Exploring the Strategy of Fusing Sucrose Synthase to Glycosyltransferase UGT76G1 in Enzymatic Biotransformation
(This article belongs to the Section Applied Biosciences and Bioengineering)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and Strains
2.2. Expression of Recombinant UGT and SuSy in E. coli
2.3. Determination of Glycosyltransferase and SuSy Activities
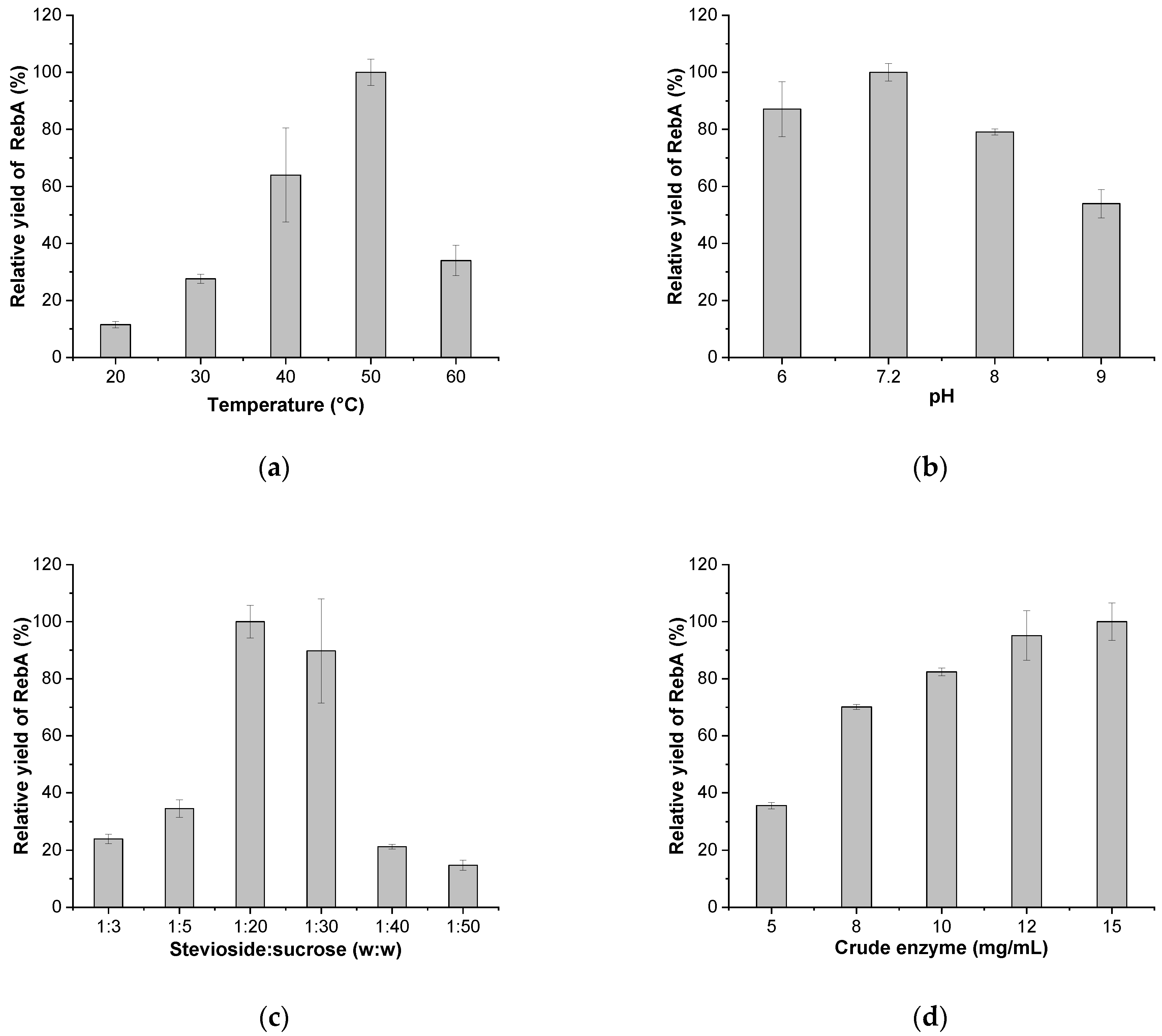
2.4. Enzymatic Synthesis of RebA
2.5. HPLC Analysis
2.6. Prediction of Protein Solubility
3. Results and Discussion
3.1. Selection of SuSys for Constructing One-Pot Two-Enzyme Systems
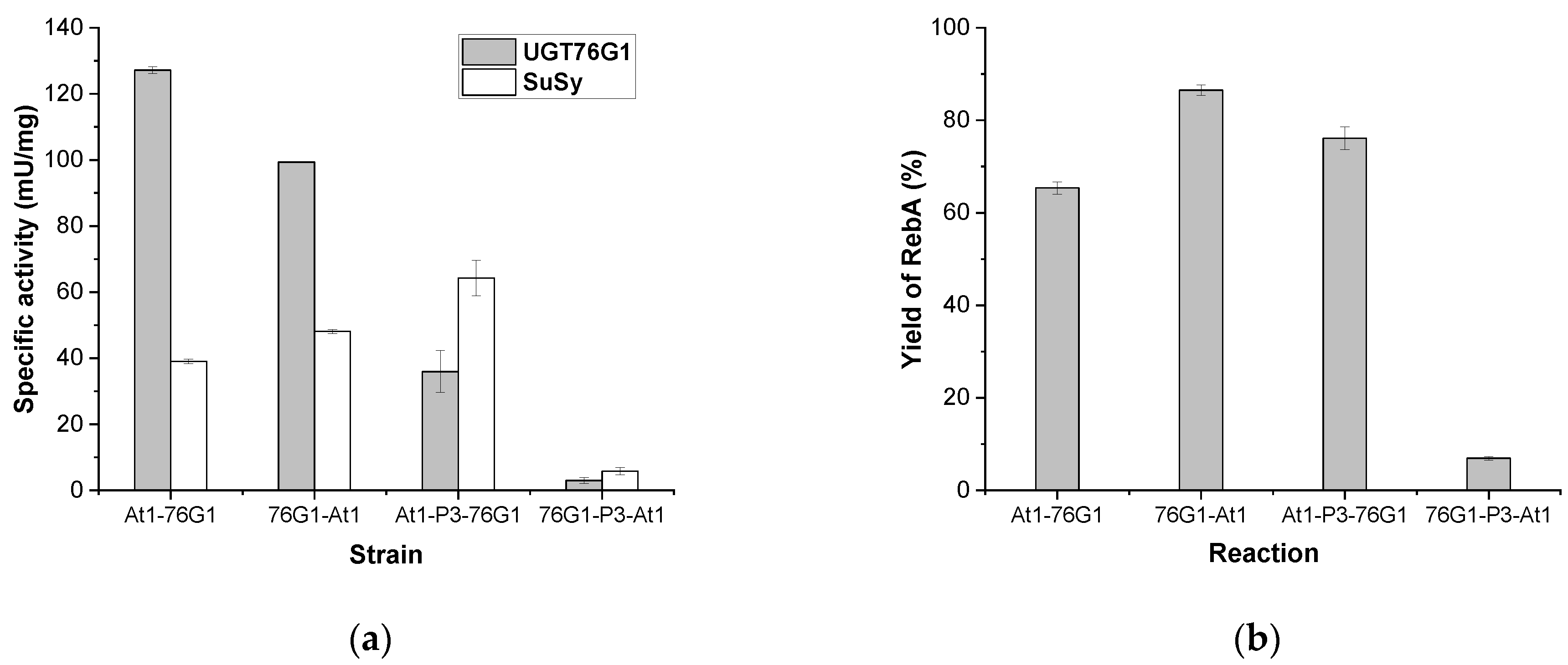
3.2. Comparison of the Construct Strategy for Expressing AtSUS1 and UGT76G1
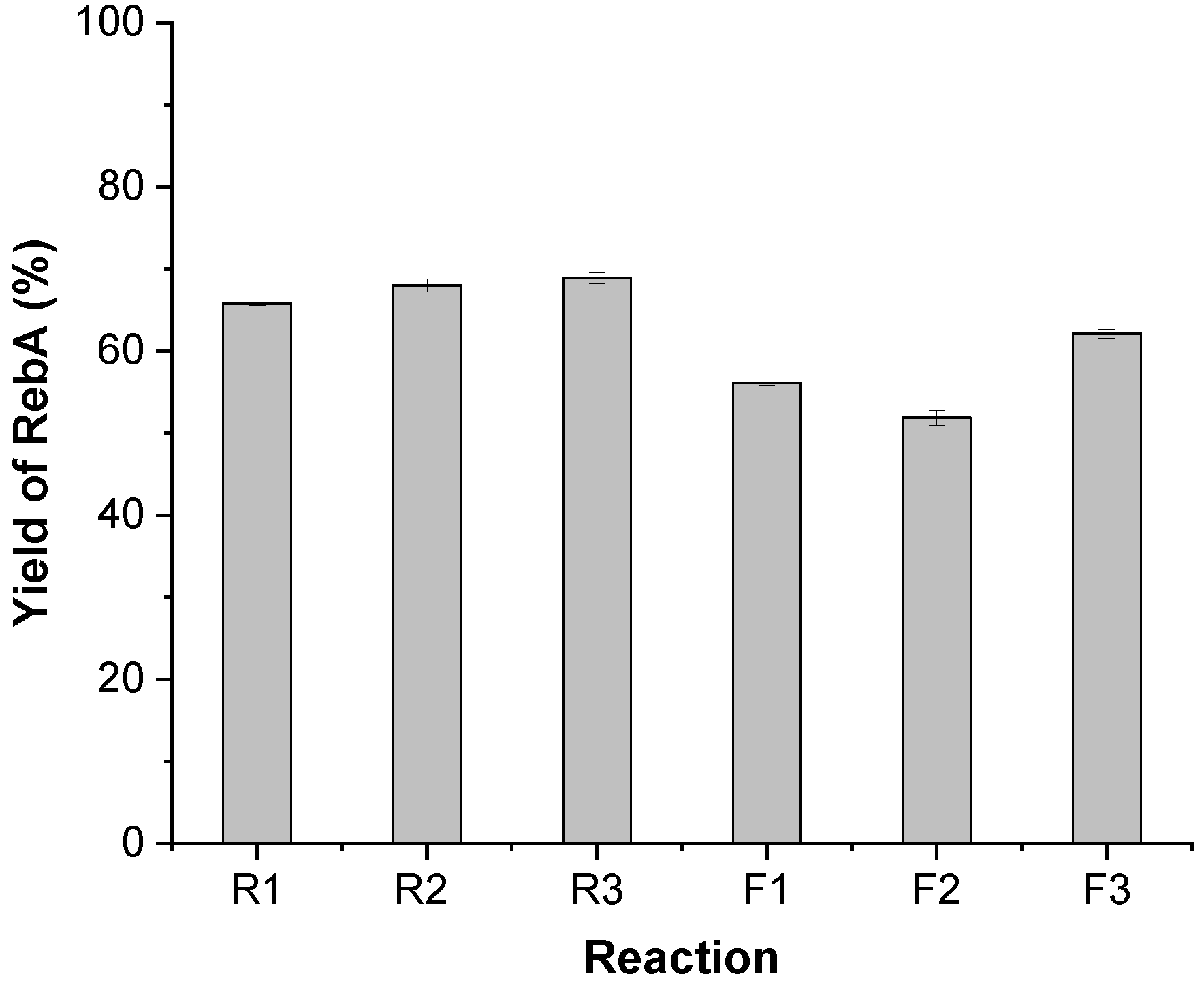
3.3. Fusion of AtSUS1 and UGT76G1 by Rigid and Flexible Linkers
3.4. Fusion of the Prokaryotic SuSy with UGT76G1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bowles, D.; Isayenkova, J.; Lim, E.K.; Poppenberger, B. Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 2005, 8, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Nidetzky, B.; Gutmann, A.; Zhong, C. Leloir glycosyltransferases as biocatalysts for chemical production. ACS Catal. 2018, 8, 6283–6300. [Google Scholar] [CrossRef]
- De Bruyn, F.; Maertens, J.; Beauprez, J.; Soetaert, W.; De Mey, M. Biotechnological advances in UDP-sugar based glycosylation of small molecules. Biotechnol. Adv. 2015, 33, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Danby, P.M.; Withers, S.G. Advances in enzymatic glycoside synthesis. ACS Chem. Biol. 2016, 11, 1784–1794. [Google Scholar] [CrossRef]
- Olsson, K.; Carlsen, S.; Semmler, A.; Simon, E.; Mikkelsen, M.D.; Moller, B.L. Microbial production of next-generation stevia sweeteners. Microb. Cell Factories 2016, 15, 207. [Google Scholar] [CrossRef] [Green Version]
- Rahimi, S.; Kim, J.; Mijakovic, I.; Jung, K.H.; Choi, G.; Kim, S.C.; Kim, Y.J. Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv. 2019, 37, 107394. [Google Scholar] [CrossRef]
- Xiao, J.B.; Muzashvili, T.S.; Georgiev, M.I. Advances in the biotechnological glycosylation of valuable flavonoids. Biotechnol. Adv. 2014, 32, 1145–1156. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhang, D.B.; Yang, D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol. Adv. 2015, 33, 717–735. [Google Scholar] [CrossRef]
- Wang, P.P.; Wang, J.L.; Zhao, G.P.; Yan, X.; Zhou, Z.H. Systematic optimization of the yeast cell factory for sustainable and high efficiency production of bioactive ginsenoside compound K. Syn. Syst. Biotechnol. 2021, 6, 69–76. [Google Scholar] [CrossRef]
- Lim, E.K.; Ashford, D.A.; Hou, B.K.; Jackson, R.G.; Bowles, D.J. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 2004, 87, 623–631. [Google Scholar] [CrossRef]
- Bungaruang, L.; Gutmann, A.; Nidetzky, B. Leloir glycosyltransferases and natural product glycosylation: Biocatalytic synthesis of the C-glucoside nothofagin, a major antioxidant of redbush herbal tea. Adv. Synth. Catal. 2013, 355, 2757–2763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmolzer, K.; Gutmann, A.; Diricks, M.; Desmet, T.; Nidetzky, B. Sucrose synthase: A unique glycosyltransferase for biocatalytic glycosylation process development. Biotechnol. Adv. 2016, 34, 88–111. [Google Scholar] [CrossRef] [PubMed]
- Diricks, M.; De Bruyn, F.; Van Daele, P.; Walmagh, M.; Desmet, T. Identification of sucrose synthase in nonphotosynthetic bacteria and characterization of the recombinant enzymes. Appl. Microbiol. Biotechnol. 2015, 99, 8465–8474. [Google Scholar] [CrossRef] [PubMed]
- Rupprath, C.; Schumacher, T.; Elling, L. Nucleotide deoxysugars: Essential tools for the glycosylation engineering of novel bioactive compounds. Curr. Med. Chem. 2005, 12, 1637–1675. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Sun, P.; Zhou, F.F.; Li, Y.; Chen, K.Q.; Jia, H.H.; Yan, M.; Gong, D.C.; Ouyang, P.K. Synthesis of rebaudioside D, using glycosyltransferase UGTSL2 and in situ UDP-glucose regeneration. Food Chem. 2018, 259, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Cai, R.; Chen, L.; Li, Y.; Jia, H.; Yan, M.; Chen, K. Natural product glycosylation: Biocatalytic synthesis of quercetin-3,4′-O-diglucoside. Appl. Biochem. Biotechnol. 2019, 190, 464–474. [Google Scholar] [CrossRef]
- Jia, F.; Narasimhan, B.; Mallapragada, S. Materials-based strategies for multi-enzyme immobilization and co-localization: A review. Biotechnol. Bioeng. 2014, 111, 209–222. [Google Scholar] [CrossRef]
- Li, Y.; Cirino, P.C. Recent advances in engineering proteins for biocatalysis. Biotechnol. Bioeng. 2014, 111, 1273–1287. [Google Scholar] [CrossRef]
- Andre, C.; Kim, S.W.; Yu, X.H.; Shanklin, J. Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2. Proc. Natl. Acad. Sci. USA 2013, 110, 3191–3196. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.P. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 2011, 29, 715–725. [Google Scholar] [CrossRef]
- Richman, A.; Swanson, A.; Humphrey, T.; Chapman, R.; McGarvey, B.; Pocs, R.; Brandle, J. Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J. 2005, 41, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Prakash, I.; Markosyan, A.; Bunders, C. Development of next generation Stevia sweetener: Rebaudioside M. Foods 2014, 3, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, L.L.; Li, Y.; Li, Y.Y.; Yan, M.; Chen, K.Q.; Hao, N.; Xu, L. Efficient enzymatic production of rebaudioside A from stevioside. Biosci. Biotechnol. Biochem. 2016, 80, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Cai, R.X.; Weng, J.Y.; Li, Y.; Jia, H.H.; Chen, K.Q.; Yan, M.; Ouyang, P.K. Production of rebaudioside D from stevioside using a UGTSL2 Asn358Phe mutant in a multi-enzyme system. Microb. Biotechnol. 2020, 13, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.D.; Elisee, C.; Newham, D.M.; Harrison, R.G. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 1999, 65, 382–388. [Google Scholar] [CrossRef]
- Liu, H.; Tegl, G.; Nidetzky, B. Glycosyltransferase co-immobilization for natural product glycosylation: Cascade biosynthesis of the C-glucoside nothofagin with efficient reuse of enzymes. Adv. Synth. Catal. 2021, 363, 2157–2169. [Google Scholar] [CrossRef]
- Chu, J.; Yue, J.; Qin, S.; Li, Y.; Wu, B.; He, B. Biocatalysis for rare ginsenoside Rh2 production in high level with co-immobilized UDP-glycosyltransferase Bs-YjiC mutant and sucrose synthase AtSuSy. Catalysts 2021, 11, 132. [Google Scholar] [CrossRef]
- Diricks, M.; Gutmann, A.; Debacker, S.; Dewitte, G.; Nidetzky, B.; Desmet, T. Sequence determinants of nucleotide binding in sucrose synthase: Improving the affinity of a bacterial sucrose synthase for UDP by introducing plant residues. Protein Eng. Des. Sel. 2017, 30, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Bieniawska, Z.; Barratt, D.H.P.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [CrossRef]
- Masada, S.; Kawase, Y.; Nagatoshi, M.; Oguchi, Y.; Terasaka, K.; Mizukami, H. An efficient chemoenzymatic production of small molecule glucosides with in situ UDP-glucose recycling. FEBS Lett. 2007, 581, 2562–2566. [Google Scholar] [CrossRef] [Green Version]
- Baroja-Fernandez, E.; Munoz, F.J.; Li, J.; Bahaji, A.; Almagro, G.; Montero, M.; Etxeberria, E.; Hidalgo, M.; Sesma, M.T.; Pozueta-Romero, J. Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. USA 2012, 109, 321–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romer, U.; Schrader, H.; Gunther, N.; Nettelstroth, N.; Frommer, W.B.; Elling, L. Expression, purification and characterization of recombinant sucrose synthase 1 from Solanum tuberosum L. for carbohydrate engineering. J. Biotechnol. 2004, 107, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Sauerzapfe, B.; Engels, L.; Elling, L. Broadening the biocatalytic properties of recombinant sucrose synthase 1 from potato (Solanum tuberosum L.) by expression in Escherichia coli and Saccharomyces cerevisiae. Enzym. Microb. Technol. 2008, 43, 289–296. [Google Scholar] [CrossRef]
- Schmolzer, K.; Lemmerer, M.; Gutmann, A.; Nidetzky, B. Integrated process design for biocatalytic synthesis by a Leloir glycosyltransferase: UDP-glucose production with sucrose synthase. Biotechnol. Bioeng. 2017, 114, 924–928. [Google Scholar] [CrossRef] [PubMed]
- Sabourin, M.; Tuzon, C.T.; Fisher, T.S.; Zakian, V.A. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast 2007, 24, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amet, N.; Wang, W.; Shen, W.C. Human growth hormone-transferrin fusion protein for oral delivery in hypophysectomized rats. J. Control. Release 2010, 141, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Amet, N.; Lee, H.F.; Shen, W.C. Insertion of the designed helical linker led to increased expression of tf-based fusion proteins. Pharm. Res. 2009, 26, 523–528. [Google Scholar] [CrossRef] [Green Version]
- Son, M.H.; Kim, B.G.; Kim, D.H.; Jin, M.; Kim, K.; Ahn, J.H. Production of flavonoid O-glucoside using sucrose synthase and flavonoid O-glucosyltransferase fusion protein. J. Microbiol. Biotechnol. 2009, 19, 709–712. [Google Scholar] [CrossRef] [Green Version]
- Argos, P. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. J. Mol. Biol. 1990, 211, 943–958. [Google Scholar] [CrossRef]
- George, R.A.; Heringa, J. An analysis of protein domain linkers: Their classification and role in protein folding. Protein Eng. Des. Sel. 2002, 15, 871–879. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.Y.; Zaro, J.L.; Shen, W.C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.Y.; Chu, J.L.; Guo, Y.L.; Li, H.; Wu, B.; He, B.F. An efficient production of high-pure xylooligosaccharides from corncob with affinity adsorption-enzymatic reaction integrated approach. Bioresour. Technol. 2017, 241, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Kusters, K.; Pohl, M.; Krauss, U.; Olcucu, G.; Albert, S.; Jaeger, K.E.; Wiechert, W.; Oldiges, M. Construction and comprehensive characterization of an EcLDCc-CatIB set-varying linkers and aggregation inducing tags. Microb. Cell Factories 2021, 20, 49. [Google Scholar] [CrossRef] [PubMed]
- Savickaite, A.; Druteika, G.; Sadauskas, M.; Malunavicius, V.; Lastauskiene, E.; Gudiukaite, R. Study of individual domains’ functionality in fused lipolytic biocatalysts based on Geobacillus lipases and esterases. Int. J. Biol. Macromol. 2021, 168, 261–271. [Google Scholar] [CrossRef]
- Grawe, A.; Ranglack, J.; Weyrich, A.; Stein, V. IFLinkC: An iterative functional linker cloning strategy for the combinatorial assembly and recombination of linker peptides with functional domains. Nucleic Acids Res. 2020, 48, e24. [Google Scholar] [CrossRef]

| Strains | Plasmids | Plasmid Description |
|---|---|---|
| At1-76G1 | pRSF-AtSUS1-76G1 | Genes encoding AtSUS1 and UGT76G1 inserted in NcoI/EcoRI and NdeI/XhoI of pRSFDuet-1, respectively |
| St1-76G1 | pRSF-StSUS1-76G1 | The coding region of AtSUS1 was replaced by that of StSUS1 in pRSF-AtSUS1-76G1 |
| Ac-76G1 | pRSF-AcSuSy-76G1 | The coding region of AtSUS1 was replaced by that of AcSuSy in pRSF-AtSUS1-76G1 |
| At3-76G1 | pRSF-AtSUS3-76G1 | The coding region of AtSUS1 was replaced by that of AtSUS3 in pRSF-AtSUS1-76G1 |
| 76G1-At1 | pRSF-76G1-AtSUS1 | Genes encoding UGT76G1 and AtSUS1 inserted in NcoI/EcoRI and NdeI/XhoI of pRSFDuet-1, respectively |
| At1-P3-76G1 | pRSF-AtSUS1-P3-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with a linker of PPP in pRSF-AtSUS1-76G1 |
| 76G1-P3-At1 | pRSF-76G1-P3-AtSUS1 | The coding regions of UGT76G1 and AtSUS1 are fused with a linker of PPP in pRSF-76G1-AtSUS1 |
| R1 | pRSF-AtSUS1-R1-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with an R1 linker in pRSF-AtSUS1-76G1 |
| R2 | pRSF-AtSUS1-R2-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with an R2 linker in pRSF-AtSUS1-76G1 |
| R3 | pRSF-AtSUS1-R3-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with an R3 linker in pRSF-AtSUS1-76G1 |
| F1 | pRSF-AtSUS1-F1-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with an F1 linker in pRSF-AtSUS1-76G1 |
| F2 | pRSF-AtSUS1-F2-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with an F2 linker in pRSF-AtSUS1-76G1 |
| F3 | pRSF-AtSUS1-F3-76G1 | The coding regions of AtSUS1 and UGT76G1 are fused with an F3 linker in pRSF-AtSUS1-76G1 |
| Acm-76G1 | pRSF-AcSuSym-76G1 | Mutations of L637M and T640V in the encoding region of AcSuSy (AcSuSym) were made in pRSF-AcSuSy-76G1 |
| Acm-R3-76G1 | pRSF-AcSuSym-R3-76G1 | The coding regions of AcSuSym and UGT76G1 are fused with an R3 linker in pRSF-AcSuSym-76G1 |
| Linker | Amino Acids | Encoding Sequence (5′-3′) |
|---|---|---|
| P3 | PPP | CACCACCACCACCACCAC |
| R1 | EAAAK | GAGGCGGCGGCGAAG |
| R2 | (EAAAK)2 | GAGGCTGCTGCGAAGGAAGCGGCGGCGAAA |
| R3 | (EAAAK)3 | GAGGCTGCTGCGAAGGAAGCGGCGGCGAAAGAGGCGGCGGCGAAG |
| F1 | GGGGS | GGTGGCGGTGGCAGC |
| F2 | (GGGGS)2 | GGTGGCGGTGGCAGCGGTGGCGGTGGCAGC |
| F3 | (GGGGS)3 | GGTGGCGGTGGCAGCGGTGGCGGTGGCAGCGGTGGCGGTGGCAGC |
| Strains Used for the Crude Extract Preparation | Specific Activity | RebA | ||
|---|---|---|---|---|
| UGT76G1 (mU/mg) | SuSy (mU/mg) | Concentration (g/L) | Yield (%) | |
| At1-76G1 | 125.1 ± 10.5 | 47.6 ± 0.2 | 35.2 ± 1.6 | 99.3 |
| At3-76G1 | 104.4 ± 4.8 | 13.7 ± 0.3 | 31.6 ± 2.7 | 90.5 |
| St1-76G1 | 88.8 ± 9.3 | 11.6 ± 0.6 | 24.6 ± 0.4 | 66.8 |
| Ac-76G1 | 114.7 ± 5.7 | 59.2 ± 0.4 | 0.63 ± 0.04 | 0.8 |
| Reaction | RebA (g/L) | |
|---|---|---|
| 1 h | 3 h | |
| At1-P3-76G1 | 3.5 ± 0.2 | 9.2 ± 0.5 |
| At1-R3-76G1 | 3.3 ± 0.1 | 8.8 ± 0.5 |
| Protein | MW (kDa) | Size (aa) | Probability of Solubility or Insolubility |
|---|---|---|---|
| UGT76G1 | 52.0 | 458 | 57% insoluble |
| AtSUS1 | 93.0 | 808 | 60% soluble |
| AcSuSym | 91.3 | 794 | 63% soluble |
| 76G1-P3-At1 | 147.4 | 1289 | 50% soluble |
| At1-P3-76G1 | 145.3 | 1269 | 52% soluble |
| R3 (At1-R3-76G1) | 146.4 | 1281 | 54% soluble |
| Acm-R3-76G1 | 144.6 | 1266 | 56% soluble |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Y.; Sun, P.; Cai, R.; Li, Y.; Jia, H. Exploring the Strategy of Fusing Sucrose Synthase to Glycosyltransferase UGT76G1 in Enzymatic Biotransformation. Appl. Sci. 2022, 12, 3911. https://doi.org/10.3390/app12083911
Tao Y, Sun P, Cai R, Li Y, Jia H. Exploring the Strategy of Fusing Sucrose Synthase to Glycosyltransferase UGT76G1 in Enzymatic Biotransformation. Applied Sciences. 2022; 12(8):3911. https://doi.org/10.3390/app12083911
Chicago/Turabian StyleTao, Yehui, Ping Sun, Ruxin Cai, Yan Li, and Honghua Jia. 2022. "Exploring the Strategy of Fusing Sucrose Synthase to Glycosyltransferase UGT76G1 in Enzymatic Biotransformation" Applied Sciences 12, no. 8: 3911. https://doi.org/10.3390/app12083911







