Impact of Cytochrome P450 Enzymes on the Phase I Metabolism of Drugs
Abstract
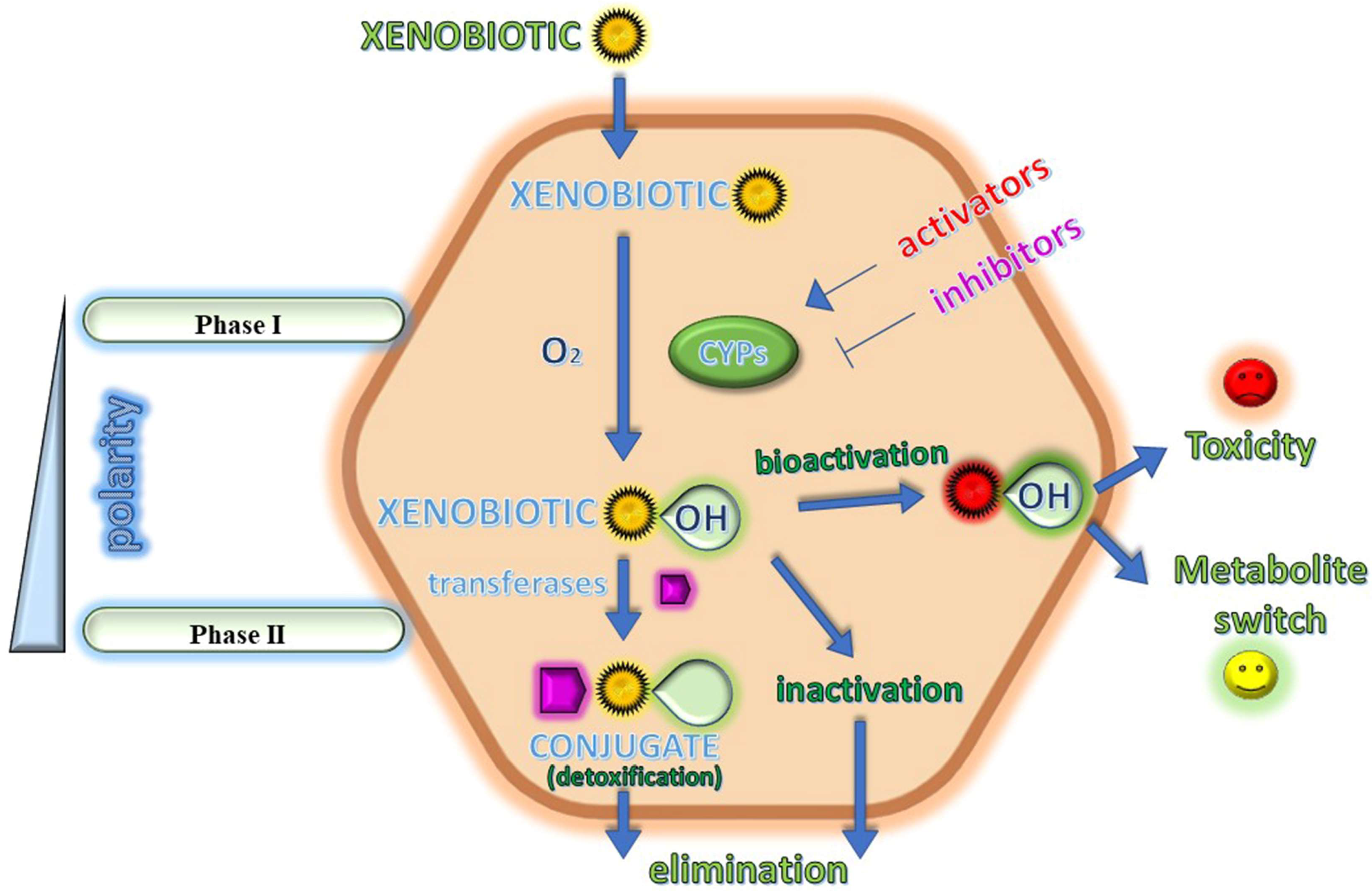
1. Introduction
2. Examples of Drugs Metabolized by CYPs
2.1. Voriconazole
2.2. Mexiletine
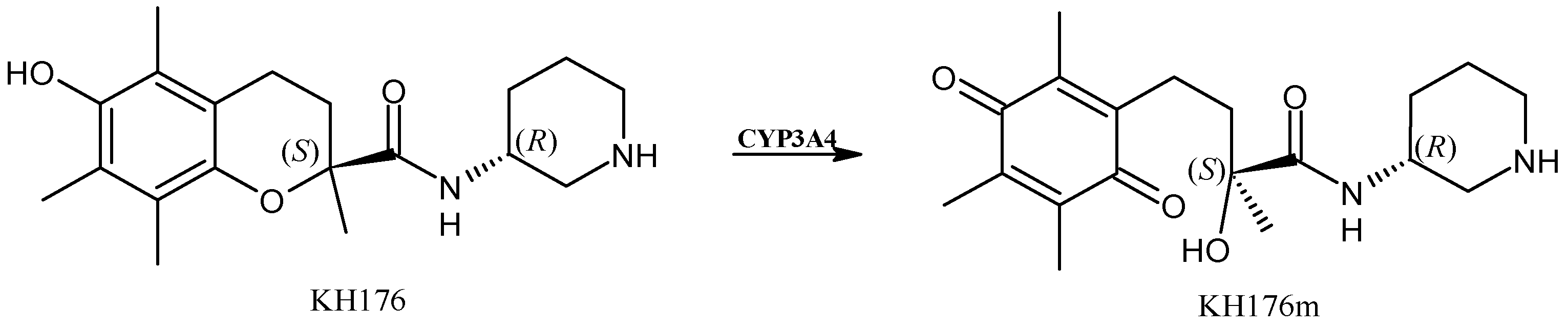
2.3. Sonlicromanol
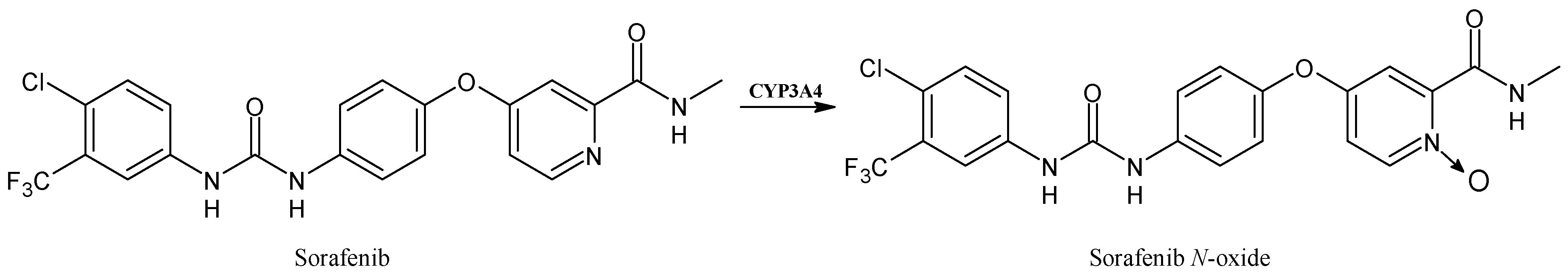
2.4. Sorafenib
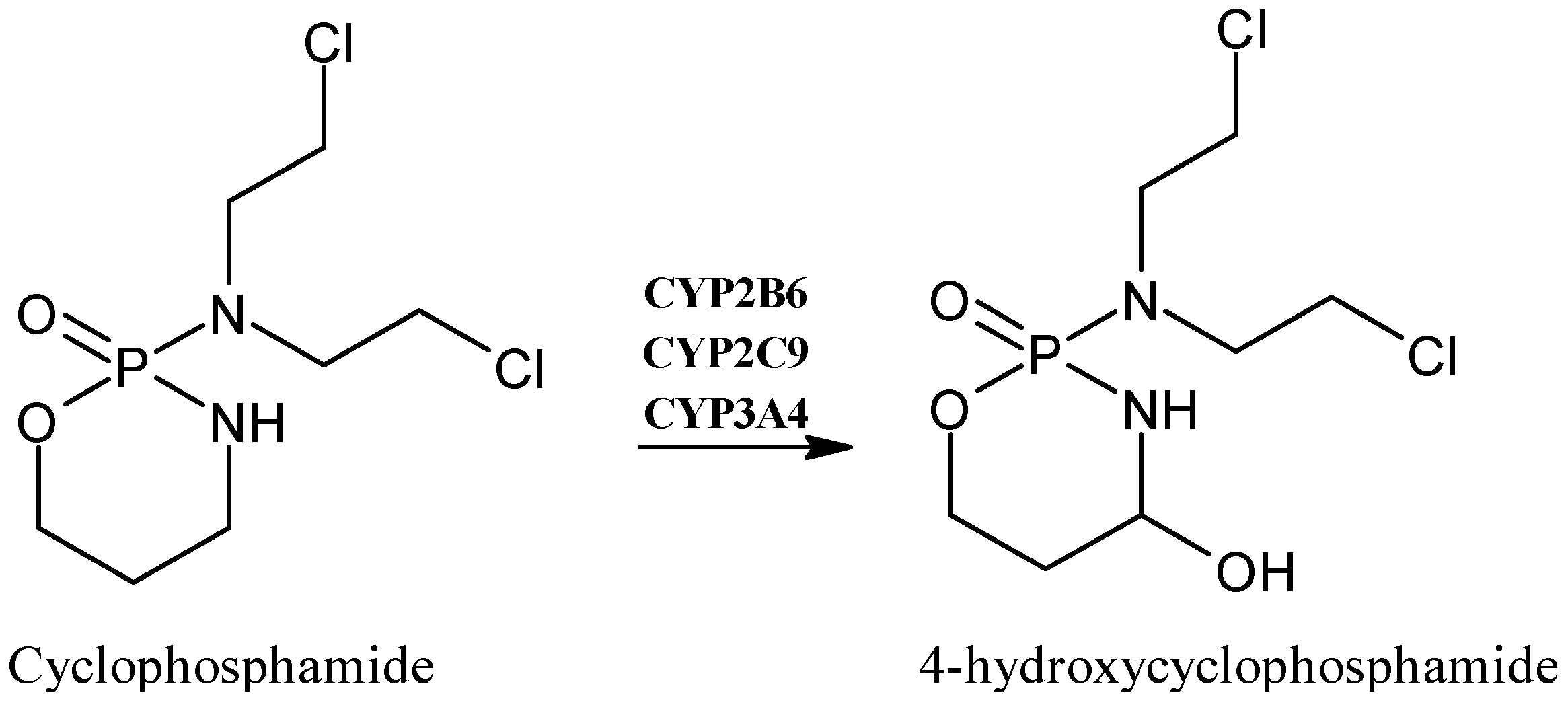
2.5. Cyclophosphamide
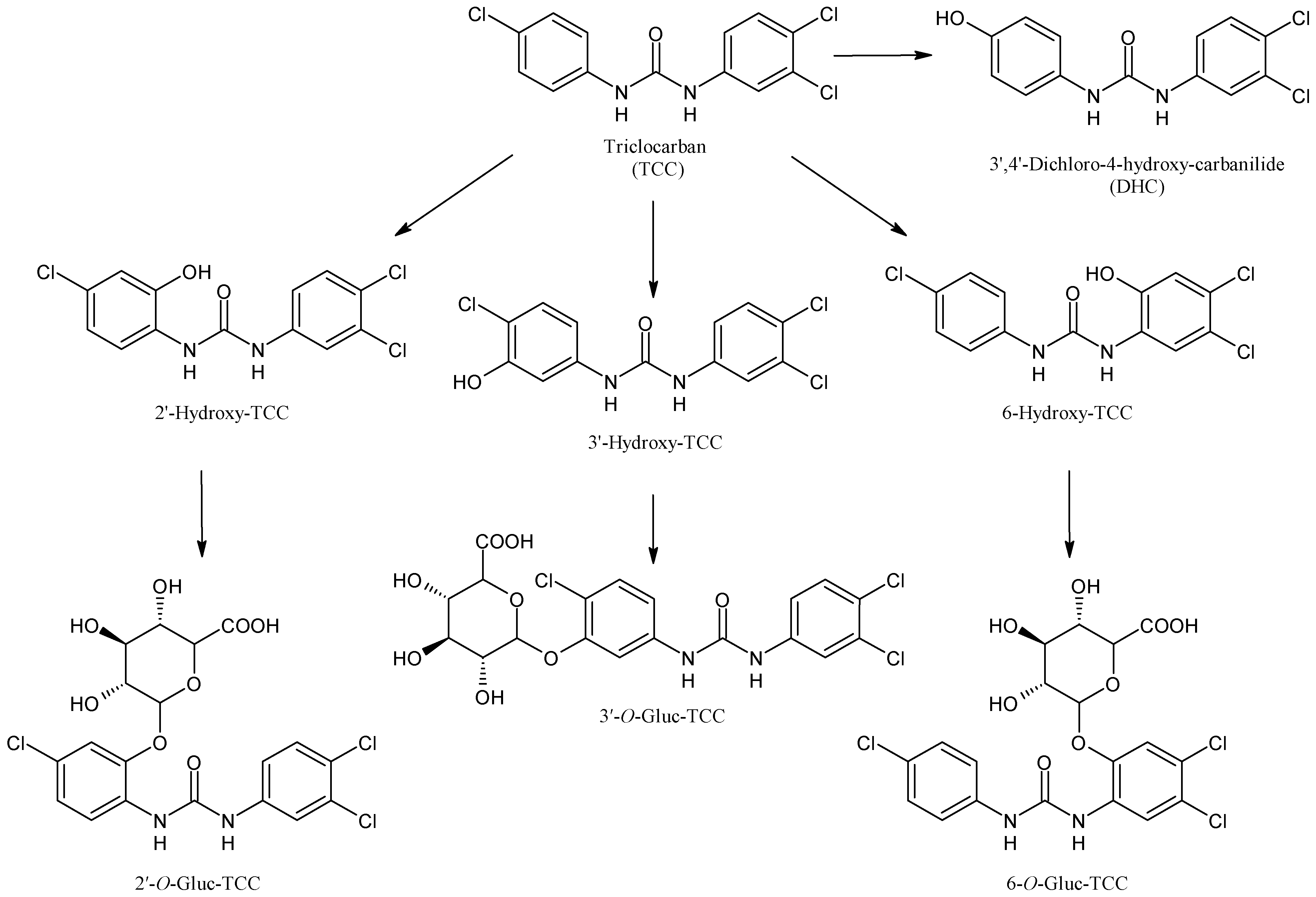
2.6. Triclocarban
2.7. Sunitinib
2.8. Hydroxychloroquine
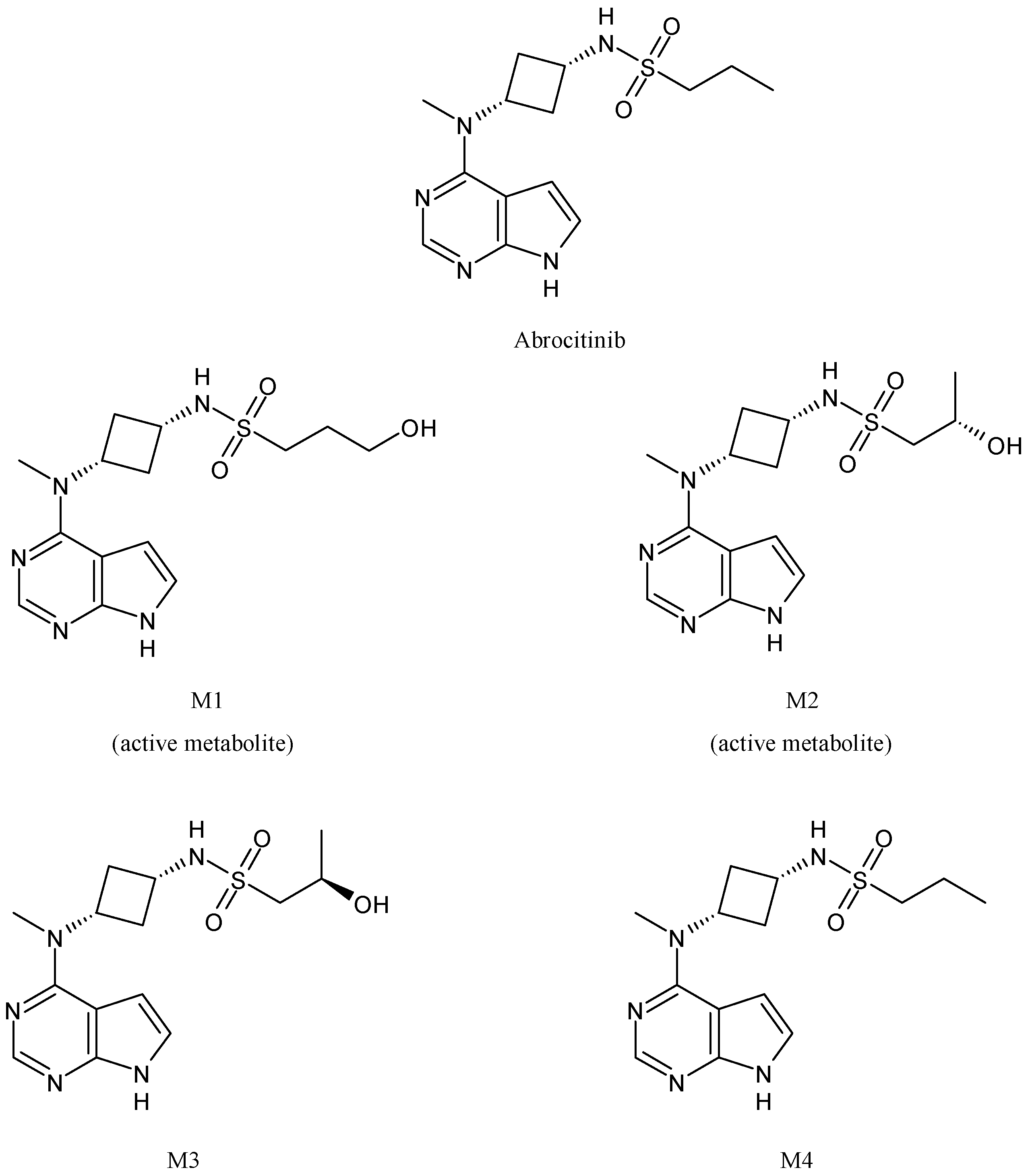
2.9. Abrocitinib
2.10. Omeprazole
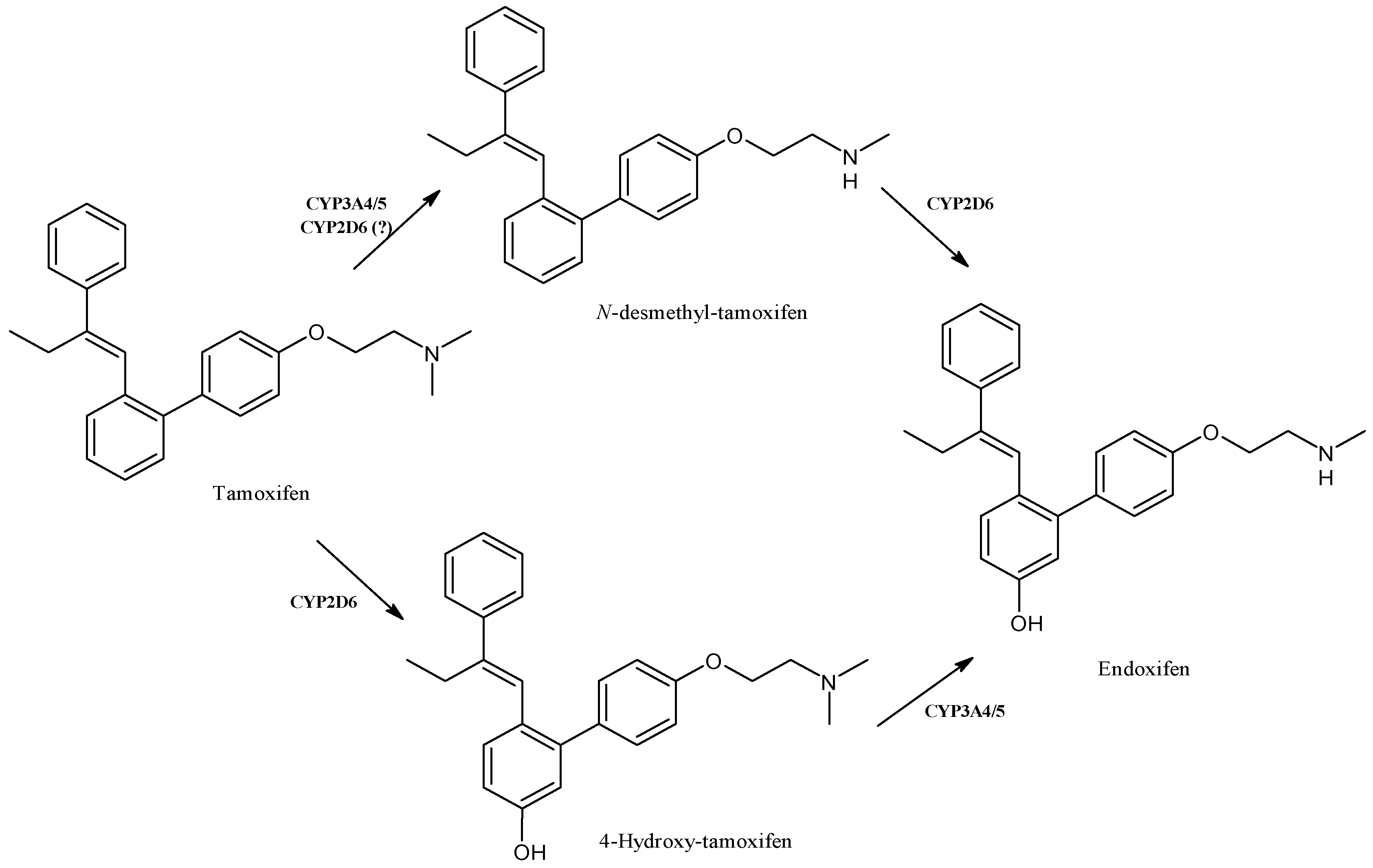
2.11. Tamoxifen
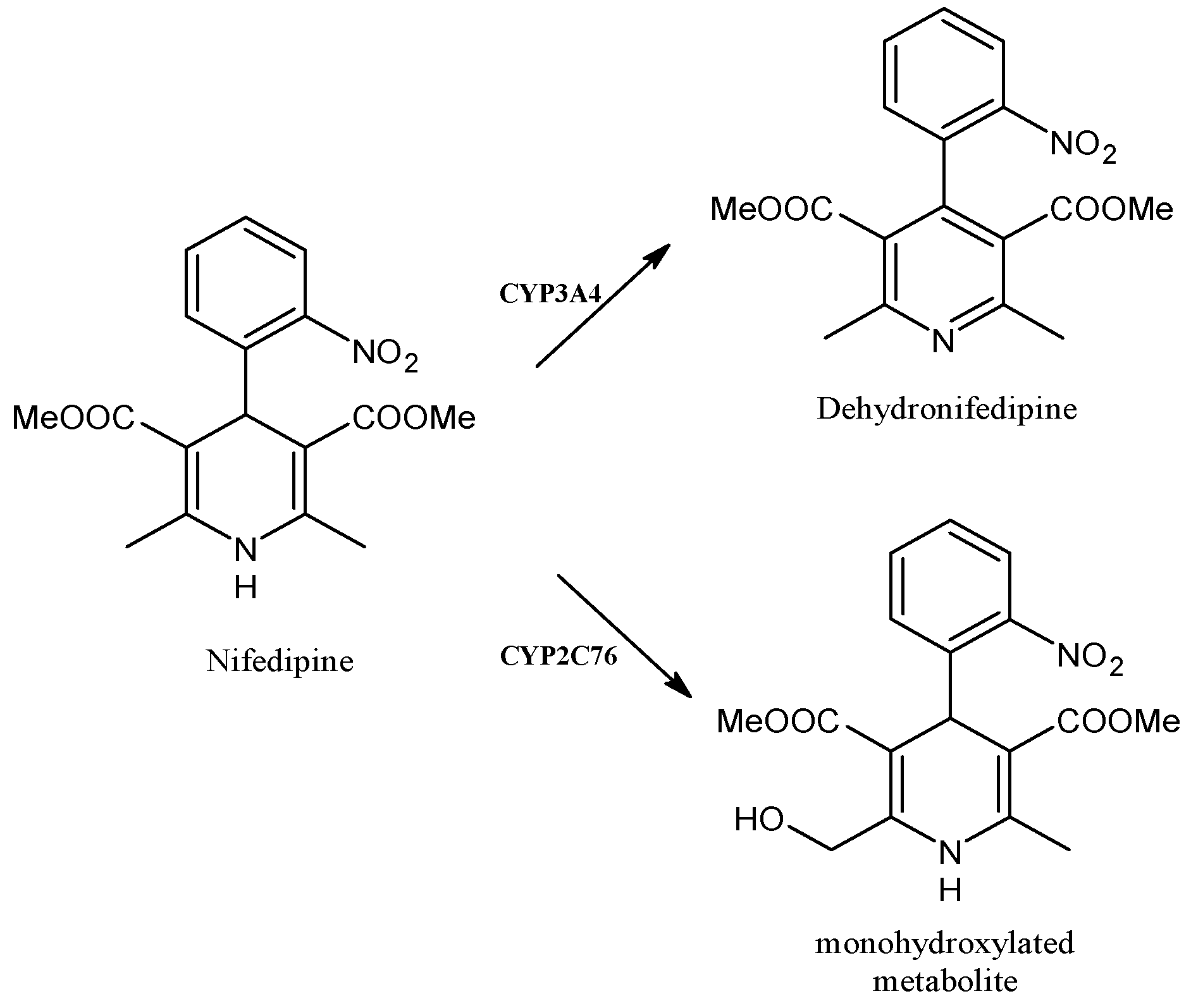
2.12. Nifedipine
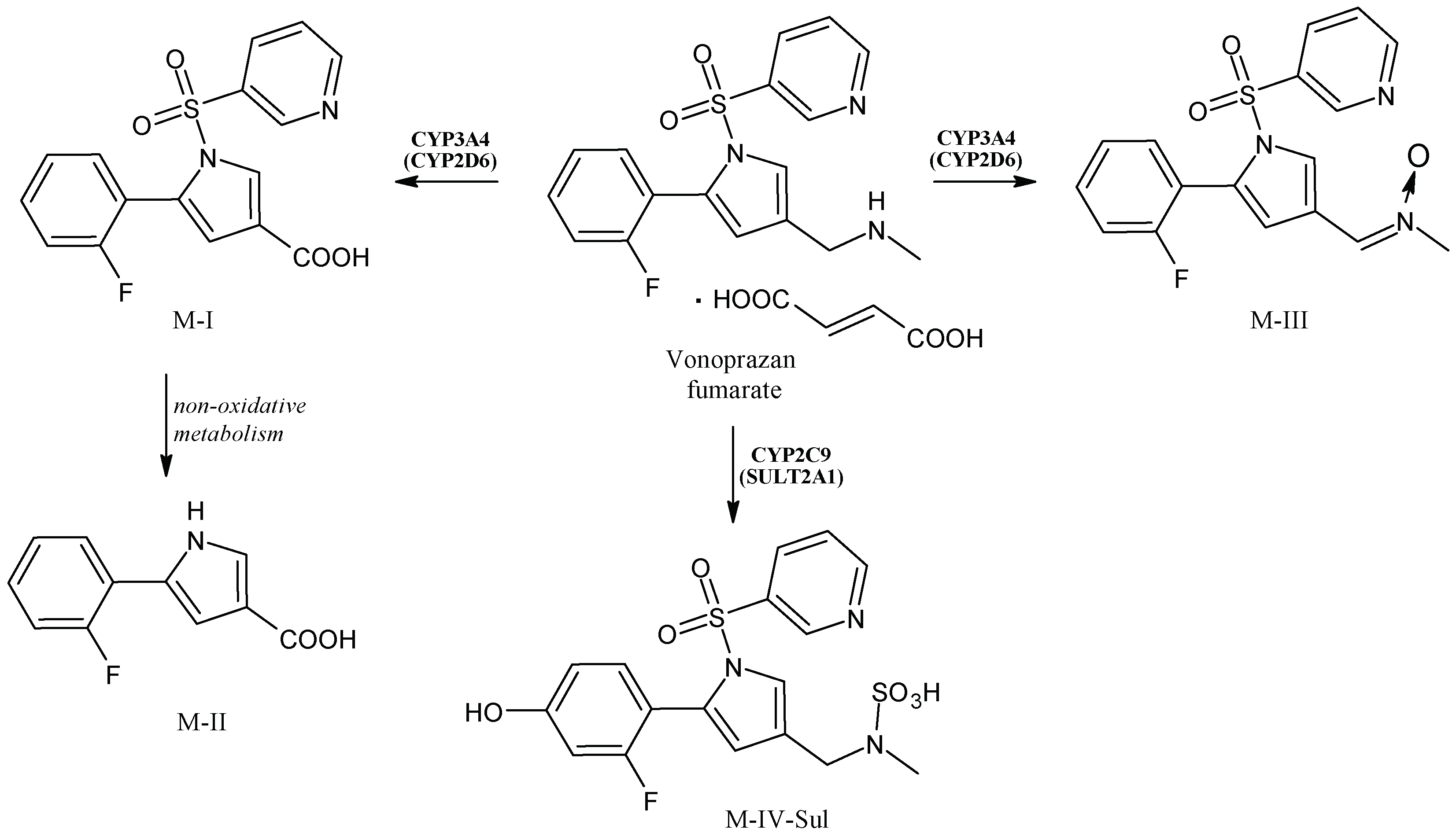
2.13. Vonoprazan
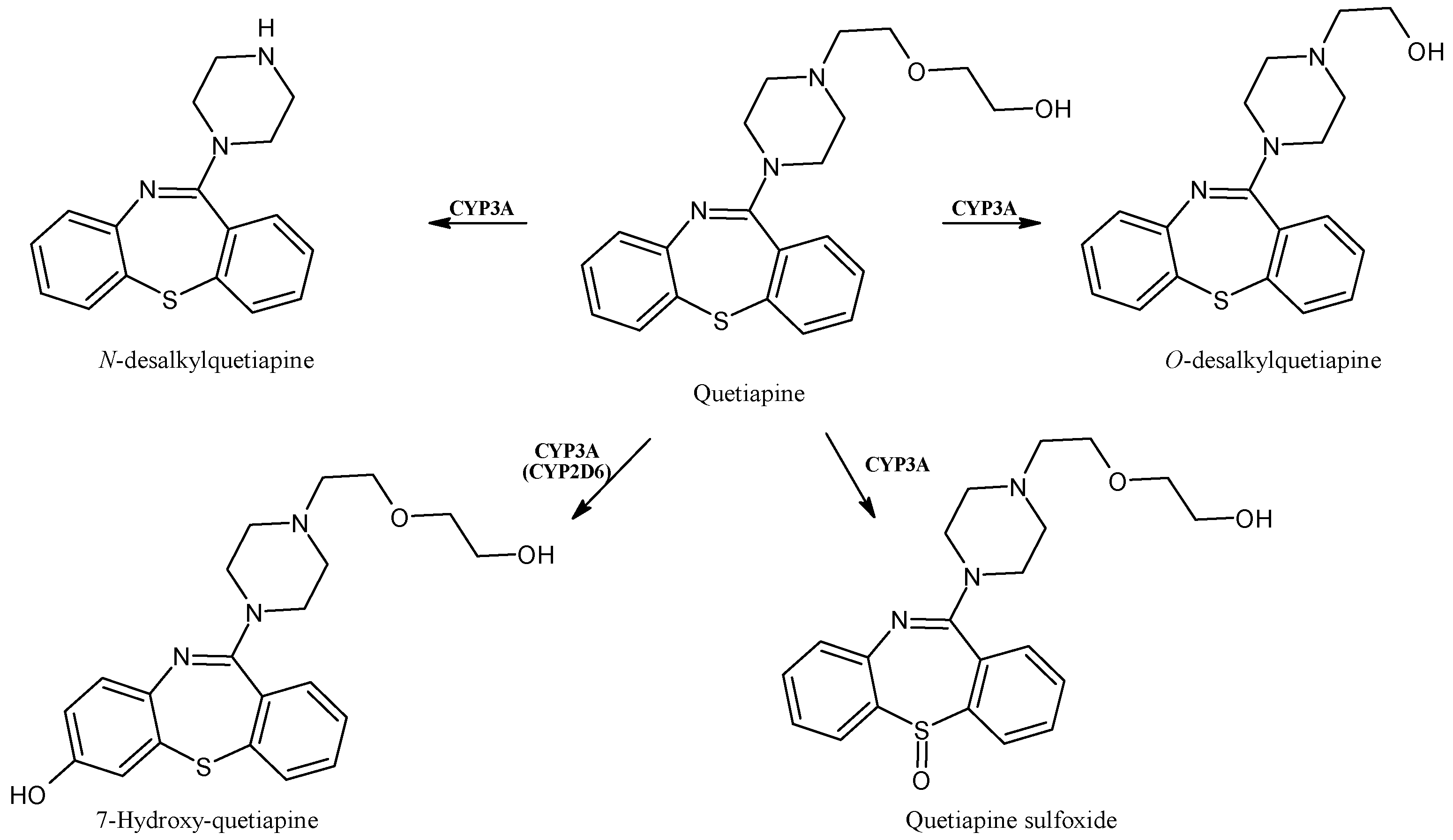
2.14. Quetiapine
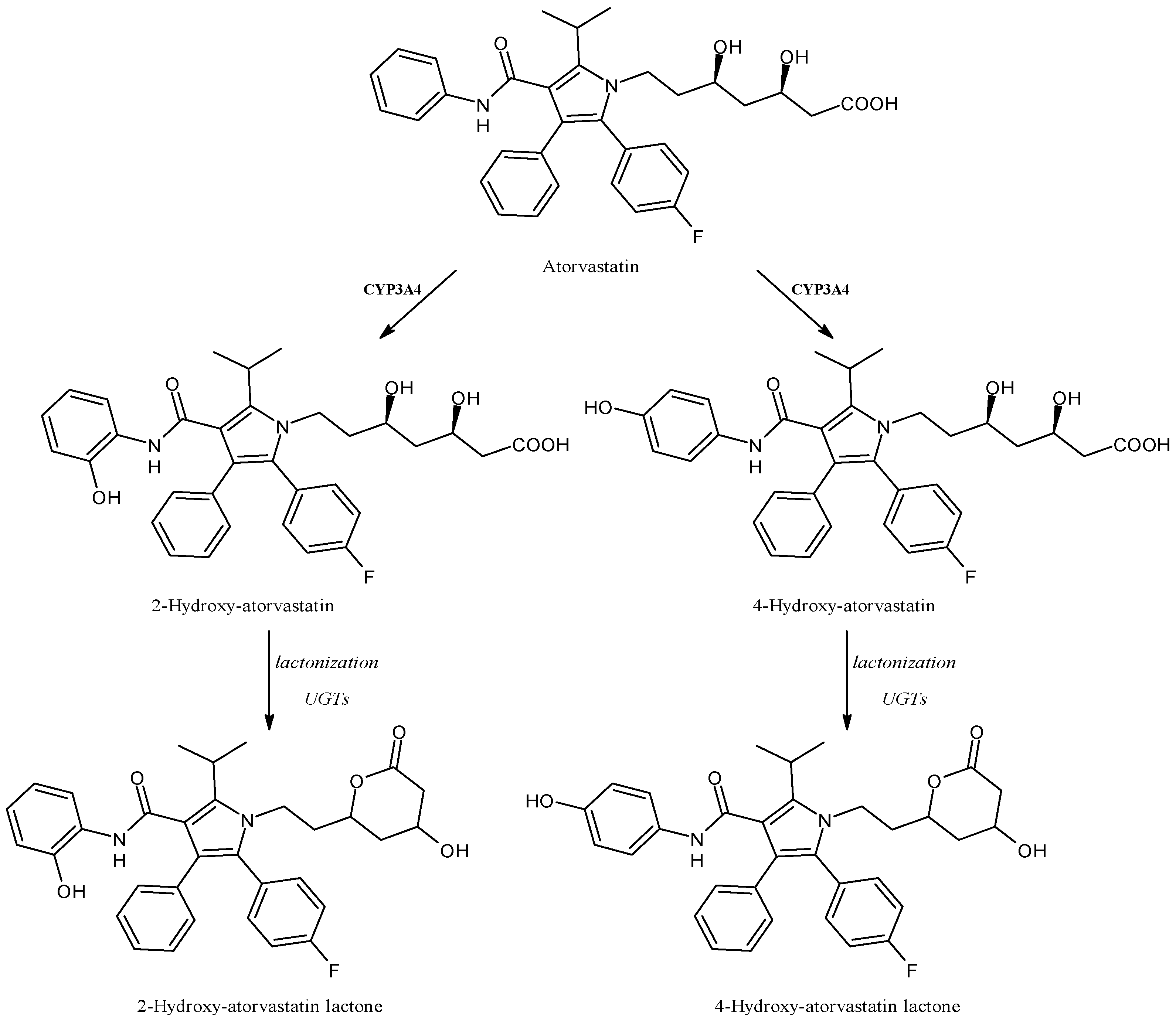
2.15. Atorvastatin
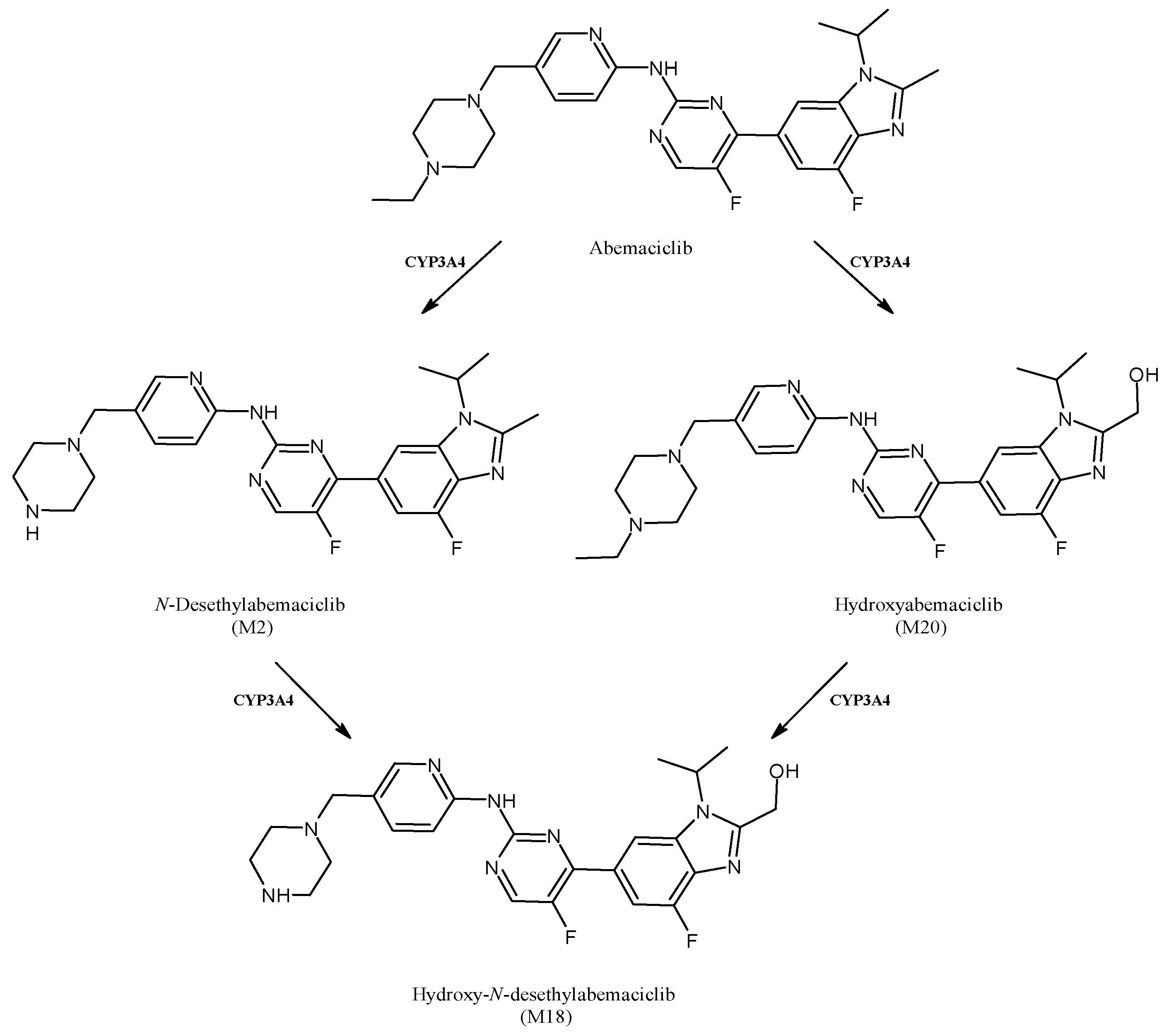
2.16. Abemaciclib
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Durairaj, P.; Fan, L.; Du, W.; Ahmad, S.; Mebrahtu, D.; Sharma, S.; Ashraf, R.A.; Liu, J.; Liu, Q.; Bureik, M. Functional expression and activity screening of all human cytochrome P450 enzymes in fission yeast. FEBS Lett. 2019, 593, 1372–1380. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Guan, X.; Dai, Z.; He, R.; Ding, X.; Yang, L.; Ge, G. Molecular probes for human cytochrome P450 enzymes: Recent progress and future perspectives. Coord. Chem. Rev. 2021, 427, 213600. [Google Scholar] [CrossRef]
- Rendic, S.; Guengerich, F.P. Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem. Res. Toxicol. 2015, 28, 38–42. [Google Scholar] [CrossRef]
- Rendic, S.P.; Guengerich, F.P. Human cytochrome P450 enzymes 5–51 as targets of drugs and natural and environmental compounds: Mechanisms, induction, and inhibition–toxic effects and benefits. Drug Metab. Rev. 2018, 50, 256–342. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.T.; Novak, R.F.; Halpert, J.R.; Johnson, E.F.; Stevens, J.C. The evolution of drug metabolism and disposition: A perspective from the editors. Drug Metab. Dispos. 2023, 51, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Drug metabolism: A half-century plus of progress, continued needs, and new opportunities. Drug Metab. Dispos. 2023, 51, 99–104. [Google Scholar] [CrossRef]
- Talevi, A.; Bellera, C.L. Drug Metabolism. In The ADME Encyclopedia; Talevi, A., Ed.; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Hinds, T.D.; Valoti, M.; Lai, Y.; Dorlo, T.P.; Li, Y. Emerging talents in Frontiers in Pharmacology: Drug metabolism and transport 2022. Front. Pharmacol. 2022, 13, 1083163. [Google Scholar] [CrossRef]
- Zuo, H.-L.; Huang, H.-Y.; Lin, Y.-C.-D.; Cai, X.-X.; Kong, X.-J.; Luo, D.-L.; Zhou, Y.-H.; Huang, H.-D. Enzyme activity of natural products on cytochrome P450. Molecules 2022, 27, 515. [Google Scholar] [CrossRef]
- Becker, D.; Bharatam, P.V.; Gohlke, H. F/g region rigidity is inversely correlated to substrate promiscuity of human cyp isoforms involved in metabolism. J. Chem. Inf. Model. 2021, 61, 4023–4030. [Google Scholar] [CrossRef]
- Manikandan, P.; Nagini, S. Cytochrome P450 structure, function and clinical significance: A review. Curr. Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, V.; Parkar, H.; Dasgupta, A. Utility of therapeutic drug monitoring in identifying clinically significant interactions between St. John’s Wort and prescription drugs. Ther. Drug Monit. 2023, 45, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Wigner, P.; Bijak, M.; Saluk-Bijak, J. Clinical potential of fruit in bladder cancer prevention and treatment. Nutrients 2022, 14, 1132. [Google Scholar] [CrossRef]
- Srinivas, N.R. Is pomegranate juice a potential perpetrator of clinical drug-drug interactions? Review of the in vitro, preclinical and clinical evidence. Eur. J. Drug Metab. Pharmacokinet. 2013, 38, 223–229. [Google Scholar] [CrossRef]
- Gjestad, C.; Hole, K.; Haslemo, T.; Diczfalusy, U.; Molden, E. Effect of grapefruit juice intake on serum level of the endogenous CYP3A4 metabolite 4β-hydroxycholesterol—An interaction study in healthy volunteers. AAPS J. 2019, 21, 58. [Google Scholar] [CrossRef]
- Tracy, T.S.; Chaudhry, A.S.; Prasad, B.; Thummel, K.E.; Schuetz, E.G.; Zhong, X.-B.; Tien, Y.-C.; Jeong, H.; Pan, X.; Shireman, L.M.; et al. Interindividual Variability in Cytochrome P450-Mediated Drug Metabolism. Drug Metab. Dispos. 2016, 44, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. A history of the roles of cytochrome P450 enzymes in the toxicity of drugs. Toxicol. Res. 2021, 37, 1–23. [Google Scholar] [CrossRef]
- Tucker, G.T. Chiral switches. Lancet 2000, 355, 1085–1087. [Google Scholar] [CrossRef]
- Hancu, G.; Modroiu, A. Chiral switch: Between therapeutical benefit and marketing strategy. Pharmaceuticals 2022, 15, 240. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Dai, L.; Tang, Y.; Zhou, W.; Dang, Y.; Sun, Q.; Tang, Z.; Zhu, M.; Ji, G. Gut microbiota and related metabolites were disturbed in ulcerative colitis and partly restored after mesalamine treatment. Front. Pharmacol. 2021, 11, 2337. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, H.; Yoon, H.Y.; Yee, J.; Gwak, H.S. Association of P450 oxidoreductase gene polymorphism with tacrolimus pharmacokinetics in renal transplant recipients: A systematic review and meta-analysis. Pharmaceutics 2022, 14, 261. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Jiang, G.; Gardner, L.; Xue, G.; Philips, S.; Ly, R.; Wilberforce, O.; Wu, X.; Cantor, E.; Dang, C.; et al. Abstract P1-08-02: Cytochrome P450 reductase gene, POR, associated with paclitaxel induced peripheral neuropathy in patients of European ancestry from the adjuvant breast cancer trial, ECOG-ACRIN E5103. Cancer Res. 2022, 82 (Suppl. 4), P1-08-02. [Google Scholar] [CrossRef]
- Singh, R.D.; Avadhesh, A.; Sharma, G.; Dholariya, S.; Shah, R.B.; Goyal, B.; Gupta, S.C. Potential of cytochrome P450, a family of xenobiotic metabolizing enzymes, in cancer therapy. Antiox. Red. Signal. 2022, 38, 853–876. [Google Scholar] [CrossRef] [PubMed]
- Tice, A.L.; Laudato, J.A.; Gordon, B.S.; Steiner, J.L. Alcohol intoxication stimulates antioxidant gene expression in skeletal muscle in a time dependent manner. FASEB J. 2022, 36 (Suppl. 1). in press. [Google Scholar] [CrossRef]
- Li, Q.S.; Jia, Y.J. Ferroptosis: A critical player and potential therapeutic target in traumatic brain injury and spinal cord injury. Neural Regen. Res. 2023, 18, 506–512. [Google Scholar] [CrossRef] [PubMed]
- McColl, E.R.; Croyle, M.A.; Zamboni, W.C.; Honer, W.G.; Heise, M.; Piquette-Miller, M.; Goralski, K.B. COVID-19 vaccines and the virus: Impact on drug metabolism and pharmacokinetics. Drug Metab. Dispos. 2023, 51, 130–141. [Google Scholar] [CrossRef]
- Iacopetta, D.; Ceramella, J.; Catalano, A.; Saturnino, C.; Pellegrino, M.; Mariconda, A.; Longo, P.; Sinicropi, M.S.; Aquaro, S. COVID-19 at a glance: An up-to-date overview on variants, drug design and therapies. Viruses 2022, 14, 573. [Google Scholar] [CrossRef]
- Wang, G.; Xiao, B.; Deng, J.; Gong, L.; Li, Y.; Li, J.; Zhong, Y. The role of cytochrome P450 enzymes in COVID-19 pathogenesis and therapy. Front. Pharmacol. 2022, 13, 791922. [Google Scholar] [CrossRef]
- Johnson, L.B.; Kauffman, C.A. Voriconazole: A New Triazole Antifungal Agent. Clin. Infect. Dis. 2003, 36, 630–637. [Google Scholar] [CrossRef]
- Hyland, R.; Jones, B.C.; Smith, D.A. Identification of the cytochrome P450 enzymes involved in the N-oxidation of voriconazole. Drug Metab. Dispos. 2003, 31, 540–547. [Google Scholar] [CrossRef]
- Barbarino, J.M.; Owusu-Obeng, A.; Klein, T.E.; Altman, R.B. PharmGKB summary: Voriconazole pathway, pharmacokinetics. Pharmacogenet. Genom. 2017, 27, 201. [Google Scholar] [CrossRef] [PubMed]
- Amsden, J.R.; Gubbins, P.O. Pharmacogenomics of triazole antifungal agents: Implications for safety, tolerability and efficacy. Expert Opin. Drug Metab. Toxicol. 2017, 13, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Le Daré, B.; Boglione-Kerrien, C.; Reizine, F.; Gangneux, J.P.; Bacle, A. Toward the personalized and integrative management of voriconazole dosing during COVID-19-associated pulmonary aspergillosis. Crit. Care 2021, 25, 152. [Google Scholar] [CrossRef] [PubMed]
- Schulz, J.; Kluwe, F.; Mikus, G.; Michelet, R.; Kloft, C. Novel insights into the complex pharmacokinetics of voriconazole: A review of its metabolism. Drug Metab. Rev. 2019, 51, 247–265. [Google Scholar] [CrossRef]
- Gautier-Veyret, E.; Truffot, A.; Bailly, S.; Fonrose, X.; Thiebaut-Bertrand, A.; Tonini, J.; Cahn, J.; Stanke-Labesque, F. Inflammation is a potential risk factor of voriconazole overdose in hematological patients. Fundam. Clin. Pharmacol. 2018, 33, 232–238. [Google Scholar] [CrossRef]
- Moorthy, G.S.; Vedar, C.; Zane, N.; Prodell, J.L.; Zuppa, A.F. Development and validation of a volumetric absorptive microsampling assay for analysis of voriconazole and voriconazole N-oxide in human whole blood. J. Chromatogr. B 2019, 1105, 67–75. [Google Scholar] [CrossRef]
- Blakely, K.M.; Drucker, A.M.; Rosen, C.F. Drug-induced photosensitivity—An update: Culprit drugs, prevention and management. Drug Saf. 2019, 42, 827–847. [Google Scholar] [CrossRef]
- Lee, V.; Gober, M.D.; Bashir, H.; O’Day, C.; Blair, I.A.; Mesaros, C.; Weng, L.; Huang, A.; Chen, A.; Tang, R.; et al. Voriconazole enhances UV-induced DNA damage by inhibiting catalase and promoting oxidative stress. Exp. Dermatol. 2020, 29, 29–38. [Google Scholar] [CrossRef]
- Tanaka, H.; Okuma, M.; Ishii, T. Occurrence of voriconazole-induced cutaneous squamous cell carcinoma in Japan: Data mining from different national pharmacovigilance databases. Pharmazie 2022, 77, 307–310. [Google Scholar] [CrossRef]
- Nobeyama, Y.; Umezawa, Y.; Asahina, A. Risk of squamous cell carcinoma in immunosuppressed patients with voriconazole-related actinic keratosis. J. Dermatol. 2022, 49, 1168–1172. [Google Scholar] [CrossRef]
- Zhorov, B.S. Molecular modeling of cardiac sodium channel with mexiletine. Membranes 2022, 12, 1252. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Talon, S.; De Bellis, M.; Desaphy, J.-F.; Franchini, C.; Lentini, G.; Catalano, A.; Corbo, F.; Tortorella, V.; Conte-Camerino, D. Inhibition of skeletal muscle sodium currents by mexiletine analogues: Specific hydrophobic interactions rather than lipophilia per se account for drug therapeutic profile. Naunyn Schmiedeberg’s Arch. Pharmacol. 2003, 367, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, R.; Fort, R.M.; Rydberg, P.; Harvey, J.N.; Mulholland, A.J. Quantum mechanics/molecular mechanics modeling of drug metabolism: Mexiletine N-hydroxylation by cytochrome P450 1A2. Chem. Res. Toxicol. 2016, 29, 963–971. [Google Scholar] [CrossRef]
- Catalano, A.; Carocci, A.; Fracchiolla, G.; Franchini, C.; Lentini, G.; Tortorella, V.; De Luca, A.; De Bellis, J.; Desaphy, J.-F.; Conte Camerino, D. Stereospecific synthesis of “para-hydroxymexiletine” and sodium channel blocking activity evaluation. Chirality 2004, 16, 72–78. [Google Scholar] [CrossRef] [PubMed]
- De Bellis, M.; De Luca, A.; Rana, F.; Cavalluzzi, M.M.; Catalano, A.; Lentini, G.; Franchini, C.; Tortorella, V.; Conte Camerino, D. Evaluation of the pharmacological activity of the major mexiletine metabolites on skeletal muscle sodium currents. Br. J. Pharmacol. 2006, 149, 300–310. [Google Scholar] [CrossRef]
- Catalano, A.; Desaphy, J.F.; Lentini, G.; Carocci, A.; Di Mola, A.; Bruno, C.; Carbonara, R.; de Palma, A.; Budriesi, R.; Ghelardini, C.; et al. Synthesis and toxicopharmacological evaluation of m-hydroxymexiletine, the first metabolite of mexiletine more potent than the parent compound on voltage-gated sodium channels. J. Med. Chem. 2012, 55, 1418–1422. [Google Scholar] [CrossRef] [PubMed]
- Beyrath, J.; Pellegrini, M.; Renkema, H.; Houben, L.; Pecheritsyna, S.; van Zandvoort, P.; van den Broek, P.; Bekel, A.; Eftekhari, P.; Smeitink, J.A.M. KH176 safeguards mitochondrial diseased cells from redox stress-induced cell death by interacting with the thioredoxin system/peroxiredoxin enzyme machinery. Sci. Rep. 2018, 8, 6577. [Google Scholar] [CrossRef] [PubMed]
- Khondrion. Khondrion Granted Orphan Drug Designation for KH176 for the Treatment of MIDD from European Commission. 2019. Available online: https://www.khondrion.com/khondrion-granted-orphan-drug-designation-for-kh176-for-the-treatment-of-midd-from-european-commission/ (accessed on 11 July 2020).
- Xiao, Y.; Yim, K.; Zhang, H.; Bakker, D.; Nederlof, R.; Smeitink, J.A.; Renkema, H.; Hollmann, M.W.; Weber, N.C.; Zuurbier, C.J. The redox modulating sonlicromanol active metabolite KH176m and the antioxidant MPG protect against short-duration cardiac ischemia-reperfusion injury. Cardiovasc. Drugs Ther. 2021, 35, 745–758. [Google Scholar] [CrossRef]
- Ceramella, J.; Iacopetta, D.; Franchini, A.; De Luca, M.; Saturnino, C.; Andreu, I.; Sinicropi, M.S.; Catalano, A. A look at the importance of chirality in drug activity: Some significative examples. Appl. Sci. 2022, 12, 10909. [Google Scholar] [CrossRef]
- Janssen, M.C.; Koene, S.; De Laat, P.; Hemelaar, P.; Pickkers, P.; Spaans, E.; Beukema, R.; Beyrath, J.; Groothuis, J.; Verhaak, C.; et al. The KHENERGY study: Safety and efficacy of KH 176 in mitochondrial m.3243A>G spectrum disorders. Clin. Pharmacol. Ther. 2019, 105, 101–111. [Google Scholar] [CrossRef]
- Koene, S.; Spaans, E.; Van Bortel, L.; Van Lancker, G.; Delafontaine, B.; Badilini, F.; Beyrath, J.; Smeitink, J. KH176 under development for rare mitochondrial disease: A first in man randomized controlled clinical trial in healthy male volunteers. Orphanet J. Rare Dis. 2017, 12, 163. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Renkema, H.; Smeitink, J.; Beyrath, J. Sonlicromanol’s active metabolite KH176m normalizes prostate cancer stem cell mPGES-1 overexpression and inhibits cancer spheroid growth. PLoS ONE 2021, 16, e0254315. [Google Scholar] [CrossRef] [PubMed]
- Catalano, A.; Iacopetta, D.; Sinicropi, M.S.; Franchini, C. Diarylureas as antitumor agents. Appl. Sci. 2021, 11, 374. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Pellegrino, M.; Aquaro, S.; Franchini, C.; Sinicropi, M. Diarylureas: Repositioning from antitumor to antimicrobials or multi-target agents against new pandemics. Antibiotics 2021, 10, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, T.M.; Luo, J.Z.; Lan, C.L.; Wei, Z.L.; Fu, T.H.; Liao, X.W.; Zhu, G.Z.; Ye, X.P.; Peng, T. CYP2C8 suppress proliferation, migration, invasion and sorafenib resistance of hepatocellular carcinoma via PI3K/Akt/p27kip1 axis. J. Hepatocell. Carcinoma 2021, 8, 1323. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.C.; Gillani, T.B.; Rawling, T.; Murray, M. Differential inhibition of human CYP2C8 and molecular docking interactions elicited by sorafenib and its major N-oxide metabolite. Chem. Biol. Interact. 2021, 338, 109401. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Mariconda, A.; Rosano, C.; Scumaci, D.; Saturnino, C.; Longo, C.; Sinicropi, M.S. New achievements for the treatment of triple-negative breast cancer. Appl. Sci. 2022, 12, 5554. [Google Scholar] [CrossRef]
- Huang, L.; Winger, B.A.; Cheah, V.; Gingrich, D.; Marzan, F.; Lu, Y.; Cooper, J.C.; Aweeka, F.; Long-Boyle, J. Quantification of N, N’N’’-triethylenethiophosphoramide, N, N’N’’-triethylenephosphoramide, cyclophosphamide, and 4-hydroxy-cyclophosphamide in microvolume human plasma to support neonatal and pediatric drug studies. J. Chromatogr. Open 2022, 2, 100054. [Google Scholar] [CrossRef]
- Iacopetta, D.; Catalano, A.; Ceramella, J.; Saturnino, C.; Salvagno, L.; Ielo, I.; Drommi, D.; Scali, E.; Plutino, M.R.; Rosace, G.; et al. The different facets of triclocarban: A review. Molecules 2021, 26, 2811. [Google Scholar] [CrossRef]
- Yun, H.; Liang, B.; Kong, D.; Li, X.; Wang, A. Fate, risk and removal of triclocarban: A critical review. J. Hazard. Mater. 2020, 387, 121944. [Google Scholar] [CrossRef]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Scumaci, D.; Giuzio, F.; Saturnino, C.; Aquaro, S.; Rosano, C.; Sinicropi, M.S. Multidrug resistance (MDR): A widespread phenomenon in pharmacological therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef] [PubMed]
- Baumann, A.; Lohmann, W.; Rose, T.; Ahn, K.C.; Hammock, B.D.; Karst, U.; Schebb, N.H. Electrochemistry-mass spectrometry unveils the formation of reactive triclocarban metabolites. Drug Metab. Dispos. 2010, 38, 2130–2138. [Google Scholar] [CrossRef] [PubMed]
- Schebb, N.H.; Muvvala, J.B.; Morin, D.; Buckpitt, A.R.; Hammock, B.D.; Rice, R.H. Metabolic activation of the antibacterial agent triclocarban by cytochrome P450 1A1 yielding glutathione adducts. Drug Metab. Dispos. 2014, 42, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lu, Y.; Liang, Y.; Jiang, L.; Cai, Z. Triclocarban-induced responses of endogenous and xenobiotic metabolism in human hepatic cells: Toxicity assessment based on nontargeted metabolomics approach. J. Hazard. Mater. 2020, 392, 122475. [Google Scholar] [CrossRef]
- Scrivano, L.; Parisi, O.I.; Iacopetta, D.; Ruffo, M.; Ceramella, J.; Sinicropi, M.S.; Puoci, F. Molecularly imprinted hydrogels for sustained release of sunitinib in breast cancer therapy. Polym. Adv. Technol. 2019, 30, 743–748. [Google Scholar] [CrossRef]
- Speed, B.; Bu, H.Z.; Pool, W.F.; Peng, G.W.; Wu, E.Y.; Patyna, S.; Bello, C.; Kang, P. Pharmacokinetics, distribution, and metabolism of [14C] sunitinib in rats, monkeys, and humans. Drug Metab Dispos. 2012, 40, 539–555. [Google Scholar] [CrossRef]
- Patel, N.D.; Chakrabory, K.; Messmer, G.; Krishnan, K.; Bossaer, J.B. Severe sunitinib-induced myelosuppression in a patient with a CYP3A4 polymorphism. J. Oncol. Pharm. Pract. 2018, 24, 623–626. [Google Scholar] [CrossRef]
- Amaya, G.M.; Durandis, R.; Bourgeois, D.S.; Perkins, J.A.; Abouda, A.A.; Wines, K.J.; Mohamud, M.; Starks, S.A.; Daniels, R.N.; Jackson, K.D. Cytochromes P4501A2 and 3A4 catalyze the metabolic activation of sunitinib. Chem. Res. Toxicol. 2018, 31, 570–584. [Google Scholar] [CrossRef]
- Wang, T.F.; Lim, W. What is the role of hydroxychloroquine in reducing thrombotic risk in patients with antiphospholipid antibodies? Hematol. Am. Soc. Hematol. Educ. Progr. 2016, 2016, 714–716. [Google Scholar] [CrossRef]
- Alarcon, G.S.; McGwin, G.; Bertoli, A.M.; Fessler, B.J.; Calvo-Alen, J.; Bastian, H.M.; Vila, L.M.; Reveille, J.D.; Group, L.S. Effect of hydroxychloroquine on the survival of patients with systemic lupus erythematosus: Data from LUMINA, a multiethnic US cohort (LUMINA L). Ann. Rheum. Dis. 2007, 66, 1168–1172. [Google Scholar] [CrossRef]
- Paludetto, M.N.; Kurkela, M.; Kahma, H.; Backman, J.T.; Niemi, M.; Filppula, A.M. Hydroxychloroquine is metabolized by CYP2D6, CYP3A4, and CYP2C8, and inhibits CYP2D6, while its metabolites also inhibit CYP3A in vitro. Drug Metab. Dispos. 2022, 51, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Skipper, C.; Pastick, K.A.; Engen, N.W.; Bangdiwala, A.S.; Abassi, M.; Lofgren, S.M.; Williams, D.A.; Okafor, E.C.; Pullen, M.F.; Nicol, M.R.; et al. Hydroxychloroquine in nonhospitalized adults with early COVID-19: A randomized trial. Ann. Intern. Med. 2020, 173, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Ceramella, J.; Iacopetta, D.; Sinicropi, M.S.; Andreu, I.; Mariconda, A.; Saturnino, C.; Giuzio, F.; Longo, P.; Aquaro, S.; Catalano, A. Drugs for COVID-19: An update. Molecules 2022, 27, 8562. [Google Scholar] [CrossRef] [PubMed]
- Biswas, M.; Roy, D.N. Potential clinically significant drug-drug interactions of hydroxychloroquine used in the treatment of COVID-19. Int. J. Clin. Pract. 2021, 75, e14710. [Google Scholar] [CrossRef]
- Silverberg, J.I.; Simpson, E.L.; Thyssen, J.P.; Gooderham, M.; Chan, G.; Feeney, C.; Biswas, P.; Valdez, H.; DiBonaventura, M.; Nduaka, C.; et al. Efficacy and safety of abrocitinib in patients with moderate-to-severe atopic dermatitis: A randomized clinical trial. JAMA Dermatol. 2020, 156, 863–873. [Google Scholar] [CrossRef]
- Dowty, M.; Yang, X.; Lin, J.; Bauman, J.; Doran, A.; Goosen, T.; Wood, L.; Johnson, J.; Banfield, C.; Gupta, P.; et al. P190—The effect of CYP2C9 and CYP2C19 genotype on the pharmacokinetics of PF 04965842, a JAK1 inhibitor in clinical development. Drug Metab. Pharmacokinet. 2020, 35 (Suppl. 1), S80. [Google Scholar] [CrossRef]
- Wang, E.Q.; Le, V.; O’Gorman, M.; Tripathy, S.; Dowty, M.E.; Wang, L.; Malhotra, B.K. Effects of hepatic impairment on the pharmacokinetics of abrocitinib and its metabolites. J. Clin. Pharmacol. 2021, 61, 1311–1323. [Google Scholar] [CrossRef]
- Tripathy, S.; Wentzel, D.; Wan, X.K.; Kavetska, O. Validation of enantioseparation and quantitation of an active metabolite of abrocitinib in human plasma. Bioanalysis 2021, 13, 1477–1486. [Google Scholar] [CrossRef]
- Wang, E.Q.; Le, V.; Winton, J.A.; Tripathy, S.; Raje, S.; Wang, L.; Dowty, M.E.; Malhotra, B.K. Effects of renal impairment on the pharmacokinetics of abrocitinib and its metabolites. J. Clin. Pharmacol. 2022, 62, 505–519. [Google Scholar] [CrossRef]
- Wang, X.; Dowty, M.E.; Wouters, A.; Tatulych, S.; Connell, C.A.; Le, V.H.; Tripathy, S.; O’Gorman, M.T.; Winton, J.A.; Yin, N.; et al. Assessment of the effects of inhibition or induction of CYP2C19 and CYP2C9 enzymes, or inhibition of OAT3, on the pharmacokinetics of abrocitinib and its metabolites in healthy individuals. Eur. J. Drug Metab. Pharmacokinet. 2022, 47, 419–429. [Google Scholar] [CrossRef]
- Liu, Z.; Xiong, J.; Gao, S.; Zhu, M.X.; Sun, K.; Li, M.; Zhang, G.; Li, Y. Ameliorating cancer cachexia by inhibiting cancer cell release of Hsp70 and Hsp90 with omeprazole. J. Cachexia Sarcopenia Muscle 2021, 13, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Hedge, D.D. Esomeprazole: A clinical review. Am. J. Health Syst. Pharm. 2002, 59, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Shirasaka, Y.; Sager, J.E.; Lutz, J.D.; Davis, C.; Isoherranen, N. Inhibition of CYP2C19 and CYP3A4 by omeprazole metabolites and their contribution to drug-drug interactions. Drug Metab. Dispos. 2013, 41, 1414–1424. [Google Scholar] [CrossRef]
- Li, X.Q.; Weidolf, L.; Simonsson, R.; Andersson, T.B. Enantiomer/enantiomer interactions between the S- and R- isomers of omeprazole in human cytochrome P450 enzymes: Major role of CYP2C19 and CYP3A4. J. Pharmacol. Exp. Ther. 2005, 315, 777–787. [Google Scholar] [CrossRef]
- Park, S.; Hyun, Y.J.; Kim, Y.R.; Lee, J.H.; Ryu, S.; Kim, J.M.; Oh, W.-Y.; Na, H.S.; Lee, J.G.; Seo, D.W. Effects of CYP2C19 genetic polymorphisms on PK/PD responses of omeprazole in Korean healthy volunteers. J. Korean Med. Sci. 2017, 32, 729. [Google Scholar] [CrossRef]
- Yoshinari, K.; Ueda, R.; Kusano, K.; Yoshimura, T.; Nagata, K.; Yamazoe, Y. Omeprazole transactivates human CYP1A1 and CYP1A2 expression through the common regulatory region containing multiple xenobiotic-responsive elements. Biochem. Pharmacol. 2008, 76, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Iacopetta, D.; Ceramella, J.; Baldino, N.; Sinicropi, M.S.; Catalano, A. Targeting breast cancer: An overlook on current strategies. Int. J. Mol. Sci. 2023, 24, 3643. [Google Scholar] [CrossRef] [PubMed]
- Khor, C.C.; Winter, S.; Sutiman, N.; Mürdter, T.E.; Chen, S.; Lim, J.S.L.; Li, Z.; Li, J.; Sim, K.S.; Ganchev, B.; et al. Cross-ancestry GWAS defines the extended CYP2D6 locus as the principal genetic determinant of endoxifen plasma concentrations. Clin. Pharmacol. Ther. 2023, 113, 712–723. [Google Scholar] [CrossRef]
- Helland, T.; Alsomairy, S.; Lin, C.; Søiland, H.; Mellgren, G.; Hertz, D.L. Generating a precision endoxifen prediction algorithm to advance personalized tamoxifen treatment in patients with breast cancer. J. Personal. Med. 2021, 11, 201. [Google Scholar] [CrossRef]
- Madlensky, L.; Natarajan, L.; Tchu, S.; Pu, M.; Mortimer, J.; Flatt, S.W.; Nikoloff, D.M.; Hillman, G.; Fontecha, M.R.; Lawrence, H.J.; et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin. Pharmacol. 2011, 89, 718–725. [Google Scholar] [CrossRef]
- Chan, C.W.H.; Law, B.M.H.; So, W.K.W.; Chow, K.M.; Waye, M.M.Y. Pharmacogenomics of breast cancer: Highlighting CYP2D6 and tamoxifen. J. Cancer Res. Clin. Oncol. 2020, 146, 1395–1404. [Google Scholar] [CrossRef]
- Klein, D.J.; Thorn, C.F.; Desta, Z.; Flockhart, D.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Tamoxifen pathway, pharmacokinetics. Pharm. Genom. 2013, 23, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Kanji, C.R.; Nyabadza, G.; Nhachi, C.; Masimirembwa, C. Pharmacokinetics of tamoxifen and its major metabolites and the effect of the African ancestry specific CYP2D6* 17 variant on the formation of the active metabolite, endoxifen. J. Personal. Med. 2023, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.M.; Schaefer, T.J. Nifedipine. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK537052/ (accessed on 14 February 2023).
- Mokhtari, B.; Nematollahi, D.; Salehzadeh, H. Electrochemical simultaneous determination of nifedipine and its main metabolite dehydronifedipine using MWCNT modified glassy carbon electrode. J. Mol. Liq. 2018, 264, 543–549. [Google Scholar] [CrossRef]
- Hosaka, S.; Murayama, N.; Satsukawa, M.; Shimizu, M.; Uehara, S.; Fujino, H.; Iwasaki, K.; Iwano, S.; Uno, Y.; Yamazaki, H. Evaluation of 89 compounds for identification of substrates for cynomolgus monkey CYP2C76, a new bupropion/nifedipine oxidase. Drug Metab. Dispos. 2015, 43, 27–33. [Google Scholar] [CrossRef]
- Uno, Y.; Yamazaki, H. mRNA levels of drug-metabolizing enzymes in 11 brain regions of cynomolgus macaques. Drug Metab. Pharmacokinet. 2020, 35, 248–252. [Google Scholar] [CrossRef]
- Kuroha, M.; Kayaba, H.; Kishimoto, S.; Khalil, W.F.; Shimoda, M.; Kokue, E. Effect of oral ketoconazole on first-pass effect of nifedipine after oral administration in dogs. J. Pharm. Sci. 2002, 91, 868–873. [Google Scholar] [CrossRef]
- Moore, L.B.; Goodwin, B.; Jones, S.A.; Wisely, G.B.; Serabjit-Singh, C.J.; Willson, T.M.; Collins, J.L.; Kliewer, S.A. St. John’s wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc. Natl. Acad. Sci. USA 2000, 97, 7500–7502. [Google Scholar] [CrossRef]
- Dahan, A.; Altman, H. Food-drug interaction: Grapefruit juice augments drug bioavailability—Mechanism, extent and relevance. Eur. J. Clin. Nutr. 2004, 58, 1–9. [Google Scholar] [CrossRef]
- Greenblatt, D.J.; von Moltke, L.L.; Harmatz, J.S.; Chen, G.; Weemhoff, J.L.; Jen, C.; Kelley, C.J.; LeDuc, B.W.; Zinny, M.A. Time course of recovery of cytochrome p450 3A function after single doses of grapefruit juice. Clin. Pharmacol. Ther. 2003, 74, 121–129. [Google Scholar] [CrossRef]
- Veronese, M.L.; Gillen, L.P.; Burke, J.P.; Dorval, E.P.; Hauck, W.W.; Pequignot, E.; Waldman, S.A.; Greenberg, H.E. Exposure-dependent inhibition of intestinal and hepatic CYP3A4 in vivo by grapefruit juice. J. Clin. Pharmacol. 2003, 43, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Rashid, T.J.; Martin, U.; Clarke, H.; Waller, D.G.; Renwick, A.G.; George, C.F. Factors affecting the absolute bioavailability of nifedipine. Br. J. Clin. Pharmacol. 1995, 40, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Odou, P.; Ferrari, N.; Barthelemy, C.; Brique, S.; Lhermitte, M.; Vincent, A.; Libersa, C.; Robert, H. Grapefruit juice-nifedipine interaction: Possible involvement of several mechanisms. J. Clin. Pharm. Ther. 2005, 30, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Garnock-Jones, K.P. Vonoprazan: First global approval. Drugs 2015, 75, 439–443. [Google Scholar] [CrossRef]
- Sugano, K. Vonoprazan fumarate, a novel potassium-competitive acid blocker, in the management of gastroesophageal reflux disease: Safety and clinical evidence to date. Ther. Adv. Gastroenterol. 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, C.; Wang, S.; Zhou, Q.; Dai, D.; Shi, J.; Xu, X.; Luo, Q. Cytochrome P450-based drug-drug interactions of vonoprazan in vitro and in vivo. Front. Pharmacol. 2020, 11, 53. [Google Scholar] [CrossRef]
- Echizen, H. The first-in-class potassium-competitive acid blocker, vonoprazan fumarate: Pharmacokinetic and pharmacodynamic considerations. Clin. Pharmacokinet. 2016, 55, 409–418. [Google Scholar] [CrossRef]
- Yoneyama, T.; Teshima, K.; Jinno, F.; Kondo, T.; Asahi, S. A validated simultaneous quantification method for vonoprazan (TAK-438F) and its 4 metabolites in human plasma by the liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 2016, 1015, 42–49. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Dai, D.; Cai, J.; Wang, S.; Hong, Y.; Zhou, S.; Zhao, F.; Zhou, Q.; Geng, P.; et al. Evaluation of commonly used cardiovascular drugs in inhibiting vonoprazan metabolism in vitro and in vivo. Front. Pharmacol. 2022, 13, 909168. [Google Scholar] [CrossRef]
- Graham, D.Y.; Dore, M.P. Update on the use of vonoprazan: A competitive acid blocker. Gastroenterology 2018, 154, 462–466. [Google Scholar] [CrossRef]
- Zargar, S.; Wani, T.A.; Alsaif, N.A.; Khayyat, A.I.A. A comprehensive investigation of interactions between antipsychotic drug quetiapine and human serum albumin using multi-spectroscopic, biochemical, and molecular modeling approaches. Molecules 2022, 27, 2589. [Google Scholar] [CrossRef] [PubMed]
- Højlund, M.; Andersen, K.; Ernst, M.T.; Correll, C.U.; Hallas, J. Use of low-dose quetiapine increases the risk of major adverse cardiovascular events: Results from a nationwide active comparator-controlled cohort study. World Psychiatry 2022, 21, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Bakken, G.V.; Rudberg, I.; Christensen, H.; Molden, E.; Refsum, H.; Hermann, M. Metabolism of quetiapine by CYP3A4 and CYP3A5 in presence or absence of cytochrome B5. Drug Metab. Dispos. 2009, 37, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Li, K.Y.; Li, X.; Cheng, Z.N.; Zhang, B.K.; Peng, W.X.; Li, H. De Effect of erythromycin on metabolism of quetiapine in Chinese suffering from schizophrenia. Eur. J. Clin. Pharmacol. 2005, 60, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Babu, D.; Tran, N.; Reiz, B.; Siraki, A.G. Neutrophil myeloperoxidase-mediated n-demethylation of quetiapine leads to N-desalkylquetiapine, a pharmacologically active cytochrome P450 metabolite. Chem. Res. Toxicol. 2022, 35, 1001–1010. [Google Scholar] [CrossRef]
- Castberg, I.; Skogvoll, E.; Spigset, O. Quetiapine and drug interactions: Evidence from a routine therapeutic drug monitoring service. J. Clin. Psychiatry 2007, 68, 1540–1545. [Google Scholar] [CrossRef]
- Sheridan, A.; Wheeler-Jones, C.P.; Gage, M.C. The immunomodulatory effects of statins on macrophages. Immuno 2022, 2, 317–343. [Google Scholar] [CrossRef]
- Motoji, Y.; Fukazawa, R.; Matsui, R.; Nagi-Miura, N.; Miyagi, Y.; Itoh, Y.; Ishii, Y. Kawasaki disease-like vasculitis facilitates atherosclerosis, and statin shows a significant antiatherosclerosis and anti-inflammatory effect in a Kawasaki disease model mouse. Biomedicines 2022, 10, 1794. [Google Scholar] [CrossRef]
- Palleria, C.; Roberti, R.; Iannone, L.F.; Tallarico, M.; Barbieri, M.A.; Vero, A.; Manti, A.; De Sarro, G.; Spina, E.; Russo, E. Clinically relevant drug interactions between statins and antidepressants. J. Clin. Pharm. Ther. 2019, 45, 227–239. [Google Scholar] [CrossRef]
- Partani, P.; Verma, S.M.; Gurule, S.; Khuroo, A.; Monif, T. Simultaneous quantitation of atorvastatin and its two active metabolites in human plasma by liquid chromatography electrospray tandem mass spectrometry. J. Pharm. Anal. 2014, 4, 26–36. [Google Scholar] [CrossRef]
- Turner, R.M.; Fontana, V.; Bayliss, M.; Whalley, S.; Castelazo, A.S.; Pirmohamed, M. Development, validation and application of a novel HPLC-MS/MS method for the quantification of atorvastatin, bisoprolol and clopidogrel in a large cardiovascular patient cohort. J. Pharm. Biomed. Anal. 2018, 159, 272–281. [Google Scholar] [CrossRef]
- Turner, R.M.; Pirmohamed, M. Statin-related myotoxicity: A comprehensive review of pharmacokinetic, pharmacogenomic and muscle components. J. Clin. Med. 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Hukkanen, J.; Puurunen, J.; Hyötyläinen, T.; Savolainen, M.J.; Ruokonen, A.; Morin-Papunen, L.; Orešič, M.; Piltonen, T.T.; Tapanainen, J.S. The effect of atorvastatin treatment on serum oxysterol concentrations and cytochrome P450 3A4 activity. Br. J. Clin. Pharmacol. 2015, 80, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.; Kim, H.; Heo, Y.; Yoon, H.Y.; Song, G.; Gwak, H.S. Association between CYP3A5 polymorphism and statin-induced adverse events: A systemic review and meta-analysis. J. Pers. Med. 2021, 11, 677. [Google Scholar] [CrossRef]
- Martinez-Chavez, A.; Loos, N.H.; Lebre, M.C.; Tibben, M.M.; Rosing, H.; Beijnen, J.H.; Schinkel, A.H. ABCB1 and ABCG2 limit brain penetration and, together with CYP3A4, total plasma exposure of abemaciclib and its active metabolites. Pharmacol. Res. 2022, 178, 105954. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Chavez, A.; Tibben, M.; de Jong, K.A.M.; Rosing, H.; Schinkel, A.H.; Beijnen, J.H. Simultaneous quantification of abemaciclib and its active metabolites in human and mouse plasma by UHLC-MS/MS. J. Pharm. Biomed. Anal. 2021, 203, 114225. [Google Scholar] [CrossRef]
- Turner, P.K.; Hall, S.D.; Chapman, S.C.; Rehmel, J.L.; Royalty, J.E.; Guo, Y.; Kulanthaivel, P. Abemaciclib does not have a clinically meaningful effect on pharmacokinetics of CYP1A2, CYP2C9, CYP2D6, and CYP3A4 substrates in patients with cancer. Drug Metab. Dispos. 2020, 48, 796–803. [Google Scholar] [CrossRef]
- Goldwaser, E.; Laurent, C.; Lagarde, N.; Fabrega, S.; Nay, L.; Villoutreix, B.O.; Jelsch, C.; Nicot, A.B.; Loriot, M.A.; Miteva, M.A. Machine learning-driven identification of drugs inhibiting cytochrome P450 2C9. PLoS Comput. Biol. 2022, 18, e1009820. [Google Scholar] [CrossRef]





| Drug | Activity | CYPs Biotransforming the Drug | CYPs Inhibited by the Drug | CYPs Induced by the Drug |
|---|---|---|---|---|
| Voriconazole | Antifungal | CYP2C19, CYP2C9, CYP3A4 | ||
| Mexiletine | Antiarrhythmic | CYP2E1, CYP2D6, CYP1A2 | ||
| Sonlicromanol | Antioxidant | CYP3A4 | ||
| Sorafenib | Anticancer | CYP3A4 | CYP2C8 | |
| Cyclophosphamide | Anticancer | CYP2B6, CYP2C9, CYP3A4 | ||
| Triclocarban | Antibacterial | CYP1A1 | ||
| Sunitinib | Anticancer | CYP1A2, CYP3A4 | ||
| Hydroxychloroquine | Antimalarial | CYP3A4, CYP2D6, CYP2C8 | CYP2J2, CYP3A4 | |
| Abrocitinib | Antipsoriatic | CYP2C19, CYP2C9, CYP3A4, CYP2B6 | ||
| Omeprazole | Proton-pump inhibitor | CYP2C19, CYP3A4 | CYP1A1, CYP1A2 | |
| Tamoxifen | Anticancer | CYP3A4, CYP3A5, CYP2D6 | ||
| Nifedipine | Antihyperthensive | CYP3A4, CYP2C76 | ||
| Vonoprazan | Acid blocker | CYP3A4, CYP2B6, CYP2C19, CYP2C9, CYP2D6 | ||
| Quetiapine | Antipsychotic | CYP3A4, CYP3A5 | ||
| Atorvastatin | Antihyperlipidemic | CYP3A4, CYP3A5 | ||
| Abemaciclib | Anticancer | CYP3A4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iacopetta, D.; Ceramella, J.; Catalano, A.; Scali, E.; Scumaci, D.; Pellegrino, M.; Aquaro, S.; Saturnino, C.; Sinicropi, M.S. Impact of Cytochrome P450 Enzymes on the Phase I Metabolism of Drugs. Appl. Sci. 2023, 13, 6045. https://doi.org/10.3390/app13106045
Iacopetta D, Ceramella J, Catalano A, Scali E, Scumaci D, Pellegrino M, Aquaro S, Saturnino C, Sinicropi MS. Impact of Cytochrome P450 Enzymes on the Phase I Metabolism of Drugs. Applied Sciences. 2023; 13(10):6045. https://doi.org/10.3390/app13106045
Chicago/Turabian StyleIacopetta, Domenico, Jessica Ceramella, Alessia Catalano, Elisabetta Scali, Domenica Scumaci, Michele Pellegrino, Stefano Aquaro, Carmela Saturnino, and Maria Stefania Sinicropi. 2023. "Impact of Cytochrome P450 Enzymes on the Phase I Metabolism of Drugs" Applied Sciences 13, no. 10: 6045. https://doi.org/10.3390/app13106045
APA StyleIacopetta, D., Ceramella, J., Catalano, A., Scali, E., Scumaci, D., Pellegrino, M., Aquaro, S., Saturnino, C., & Sinicropi, M. S. (2023). Impact of Cytochrome P450 Enzymes on the Phase I Metabolism of Drugs. Applied Sciences, 13(10), 6045. https://doi.org/10.3390/app13106045











