRef][Green Version]
- Lu, L.; Zhang, H.J.; Jiang, H. Content analysis for audio classification and segmentation. IEEE Trans. Speech Audio Process. 2002, 10, 504–516. [Google Scholar] [CrossRef]
- Vrysis, L.; Tsipas, N.; Thoidis, I.; Dimoulas, C. 1D/2D Deep CNNs vs. Temporal Feature Integration for General Audio Classification. J. Audio Eng. Soc. 2020, 68, 66–77. [Google Scholar] [CrossRef]
- Wei, P.; He, F.; Li, L.; Li, J. Research on sound classification based on SVM. Neural Comput. Appl. 2020, 32, 1593–1607. [Google Scholar] [CrossRef]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. An ensemble learning approach for brain cancer detection exploiting radiomic features. Comput. Methods Programs Biomed. 2020, 185, 105134. [Google Scholar] [CrossRef]
- Carfora, M.F.; Martinelli, F.; Mercaldo, F.; Nardone, V.; Orlando, A.; Santone, A.; Vaglini, G. A “pay-how-you-drive” car insurance approach through cluster analysis. Soft Comput. 2019, 23, 2863–2875. [Google Scholar] [CrossRef]
- Anthonisen, N.; Manfreda, J.; Warren, C.; Hershfield, E.; Harding, G.; Nelson, N. Antibiotic therapy in exacerbations of chronic obstructive pulmonary disease. Ann. Intern. Med. 1987, 106, 196–204. [Google Scholar] [CrossRef]
- Orimadegun, A.; Adepoju, A.; Myer, L. A Systematic Review and Meta-analysis of Sex Differences in Morbidity and Mortality of Acute Lower Respiratory Tract Infections among African Children. J. Pediatr. Rev. 2020, 8, 65. [Google Scholar] [CrossRef]
- Brooks, W.A. Bacterial Pneumonia. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 446–453. [Google Scholar]
- Trinh, N.T.; Bruckner, T.A.; Lemaitre, M.; Chauvin, F.; Levy, C.; Chahwakilian, P.; Cohen, R.; Chalumeau, M.; Cohen, J.F. Association between National Treatment Guidelines for Upper Respiratory Tract Infections and Outpatient Pediatric Antibiotic Use in France: An Interrupted Time–Series Analysis. J. Pediatr. 2020, 216, 88–94. [Google Scholar] [CrossRef]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Mitchell, T.M. Machine learning and data mining. Commun. ACM 1999, 42, 30–36. [Google Scholar] [CrossRef]
- Yamashita, M.; Matsunaga, S.; Miyahara, S. Discrimination between healthy subjects and patients with pulmonary emphysema by detection of abnormal respiration. In Proceedings of the 2011 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Prague, Czech Republic, 22–27 May 2011; pp. 693–696. [Google Scholar]
- Jin, F.; Krishnan, S.; Sattar, F. Adventitious sounds identification and extraction using temporal–spectral dominance-based features. IEEE Trans. Biomed. Eng. 2011, 58, 3078–3087. [Google Scholar]
- Flietstra, B.; Markuzon, N.; Vyshedskiy, A.; Murphy, R. Automated analysis of crackles in patients with interstitial pulmonary fibrosis. Pulm. Med. 2011, 2011, 590506. [Google Scholar] [CrossRef]
- Lang, R.; Lu, R.; Zhao, C.; Qin, H.; Liu, G. Graph-based semi-supervised one class support vector machine for detecting abnormal lung sounds. Appl. Math. Comput. 2020, 364, 124487. [Google Scholar] [CrossRef]
- Charleston-Villalobos, S.; Gonzalez-Camarena, R.; Chi-Lem, G.; Aljama-Corrales, T. Crackle Sounds Analysis by EprclMode Decomposition. IEEE Eng. Med. Biol. Mag. 2007, 26, 40–47. [Google Scholar]
- Rizal, A.; Anggraeni, L.; Suryani, V. Normal lung sound classification using LPC and back propagation neural network. In Proceedings of the International Seminar on Electrical Power, Electronics Communication, Brawijaya, Indonesia, 16–17 May 2006; pp. 6–10. [Google Scholar]
- Taplidou, S.A.; Hadjileontiadis, L.J. Wheeze detection based on time-frequency analysis of breath sounds. Comput. Biol. Med. 2007, 37, 1073–1083. [Google Scholar] [CrossRef]
- Rizal, A.; Hidayat, R.; Nugroho, H.A. Signal domain in respiratory sound analysis: Methods, application and future development. J. Comput. Sci. 2015, 11, 1005. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Takahashi, T.; Amagasa, T.; Kitagawa, H. Turank: Twitter user ranking based on user-tweet graph analysis. In International Conference on Web Information Systems Engineering; Springer: Berlin/Heidelberg, Germany, 2010; pp. 240–253. [Google Scholar]
- Scaffa, A.; Yao, H.; Oulhen, N.; Wallace, J.; Peterson, A.L.; Rizal, S.; Ragavendran, A.; Wessel, G.; De Paepe, M.E.; Dennery, P.A. Single-cell transcriptomics reveals lasting changes in the lung cellular landscape into adulthood after neonatal hyperoxic exposure. Redox Biol. 2021, 48, 102091. [Google Scholar] [CrossRef]
- Torre-Cruz, J.; Canadas-Quesada, F.; García-Galán, S.; Ruiz-Reyes, N.; Vera-Candeas, P.; Carabias-Orti, J. A constrained tonal semi-supervised non-negative matrix factorization to classify presence/absence of wheezing in respiratory sounds. Appl. Acoust. 2020, 161, 107188. [Google Scholar] [CrossRef]
- Acharya, J.; Basu, A. Deep neural network for respiratory sound classification in wearable devices enabled by patient specific model tuning. IEEE Trans. Biomed. Circuits Syst. 2020, 14, 535–544. [Google Scholar] [CrossRef]
- Srivastava, A.; Jain, S.; Miranda, R.; Patil, S.; Pandya, S.; Kotecha, K. Deep learning based respiratory sound analysis for detection of chronic obstructive pulmonary disease. PeerJ Comput. Sci. 2021, 7, e369. [Google Scholar] [CrossRef]
- Shi, Y.; Li, Y.; Cai, M.; Zhang, X.D. A lung sound category recognition method based on wavelet decomposition and BP neural network. Int. J. Biol. Sci. 2019, 15, 195. [Google Scholar] [CrossRef]
- Mondal, A.; Bhattacharya, P.; Saha, G. Detection of lungs status using morphological complexities of respiratory sounds. Sci. World J. 2014, 2014, 182938. [Google Scholar] [CrossRef]
- Gnitecki, J.; Moussavi, Z. The fractality of lung sounds: A comparison of three waveform fractal dimension algorithms. Chaos Solitons Fractals 2005, 26, 1065–1072. [Google Scholar] [CrossRef]
- Ayari, F.; Ksouri, M.; Alouani, A. A new scheme for automatic classification of pathologic lung sounds. Int. J. Comput. Sci. Issues (IJCSI) 2012, 9, 448. [Google Scholar]
- Alsmadi, S.S.; Kahya, Y.P. Online classification of lung sounds using DSP. In Proceedings of the Second Joint 24th Annual Conference and the Annual Fall Meeting of the Biomedical Engineering Society][Engineering in Medicine and Biology, Houston, TX, USA, 23–26 October 2002; Volume 2, pp. 1771–1772. [Google Scholar]
- Hadjileontiadis, L.J. A texture-based classification of crackles and squawks using lacunarity. IEEE Trans. Biomed. Eng. 2009, 56, 718–732. [Google Scholar] [CrossRef]
- Kahya, Y.P.; Yeginer, M.; Bilgic, B. Classifying respiratory sounds with different feature sets. In Proceedings of the 2006 International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 30 August–3 September 2006; pp. 2856–2859. [Google Scholar]
- Charleston-Villalobos, S.; Castañeda-Villa, N.; Gonzalez-Camarena, R.; Mejia-Avila, M.; Aljama-Corrales, T. Adventitious lung sounds imaging by ICA-TVAR scheme. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 1354–1357. [Google Scholar]
- Yamashita, M.; Himeshima, M.; Matsunaga, S. Robust classification between normal and abnormal lung sounds using adventitious-sound and heart-sound models. In Proceedings of the 2014 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Florence, Italy, 4–9 May 2014; pp. 4418–4422. [Google Scholar]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable Deep Learning for Pulmonary Disease and Coronavirus COVID-19 Detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]





| Model | F-Measure | Specificity | Sensitivity |
|---|---|---|---|
| kNN | 0.981 (0.993) | 0.965 (0.988) | 0.997 (0.999) |
| SVM | 0.983 (0.994) | 0.966 (0.990) | 1.000 (1.000) |
| Neural Network | 0.983 (0.991) | 0.979 (0.988) | 0.988 (0.995) |
| Logistic Regression | 0.979 (0.988) | 0.976 (0.986) | 0.982 (0.992) |
| Model | F-Measure | Specificity | Sensitivity |
|---|---|---|---|
| kNN | 0.892 (0.932) | 0.883 (0.927) | 0.908 (0.939) |
| SVM | 0.872 (0.936) | 0.890 (0.931) | 0.907 (0.938) |
| Neural Network | 0.923 (0.948) | 0.917 (0.941) | 0.931 (0.958) |
| Logistic Regression | 0.892 (0.916) | 0.886 (0.906) | 0.904 (0.929) |
| Actual Class | ||||||||
|---|---|---|---|---|---|---|---|---|
| Asthma | Be | Bl | COPD | LRTI | Pneumonia | URTI | ||
| Predicted | Asthma | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| class | Be | 0 | 6 | 1 | 0 | 0 | 0 | 0 |
| Bl | 0 | 0 | 6 | 0 | 0 | 0 | 0 | |
| COPD | 0 | 1 | 0 | 60 | 1 | 1 | 1 | |
| LRTI | 0 | 0 | 0 | 0 | 2 | 0 | 0 | |
| Pneumonia | 0 | 0 | 0 | 0 | 0 | 6 | 0 | |
| URTI | 0 | 0 | 0 | 2 | 0 | 0 | 14 |
| Class | F-Measure | Specificity | Sensitivity |
|---|---|---|---|
| Asthma | 1 | 1 | 1 |
| Be | 10.92 | 1 | 0.86 |
| Bl | 0.92 | 0.86 | 1 |
| COPD | 0.96 | 0.97 | 0.95 |
| LRTI | 0.80 | 0.67 | 1 |
| Pneumonia | 0.92 | 0.86 | 1 |
| URTI | 0.90 | 0.93 | 0.88 |
| Research | Features | Performance |
|---|---|---|
| Charleston et al. [32] | IMF | N.A. |
| Rizal et al. [33] | BP-NN | 98.33% |
| Mondal et al. [42] | ELM, SVM | 92.86% |
| Gnitecki et al. [43] | fractal | N.A. |
| Ayari et al. [44] | width | 98.3% |
| Alsmadi et al. [45] | K-NN | N.A. |
| Hadjileontiadis et al. [46] | Lacunarity | 99% |
| Kahya et al. [47] | AR coefficient | 67% |
| Charleston et al. [48] | Time-variant AR | N.A. |
| Yamashita et al. [49] | MFCC | 83% |
| Torre et al. [38] | NMF | 95% |
| Acharya et al. [39] | MEL | 71% |
| Srivastava et al. [40] | CNN | 93% |
| Shi et al. [41] | NN | 92.5% |
| Our method | CR,RMS,SC,SR,ZCR,MEL,T,P | 98% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. A Neural Network-Based Method for Respiratory Sound Analysis and Lung Disease Detection. Appl. Sci. 2022, 12, 3877. https://doi.org/10.3390/app12083877
Brunese L, Mercaldo F, Reginelli A, Santone A. A Neural Network-Based Method for Respiratory Sound Analysis and Lung Disease Detection. Applied Sciences. 2022; 12(8):3877. https://doi.org/10.3390/app12083877
Chicago/Turabian StyleBrunese, Luca, Francesco Mercaldo, Alfonso Reginelli, and Antonella Santone. 2022. "A Neural Network-Based Method for Respiratory Sound Analysis and Lung Disease Detection" Applied Sciences 12, no. 8: 3877. https://doi.org/10.3390/app12083877
APA StyleBrunese, L., Mercaldo, F., Reginelli, A., & Santone, A. (2022). A Neural Network-Based Method for Respiratory Sound Analysis and Lung Disease Detection. Applied Sciences, 12(8), 3877. https://doi.org/10.3390/app12083877







