A Fast Parallel Random Forest Algorithm Based on Spark
Abstract
:1. Introduction
2. Preliminary Knowledge
2.1. Random Forest Algorithm
2.2. Apache Spark
3. Improved Random Forest Algorithm
3.1. The New Gini Coefficient
3.2. Approximate Equal-Frequency Binning Method
| Algorithm 1: Approximate equal frequency binning algorithm |
| Input: Continuous values of feature a |
| Output: All candidate split points of feature a |
| Step 1: All feature values are sorted in ascending order; Step 2: Obtain all distinct values and the number of each value counts = {count1, count2, …, countm}; Step 3: Set the number of boxes bins = int(sqrt(num(A′))); Step 4: The size of the current bin is set to a constant S = sum(counts)/bins; Step 5: The binning operation. All feature values are processed sequentially; |
| Step 5.1 If counti >= S then a′i is treated as a large number and boxed individually, set the average of this feature value and the next feature value as the candidate split point; |
| Step 5.2 If counti < S then add the remaining feature value in order until the number of feature value is greater than or equal to S. At this time, the average of the largest feature value in the box and the next feature value not in the box is set as the candidate split point; Step 5.3 bins = bins − 1; if bins = 1, go to step 6, or jump to step 4. Step 6: Put all remaining feature value into the last bin and the algorithm ends. |
4. Parallelization of Random Forest Based on Spark
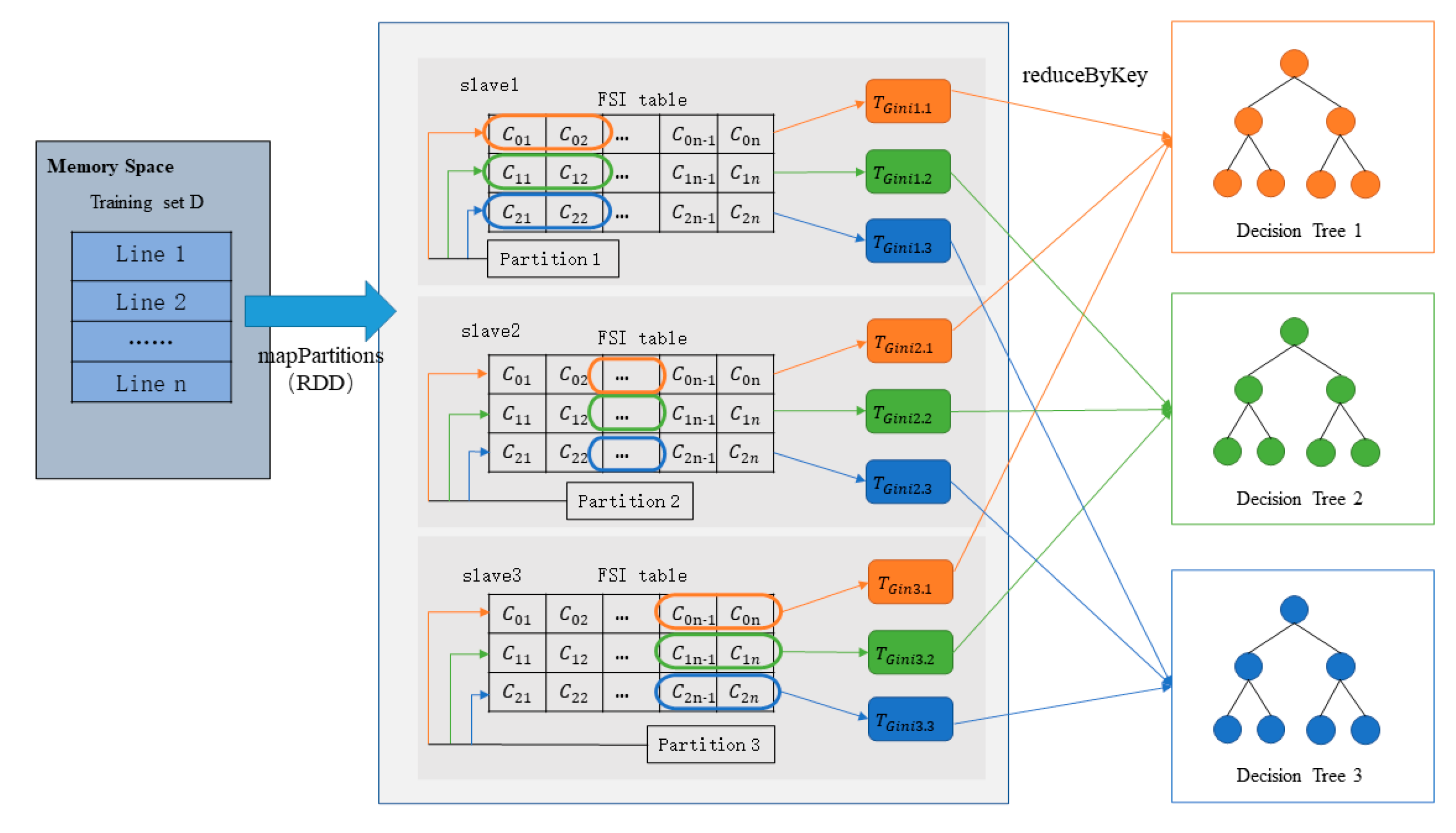
4.1. Parallel Decision Tree Training Strategy Based on a Forest Sampling Index Table
4.2. Gini Coefficient Calculation Optimization
4.3. Parallel Implementation of Improved Random Forest Algorithm
| Algorithm 2: Parallel Training Process of Optimization Random Forest |
| Input: The number of decision trees K, data set D, the FSI table |
| Output: Random forest model 1: For i = 0 to (K − 1) 2: For index in range(start, end) 3: For j, feature in enumerate(featureIndex) 4: Sc.broadcast(FSI)//Build an index table and load the training subset 5: Left = {},right = {}//Declare two dictionaries to record the number of labels for the two subsets |
| 6: LeftLen = 0,rightLen=len(node.recoeds)//Number of categories for the left and right subsets 7: The set of candidate split points is obtained based on Algorithm 1 8: For thisSplitVal in spiltNode 9: Divide the left and right subsets according to the split point 10: LeftLen + = 1,rightLen − = 1 |
| 11: Calculate the Gini coefficient according to Equation (6) 12: Take the split point with the smallest Gini coefficient 13: End for 14: End for 15: Obtain the best split feature and the best split spot, split the current node according to these two values, generate two new child nodes, and continue splitting until all nodes are leaf nodes 16: End for 17: Training to obtain a single decision tree 18: End for |
5. Experiments
5.1. Data Set Description
5.2. Evaluation of Algorithm Performance
5.3. Parallel Performance Evaluation
5.3.1. Speedup
5.3.2. Scaleup
5.3.3. Sizeup
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Dziak, J.J.; Coffman, D.L.; Lanza, S.T.; Li, R.; Jermiin, L.S. Sensitivity and specificity of information criteria. Brief. Bioinform. 2020, 21, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.S.; Orban, R.; Rajammal Ramasamy, R. A Novel Method for Survival Prediction of Hepatocellular Carcinoma Using Feature-Selection Techniques. Appl. Sci. 2022, 12, 6427. [Google Scholar] [CrossRef]
- Phan, T.N.; Kuch, V.; Lehnert, L.W. Land Cover Classification using Google Earth Engine and Random Forest Classifier—The Role of Image Composition. Remote Sens. 2020, 12, 2411. [Google Scholar] [CrossRef]
- Zheng, X.; Jia, J.; Chen, J.; Guo, S.; Sun, L.; Zhou, C.; Wang, Y. Hyperspectral Image Classification with Imbalanced Data Based on Semi-Supervised Learning. Appl. Sci. 2022, 12, 3943. [Google Scholar] [CrossRef]
- Khan, S.N.; Li, D.; Maimaitijiang, M. A Geographically Weighted Random Forest Approach to Predict Corn Yield in the US Corn Belt. Remote Sens. 2022, 14, 2843. [Google Scholar] [CrossRef]
- Memiş, S.; Enginoğlu, S.; Erkan, U. Fuzzy parameterized fuzzy soft k-nearest neighbor classifier. Neurocomputing 2022, 500, 351–378. [Google Scholar] [CrossRef]
- Zayed, Y.; Salman, Y.; Hasasneh, A. A Recommendation System for Selecting the Appropriate Undergraduate Program at Higher Education Institutions Using Graduate Student Data. Appl. Sci. 2022, 12, 12525. [Google Scholar] [CrossRef]
- Abdulsalam, H.; Skillicorn, D.B.; Martin, P. Classification using streaming random forests. IEEE Trans. Knowl. Data Eng. 2010, 23, 22–36. [Google Scholar] [CrossRef]
- Yang, S.; Guo, J.Z.; Jin, J.W. An improved Id3 algorithm for medical data classification. Comput. Electr. Eng. 2018, 65, 474–487. [Google Scholar] [CrossRef]
- Ruggieri, S. Efficient C4. 5 [classification algorithm]. IEEE Trans. Knowl. Data Eng. 2002, 14, 438–444. [Google Scholar] [CrossRef]
- Yu, S.; Li., X.; Wang, H. C_CART: An instance confidence-based decision tree algorithm for classification. Intell. Data Anal. 2021, 25, 929–948. [Google Scholar] [CrossRef]
- Lin, S.; Luo, W. A new multilevel CART algorithm for multilevel data with binary outcomes. Multivar. Behav. Res. 2019, 54, 578–592. [Google Scholar] [CrossRef] [PubMed]
- Seera, M.; Lim, C.P.; Loo, C.K. Motor fault detection and diagnosis using a hybrid FMM-CART model with online learning. J. Intell. Manuf. 2016, 27, 1273–1285. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A. Classification and regression trees. Encycl. Ecol. 2015, 57, 582–588. [Google Scholar]
- Assunçao, J.; Fernandes, P.; Lopes, L. Distributed Stochastic Aware Random Forests—Efficient Data Mining for Big Data. In Proceedings of the IEEE International Congress on Big Data, Santa Clara, CA, USA, 6–9 October 2013. [Google Scholar]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. Random forests for big data. Big Data Res. 2017, 9, 28–46. [Google Scholar] [CrossRef]
- Del Río, S.; López, V.; Benítez, J.M.; Herrera, F. On the use of MapReduce for imbalanced big data using Random Forest. Inf. Sci. 2014, 285, 112–137. [Google Scholar] [CrossRef]
- Mu, Y.; Liu, X.; Wang, L. A Pearson’s correlation coefficient based decision tree and its parallel implementation. Inf. Sci. 2018, 435, 40–58. [Google Scholar] [CrossRef]
- Xu, W.; Hoang, V.T. MapReduce-based improved random forest model for massive educational data processing and classification. Mob. Netw. Appl. 2021, 26, 191–199. [Google Scholar] [CrossRef]
- Chen, J.; Li, K.; Tang, Z. A parallel random forest algorithm for big data in a spark cloud computing environment. IEEE Trans. Parallel Distrib. Syst. 2016, 28, 919–933. [Google Scholar] [CrossRef]
- Lulli, A.; Oneto, L.; Anguita, D. Mining big data with random forests. Cogn. Comput. 2019, 11, 294–316. [Google Scholar] [CrossRef]
- Apache Spark. Spark Mllib-Random Forest. Available online: http://spark.apache.org/docs/latest/mllib-ensembles.html (accessed on 21 March 2023).
- Feng, X.; Wang, W. Survey on Hadoop and spark application scenarios. Appl. Res. Comput. 2018, 35, 2561–2566. [Google Scholar]
- University of California. Uci Machine Learning Repository. Available online: http://archive.ics.uci.edu/ml/datasets (accessed on 21 March 2023).
- Xu, Z.; Ni, W.; Ji, Y. Rotation forest based on multimodal genetic algorithm. J. Cent. South Univ. 2021, 28, 1747–1764. [Google Scholar] [CrossRef]
- Memiş, S.; Enginoğlu, S.; Erkan, U. A new classification method using soft decision-making based on an aggregation operator of fuzzy parameterized fuzzy soft matrices. Turk. J. Electr. Eng. Comput. Sci. 2022, 30, 871–890. [Google Scholar] [CrossRef]
- Leung, K.M. Naive bayesian classifier. Polytech. Univ. Dep. Comput. Sci. Financ. Risk Eng. 2007, 2007, 123–156. [Google Scholar]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Yin, L.; Qin, L.; Jiang, Z. A fast parallel attribute reduction algorithm using Apache Spark. Knowl. Based Syst. 2021, 212, 106582. [Google Scholar] [CrossRef]
- Zhu, W. Large-scale image retrieval solution based on Hadoop cloud computing platform. J. Comput. Appl. 2014, 34, 695. [Google Scholar]








| Age | Have a Job | Credit Status | Label |
|---|---|---|---|
| youth | no | generally | no |
| youth | no | good | no |
| youth | yes | good | yes |
| youth | yes | generally | yes |
| youth | no | generally | no |
| middle aged | no | generally | no |
| middle aged | no | good | no |
| middle aged | yes | good | yes |
| middle aged | no | very good | yes |
| middle aged | no | very good | yes |
| old age | no | very good | yes |
| old age | no | good | yes |
| old age | yes | good | yes |
| old age | yes | very good | yes |
| old age | no | generally | no |
| Feature Value | Category |
|---|---|
| 0.1 | 1 |
| 0.3 | 2 |
| 0.7 | 1 |
| 0.7 | 1 |
| 1.2 | 3 |
| Data Set | Instances | Features | Class |
|---|---|---|---|
| Glass | 214 | 9 | 7 |
| Wine | 178 | 13 | 3 |
| Ionosphere | 351 | 33 | 2 |
| Optical | 3823 | 64 | 10 |
| Image | 2310 | 19 | 7 |
| Letter | 20,000 | 16 | 26 |
| Adult | 48,842 | 14 | 2 |
| HT_senor | 919,438 | 11 | 100 |
| WinniPeg | 325,834 | 175 | 7 |
| Swarm | 24,017 | 2400 | 2 |
| SUSY | 5,000,000 | 18 | 2 |
| Our Algorithm | RF | MGARF | ||||
|---|---|---|---|---|---|---|
| Data Set | Acc | Time (s) | Acc | Time (s) | Acc | Time (s) |
| Wine | 0.9815 | 3.7 | 0.9455 | 3.8 | 0.9735 | 68.8 |
| Glass | 0.7538 | 4.4 | 0.7425 | 4.1 | 0.7325 | 73 |
| Image | 0.9547 | 4.7 | 0.9018 | 3.9 | 0.9679 | 34.3 |
| Ionosphere | 0.9057 | 8.3 | 0.9146 | 9.5 | 0.8928 | 25.3 |
| Letter | 0.9532 | 43.2 | 0.9321 | 46.4 | 0.9482 | 477.6 |
| Optical | 0.9564 | 51.9 | 0.9608 | 30.6 | 0.9652 | 98.2 |
| Adult | 0.8688 | 167.2 | 0.8512 | 173.8 | 0.8466 | 358.9 |
| Data Set | Our Algorithm | KNN | SVM | NB | |
|---|---|---|---|---|---|
| Wine | Accuracy | 0.9815 | 0.7782 | 0.9255 | 0.9818 |
| Precision | 0.9833 | 0.7933 | 0.9433 | 0.9867 | |
| Recall | 0.9825 | 0.7611 | 0.9278 | 0.9867 | |
| F1-value | 0.9824 | 0.7602 | 0.9245 | 0.9852 | |
| Glass | Accuracy | 0.7538 | 0.6705 | 0.6000 | 0.5692 |
| Precision | 0.8559 | 0.5316 | 0.5279 | 0.4306 | |
| Recall | 0.6846 | 0.5601 | 0.5509 | 0.4980 | |
| F1-value | 0.7058 | 0.5296 | 0.5200 | 0.4482 | |
| Image | Accuracy | 0.9547 | 0.9079 | 0.9460 | 0.7984 |
| Precision | 0.9563 | 0.9123 | 0.9510 | 0.8378 | |
| Recall | 0.9543 | 0.9134 | 0.9489 | 0.8126 | |
| F1-value | 0.9533 | 0.9105 | 0.9483 | 0.7982 | |
| Ionosphere | Accuracy | 0.9057 | 0.8784 | 0.8403 | 0.8221 |
| Precision | 0.9095 | 0.9247 | 0.8936 | 0.8224 | |
| Recall | 0.8642 | 0.8357 | 0.7870 | 0.8385 | |
| F1-value | 0.8769 | 0.8482 | 0.8022 | 0.8171 | |
| Letter | Accuracy | 0.9532 | 0.9017 | 0.8352 | 0.6437 |
| Precision | 0.9542 | 0.9053 | 0.8367 | 0.6562 | |
| Recall | 0.9531 | 0.8997 | 0.8331 | 0.6423 | |
| F1-value | 0.9532 | 0.9004 | 0.8327 | 0.6390 | |
| Optical | Accuracy | 0.9564 | 0.9538 | 0.9695 | 0.8204 |
| Precision | 0.9569 | 0.9552 | 0.9713 | 0.8613 | |
| Recall | 0.9564 | 0.9519 | 0.9688 | 0.8174 | |
| F1-value | 0.9564 | 0.9527 | 0.9691 | 0.8167 | |
| Adult | Accuracy | 0.8688 | 0.7749 | 0.7828 | 0.7983 |
| Precision | 0.8207 | 0.6811 | 0.8795 | 0.7419 | |
| Recall | 0.7792 | 0.6032 | 0.5457 | 0.6297 | |
| F1-value | 0.7962 | 0.6154 | 0.5216 | 0.6494 | |
| Average | Accuracy | 0.9106 | 0.8379 | 0.8738 | 0.7672 |
| Precision | 0.9195 | 0.8148 | 0.8576 | 0.7624 | |
| Recall | 0.8820 | 0.7893 | 0.8953 | 0.7465 | |
| F1-value | 0.8892 | 0.7881 | 0.7883 | 0.7363 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, L.; Chen, K.; Jiang, Z.; Xu, X. A Fast Parallel Random Forest Algorithm Based on Spark. Appl. Sci. 2023, 13, 6121. https://doi.org/10.3390/app13106121
Yin L, Chen K, Jiang Z, Xu X. A Fast Parallel Random Forest Algorithm Based on Spark. Applied Sciences. 2023; 13(10):6121. https://doi.org/10.3390/app13106121
Chicago/Turabian StyleYin, Linzi, Ken Chen, Zhaohui Jiang, and Xuemei Xu. 2023. "A Fast Parallel Random Forest Algorithm Based on Spark" Applied Sciences 13, no. 10: 6121. https://doi.org/10.3390/app13106121





