Inhibition of Listeria monocytogenes by Broth Cultures of Surface Microbiota of Wooden Boards Used in Cheese Ripening
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Microflora Sampling of Wooden Cheese-Ripening Boards
2.2. Establishing a Growth Curve for Listeria monocytogenes 2203
2.3. Growth Curves of the Cheese Board Surface Microflora
2.4. Growth of Listeria monocytogenes 2203 in the Presence of the Microbial Communities
2.5. 16S rRNA Analysis of the Cheese Board Inhibitory Communities
2.6. Amplification and Sequencing of Bacterial 16S rRNA
2.7. Sequence Processing and Analysis
2.8. Statistical Analyses
3. Results
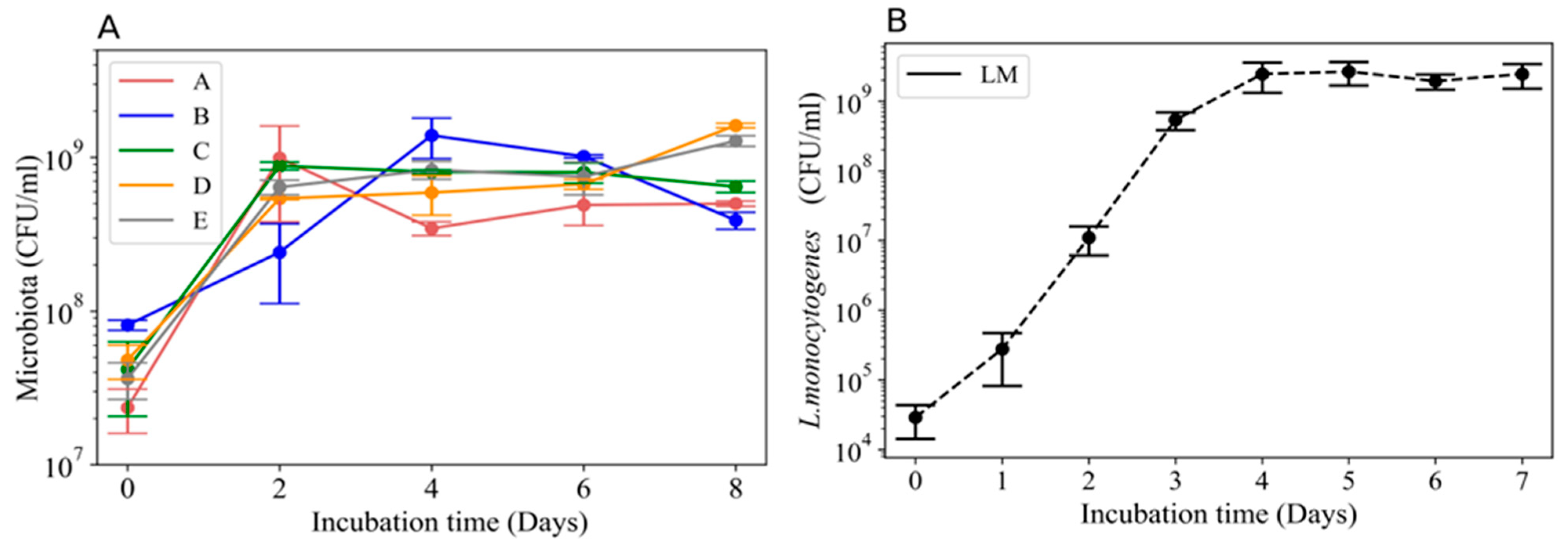
3.1. Growth Curves for the Wooden Board Microbial Communities and L. monocytogenes 2203
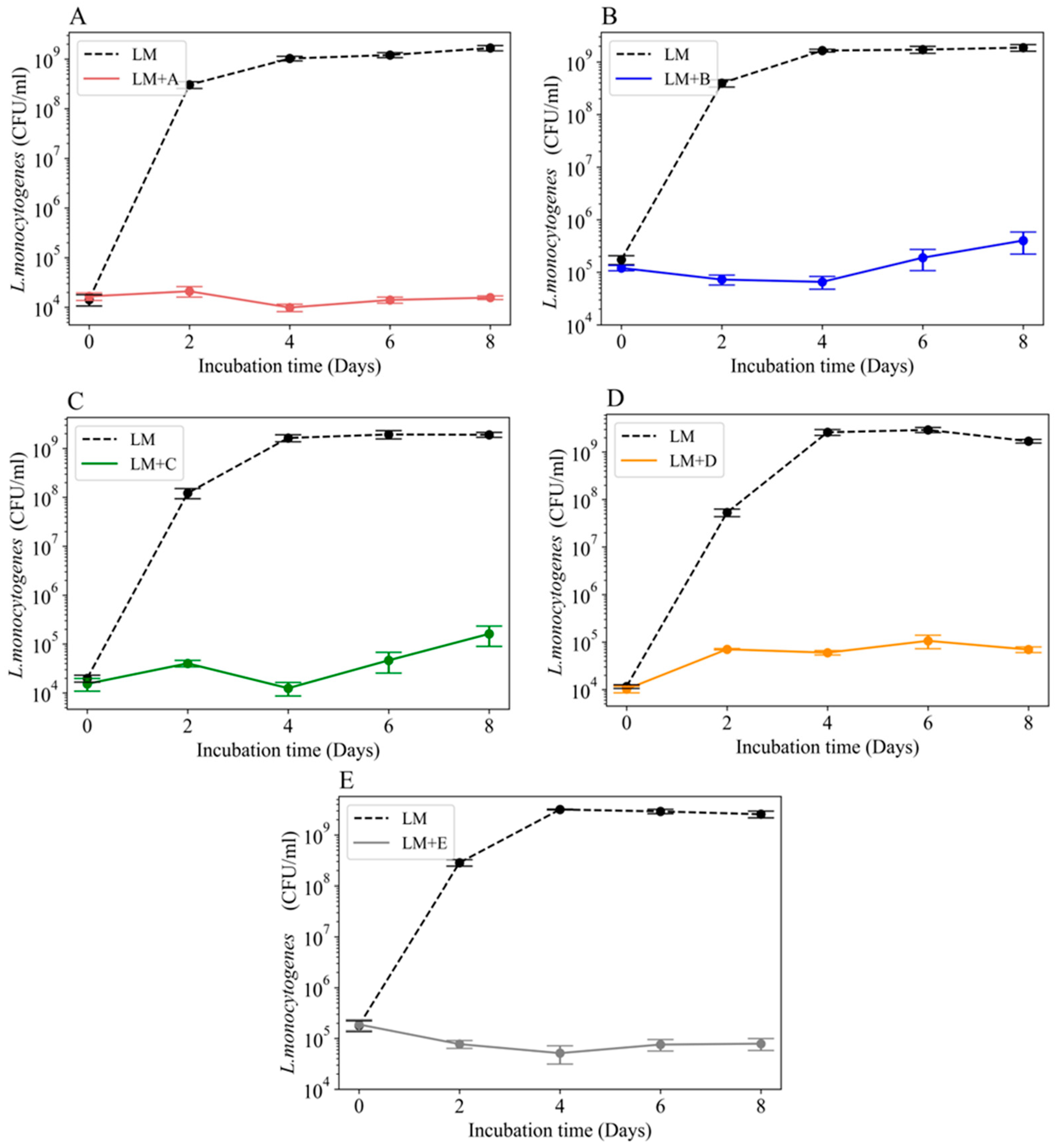
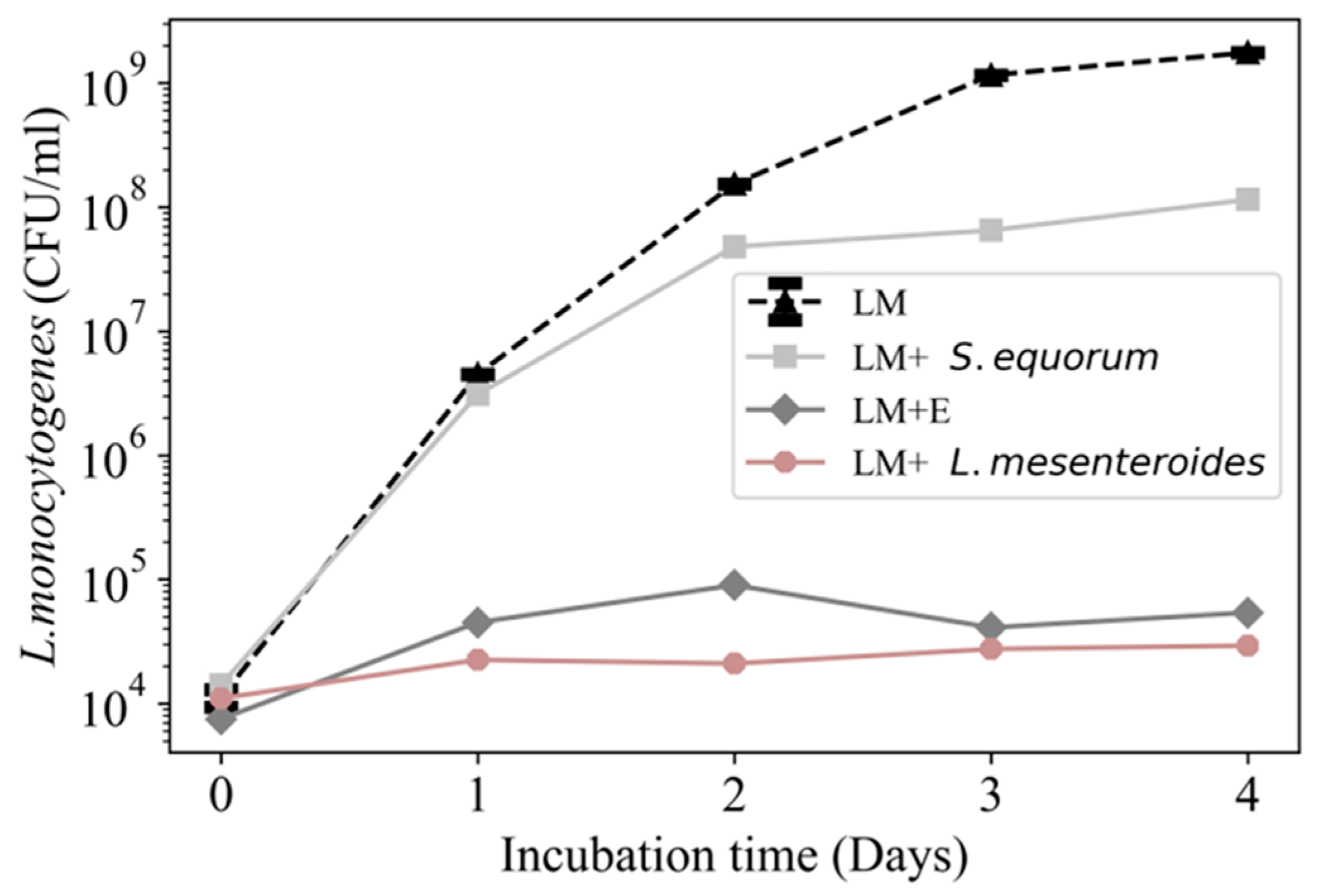
3.2. Growth of L. monocytogenes 2203 in the Presence of Microbial Communities
3.3. Sequencing Summary
3.4. Diversity and Composition of Cultured Cheese Board Bacterial Communities
3.5. Impact of L. monocytogenes 2203 on Cheese Board Bacterial Communities
3.6. Inhibition of L. monocytogenes 2203 by Specific Cheese Board Microbes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Diversity | Analysis | Sample Sets | p-Value |
|---|---|---|---|
| Shannon | Kruskal | Group | 0.06 |
| Category | 0.13 | ||
| Days | 0.5 | ||
| Chao | Kruskal | Group | 0.6 |
| Category | 0.4 | ||
| Days | 0.2 | ||
| Bray-Curtis | Betadisper | Group, days, category | >0.05 |
| PERMANOVA | Group | <0.05 * | |
| Category | 0.32 | ||
| Days | <0.05 * | ||
| Jaccard | Betadisper | Group, days, category | >0.05 |
| PERMANOVA | Group | <0.05 * | |
| Category | 0.29 | ||
| Days | <0.05 * |
| Group | Analysis: Corrected Method | Adjusted p-Value (Bray-Curtis) | Adjusted p-Value (Jaccard) |
|---|---|---|---|
| A vs. B | Pairwise Adonis: fdr | 0.023 | 0.023 |
| A vs. C | 0.239 | 0.238 | |
| A vs. D | 0.003 * | 0.003 * | |
| A vs. E | 0.003 * | 0.003 * | |
| B vs. C | 0.022 | 0.021 | |
| B vs. D | 0.003 * | 0.003 * | |
| B vs. E | 0.003 * | 0.003 * | |
| C vs. D | 0.003 * | 0.003 * | |
| C vs. E | 0.003 * | 0.003 * | |
| D vs. E | 0.003 * | 0.003 * |
References
- Aviat, F.; Gerhards, C.; Rodriguez-Jerez, J.; Michel, V.; le Bayon, I.; Ismail, R.; Federighi, M. Microbial safety of wood in contact with food: A review. Compr. Rev. Food Sci. Food Saf. 2016, 15, 491–505. [Google Scholar] [CrossRef]
- Dervisoglu, M.; Yazici, F. Ripening changes of Kulek cheese in wooden and plastic containers. J. Food Eng. 2001, 48, 243–249. [Google Scholar] [CrossRef]
- di Grigoli, A.; Francesca, N.; Gaglio, R.; Guarrasi, V.; Moschetti, M.; Scatassa, M.L.; Settanni, L.; Bonanno, A. The influence of the wooden equipment employed for cheese manufacture on the characteristics of a traditional stretched cheese during ripening. Food Microbiol. 2015, 46, 81–91. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, P.D.M.; Whitwam, R.E.; Boggs, J.D.; MacCormack, J.N.; Anderson, K.L.; Reardon, J.W.; Saah, J.R.; Graves, L.M.; Hunter, S.B.; Sobel, J. Outbreak of Listeriosis among Mexican immigrants as a result of consumption of illicitly produced Mexican-style cheese. Clin. Infect. Dis. 2005, 40, 677–682. [Google Scholar] [CrossRef]
- Melo, J.; Andrew, P.W.; Faleiro, M.L. Listeria monocytogenes in cheese and the dairy environment remains a food safety challenge: The role of stress responses. Food Res. Int. 2015, 67, 75–90. [Google Scholar] [CrossRef]
- Ak, N.O.; Cliver, D.O.; Kaspar, C.W. Decontamination of plastic and wooden cutting boards for kitchen use. J. Food Prot. 1994, 57, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Ak, N.O.; Cliver, D.O.; Kaspar, C.W. Cutting boards of plastic and wood contaminated experimentally with bacteria. J. Food Prot. 1994, 57, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Zangerl, P.; Matlschweiger, C.; Dillinger, K.; Eliskases-Lechner, F. Survival of Listeria monocytogenes after cleaning and sanitation of wooden shelves used for cheese ripening. Eur. J. Wood Wood Prod. 2009, 68, 415–419. [Google Scholar] [CrossRef]
- Mariani, C.; Oulahal, N.; Chamba, J.-F.; Dubois-Brissonnet, F.; Notz, E.; Briandet, R. Inhibition of Listeria monocytogenes by resident biofilms present on wooden shelves used for cheese ripening. Food Control 2011, 22, 1357–1362. [Google Scholar] [CrossRef]
- Bremer, P.J.; Monk, I.A.N.; Osborne, C.M. Survival of Listeria monocytogenes attached to stainless steel surfaces in the presence or absence of Flavobacterium spp. J. Food Prot. 2001, 64, 1369–1376. [Google Scholar] [CrossRef]
- Wadhawan, K.; Steinberger, A.J.; Rankin, S.A.; Suen, G.; Czuprynski, C.J. Characterizing the microbiota of wooden boards used for cheese ripening. JDS Commun. 2021, 2, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.; de Paula, T.; Lin, M.; Hall, M.B.; Suen, G. Transient changes in milk production efficiency and bacterial community composition resulting from near-total exchange of ruminal contents between high- and low-efficiency Holstein cows. J. Dairy Sci. 2017, 100, 7165–7182. [Google Scholar] [CrossRef]
- Li, W.; Edwards, A.; Riehle, C.; Cox, M.S.; Raabis, S.; Skarlupka, J.H.; Steinberger, A.J.; Walling, J.; Bickhart, D.; Suen, G. Transcriptomics analysis of host liver and meta-transcriptome analysis of rumen epimural microbial community in young calves treated with artificial dosing of rumen content from adult donor cow. Sci. Rep. 2019, 9, 790. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.D.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. 2019. Available online: http://www.R-project.org (accessed on 4 September 2022).
- Allaire, J.J. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 4 September 2022).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P. Vegan: Community Ecology Package, R package Version 2.5–7; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://CRAN.Rproject.org/package=vegan (accessed on 4 September 2022).
- Wickham, H.; Francois, R. dplyr: A Grammar of Data Manipulation, R Package Version 0.4.3.; R Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: http://CRAN.R-project.org/package=dplyr (accessed on 4 September 2022).
- Good, I.J.; Toulmin, G.H. The number of new species, and the increase in population coverage, when a sample is increased. Biometrika 1956, 43, 45–63. [Google Scholar] [CrossRef]
- Mellefont, L.A.; McMeekin, T.A.; Ross, T. Effect of relative inoculum concentration on Listeria monocytogenes growth in co-culture. Int. J. Food Microbiol. 2008, 121, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Guillier, L.; Stahl, V.; Hezard, B.; Notz, E.; Briandet, R. Modelling the competitive growth between Listeria monocytogenes and biofilm microflora of smear cheese wooden shelves. Int. J. Food Microbiol. 2008, 128, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Leisner, J.J.; Laursen, B.G.; Prévost, H.; Drider, D.; Dalgaard, P. Carnobacterium: Positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 2007, 31, 592–613. [Google Scholar] [CrossRef] [PubMed]
- Héchard, Y.; Dérijard, B.; Letellier, F.; Cenatiempo, Y. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. Microbiology 1992, 138, 2725–2731. [Google Scholar] [CrossRef] [PubMed]
- Settanni, L.; Busetta, G.; Puccio, V.; Licitra, G.; Franciosi, E.; Botta, L.; Di Gerlando, R.; Todaro, M.; Gaglio, R. In-depth investigation of the safety of wooden shelves used for traditional cheese Ripening. Appl. Environ. Microbiol. 2021, 87, e01524-21. [Google Scholar] [CrossRef]
- Gaglio, R.; Busetta, G.; Gannuscio, R.; Settanni, L.; Licitra, G.; Todaro, M. A multivariate approach to study the bacterial diversity associated to the wooden shelves used for aging traditional Sicilian cheeses. Foods 2022, 11, 774. [Google Scholar] [CrossRef]
- Licitra, G.; Caccamo, M.; Valence, F.; Lortal, S. Traditional wooden equipment used for cheesemaking and their effect on quality. In Global Cheesemaking Technology: Cheese Quality and Characteristics; Papademas, P., Bintsis, T., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 157–172. [Google Scholar]
- Ismail, R.; Aviat, F.; Gay-Perret, P.; Le Bayon, I.; Federighi, M.; Michel, V. An assessment of L. monocytogenes transfer from wooden ripening shelves to cheeses: Comparison with glass and plastic surfaces. Food Control 2017, 73, 273–280. [Google Scholar] [CrossRef]
- Imhof, R.; Schwendimann, L.; Riva Scettrini, P. Sanitising wooden boards used for cheese maturation by means of a steam-mediated heating process. J. Consum. Prot. Food Saf. 2017, 12, 255–263. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wadhawan, K.; Steinberger, A.; Rankin, S.; Suen, G.; Czuprynski, C. Inhibition of Listeria monocytogenes by Broth Cultures of Surface Microbiota of Wooden Boards Used in Cheese Ripening. Appl. Sci. 2023, 13, 5872. https://doi.org/10.3390/app13105872
Wadhawan K, Steinberger A, Rankin S, Suen G, Czuprynski C. Inhibition of Listeria monocytogenes by Broth Cultures of Surface Microbiota of Wooden Boards Used in Cheese Ripening. Applied Sciences. 2023; 13(10):5872. https://doi.org/10.3390/app13105872
Chicago/Turabian StyleWadhawan, Kirty, Andrew Steinberger, Scott Rankin, Garret Suen, and Charles Czuprynski. 2023. "Inhibition of Listeria monocytogenes by Broth Cultures of Surface Microbiota of Wooden Boards Used in Cheese Ripening" Applied Sciences 13, no. 10: 5872. https://doi.org/10.3390/app13105872
APA StyleWadhawan, K., Steinberger, A., Rankin, S., Suen, G., & Czuprynski, C. (2023). Inhibition of Listeria monocytogenes by Broth Cultures of Surface Microbiota of Wooden Boards Used in Cheese Ripening. Applied Sciences, 13(10), 5872. https://doi.org/10.3390/app13105872






