Abstract
Electroencephalographic (EEG) signal processing and machine learning can support neurologists’ work in discriminating Psychogenic Non-Epileptic Seizure (PNES) from epilepsy. PNES represents a neurological disease often misdiagnosed. Although the symptoms of PNES patients can be similar to those exhibited by epileptic patients, EEG signals during a psychogenic seizure do not show ictal patterns such as in epilepsy. Therefore, PNES diagnosis requires long-term EEG video. Applying signal processing and machine-learning methodologies could help clinicians find helpful information in the clinical diagnosis of PNES by analyzing EEG signals registered in resting conditions and in a short time. These methodologies should prevent long EEG recording sessions and avoid inducing seizures in the subjects. The aim of our study is to develop and validate several machine-learning models on a larger dataset, consisting of 225 EEGs (75 healthy, 75 PNES, and 75 subjects with epilepsy). A deep analysis of our results shows that changes in the evaluation strategy led to changes in accuracy from 45% to 83.98% for a standard Light Gradient Boosting Machine (LGBM) classifier. Our findings suggest that it is necessary to operate a very rigorous control in terms of experimental data collection (patient selection, signal acquisition) and terms of validation strategies to obtain and reproducible results.
1. Introduction
Psychogenic Non-Epileptic Seizures (PNES) and epilepsy are two distinct conditions that can present with similar symptoms, particularly during seizure activity. In fact, both PNES and epilepsy can result in episodes that resemble seizures and may involve convulsions, loss of consciousness, abnormal movements, and altered awareness. However, the underlying causes and diagnostic criteria for these conditions are different.
Electroencephalography (EEG) is a non-invasive neurophysiological technique used to measure and record the electrical activity of the brain. EEG provides valuable insights into the brain’s functioning and is widely employed in clinical, research, and diagnostic settings. EEG measures the fluctuations in electrical potentials generated by the synchronized activity of neurons in the brain. It provides a representation of the brain’s electrical activity over time. The electrical signals detected by EEG, commonly referred to as EEG signals, represent the collective activity of the brain’s neurons over time. These signals are characterized by different frequency bands, delta (0.5–4 Hz), theta (4–8 Hz), alpha (8–13 Hz), beta (13–30 Hz), and gamma (30+ Hz). Each frequency band corresponds to specific states of brain activity and provides insights into different aspects of brain function.
Analyzing EEG signals can be a valuable tool in the clinical diagnosis of PNES. In fact, PNES is considered a psychological disorder related to psychological stressors or trauma. During a psychogenic seizure, the EEG readings typically do not show any specific abnormal electrical discharges or epileptiform activity. In contrast, epilepsy is a neurological disorder characterized by abnormal electrical activity in the brain. Epileptic seizures often exhibit characteristic epileptiform discharges, such as spikes, sharp waves, or rhythmic activity in specific regions of the brain.
Since the clinical presentation of PNES can resemble epileptic seizures, this makes the distinction challenging. As a result, the patient is unaware that the seizures are not epileptic. Moreover, anti-epileptic medications, which are effective in managing epileptic seizures, generally do not alleviate PNES. Therefore PNES patients inaccurately diagnosed as epileptic initiate inappropriate drug therapy, which results in adverse reactions [1,2,3]. Different studies assess the frequency of functional or PNES seizures during the COVID-19 outbreak [4,5]. These studies evidence the possible factors associated with worsening in this population.
Although EEG alone cannot diagnose PNES because, during a PNES crisis, electrical brain activity remains normal, it can provide supportive evidence when interpreted in conjunction with other information. The most widely accepted and reliable method for diagnosing PNES is video-EEG monitoring, which is considered the gold standard in the field, and during which seizures are acquired spontaneously or provoked. Video-EEG monitoring allows simultaneous recording of both video footage and EEG signals during the occurrence of seizure events or seizure-like episodes. By capturing the patient’s physical movements, behaviors, and visible manifestations alongside the corresponding EEG patterns, video-EEG helps establish a direct correlation between the clinical manifestations and the observed electrical activity. This correlation aids healthcare professionals in distinguishing between epileptic seizures and PNES, as the manifestations in PNES are typically not associated with specific abnormal electrical discharges. Stimulation techniques are employed to trigger or induce seizures during EEG monitoring. This can include hyperventilation, photic stimulation (exposure to flashing lights), or other techniques specific to the individual patient. In PNES, the EEG remains unchanged or shows non-specific changes during the induced events. In contrast, epileptic EEG recordings show IED (interictal epileptiform discharge). However, it is challenging to distinguish IEDs often due to the muscle artefacts that overlap the signal during the EEG crisis.
In addition, prolonged EEG monitoring over an extended period, such as several hours or days, can increase the likelihood of capturing both typical and atypical events. This long-term monitoring can provide a more comprehensive assessment of the EEG patterns associated with PNES and aid in distinguishing them from epilepsy. Nevertheless, long-term monitoring and recording are expensive and time-consuming.
Machine-learning methodologies can play a crucial role in optimizing EEG analysis and reducing the need for long recording sessions while minimizing the risk of inducing seizures in subjects. One of the purpose of machine-learning techniques is to extract relevant features from the EEG signals, capturing important patterns and characteristics associated with specific conditions. These features can be used to create concise representations of the EEG data, allowing for efficient analysis and reducing the need for prolonged recording sessions. By focusing on informative features, machine learning can help identify key aspects of the EEG signals without requiring excessive recording time. Moreover, machine-learning methods, such as for instance some data augmentation approaches, can artificially expand the available dataset by generating synthetic EEG samples. This approach can be used to simulate different scenarios, including seizure activity, without the need to induce actual seizures in subjects. By incorporating augmented data into the training process, machine-learning models can learn to recognize and classify patterns associated with seizures, potentially reducing the dependence on inducing seizures during recording sessions.
Regarding identifying new methods to support medical decisions, the quantitative analysis sought to highlight differences between PNES and epileptics and healthy subjects. Specifically, some studies are based on analyzing and classifying the semiology of PNES and epilepsy [6,7]. An example is reported in [8], where the authors describe a new method for diagnosing PNESs. Fifty-five PNES video-EEG recordings were retrospectively analyzed by four epileptologists and one psychiatrist blindly and classified into four distinct groups: Hypermotor (H), Akinetic (A), Focal Motor (FM), and Subjective Symptoms(SS). Eleven signs and symptoms frequently found in PNESs were chosen for statistical validation of the classification. Agreement between ANN classification and visual classification reached 83.3%. In [9], the researchers present a pilot feasibility study with “Digital Semiology” (DS), a novel seizure encoding software. It allows semi-automated annotation of the videos of suspected events from a predetermined, hierarchical set of options, with highly detailed semiologic descriptions, somatic localization, and timing. Sixty episodes from a mixed adult and pediatric cohort from one level 4 epilepsy centre VEM archives were analyzed using DS. The reports were compared with the standard free-form ones written by the same epileptologists. The behavioural characteristics in the DS and free-form reports overlapped by 78–80%. The present study represents an important step toward forming an annotated video archive for machine-learning purposes.
Many other literature studies focus on EEG signal processing and machine learning. In [10], the authors analyze short-term EEG data for classifying epilepsy and PNES subjects, functional network, and EEG microstate features. Their results showed that the beta-band is the most helpful EEG frequency sub-band as it performs best for classifying subjects. The work was evaluated through 25 pairs of cross-validation. In [11], an automated discrimination method from EEG signals is proposed to eliminate the misdiagnosis and long inspection time of EEG recordings in PNES diagnosis. For this purpose, subbands of EEG signals are determined from discrete wavelet transform (DWT), and then classification is performed using ensemble classifiers fed with energy features extracted from the subbands. Results show that in the TLE (Temporal Lobe Epilepsy), PNES, and healthy epoch classification, performance evaluations were realized by using five-fold cross-validation method and the highest accuracy of 97.2%, the sensitivity of 97.9%, and specificity of 98.1% were achieved by applying the adaptive boosting method, and the highest accuracy of 87.1%, the sensitivity of 86.0%, and specificity of 93.6% were attained using random under sampling (RUS) boosting method in TLE patients, PNES patients, and healthy subject discrimination. The study presented in [12] investigates the quantitative electroencephalography (QEEG) features for PNES by evaluating the resting EEG spectral power changes during the periods between seizures. Using Fast Fourier transformation (FFT), a spectral power analysis was calculated for different EEG subbands from the EEG of 39 patients. As a result, six separate EEG band powers were found (C3-high beta, C3-gamma, C3-gamma-1, C3-gamma-2, P3-gamma, and P3 gamma-1) to be higher in the patients diagnosed with PNES than in the control group.
Innovative diagnostic tools that exploit non-linear EEG analysis and deep learning (DL) could provide essential support to physicians for clinical diagnosis. In [13], 18 patients with new-onset ES and 18 patients with video-recorded PNES with normal interictal EEG at visual inspection were enrolled. None of them were taking psychotropic drugs. A convolutional neural network (CNN) scheme using DL classification was designed to classify the two categories of subjects (ES vs. PNES). The proposed architecture performs an EEG time-frequency transformation and a classification step with a CNN. The CNN was able to classify the EEG recordings of subjects with ES vs. subjects with PNES with 94.4% accuracy, and a leave-one-patient-out cross-validation, providing high performance in the assigned binary classification compared to standard learning algorithms. In addition, a theoretical information analysis was carried out to interpret how CNN achieved this performance. Specifically, the feature maps’ permutation entropy (PE) was evaluated and compared in the two classes. In [14], the authors investigated the power spectrum density (PSD) in resting-state EEGs to evaluate the abnormalities in PNES-affected brains. Additionally, they used functional connectivity tools, such as phase lag index (PLI) and graph-derived metrics, to observe better the integration of distributed information of regular and synchronized multi-scale communication within and across inter-regional brain areas. Three classification models, namely, support vector machine (SVM), linear discriminant analysis (LDA), and multilayer perceptron (MLP), were used to model the relationship between the functional connectivity features. The best performance for the participants’ discrimination was obtained using the MLP classifier with an accuracy of 91.02% and a leave-one-out cross-validation.
Faiman et al., stated that quantitative analysis from resting-state electroencephalogram (EEG) provides helpful information to support the diagnosis of seizure disorders [15]. Studies were selected from five databases and evaluated using the Quality Assessment of Diagnostic Accuracy Studies-2. Results suggest that oscillations along the theta frequency (4–8 Hz) may have a relevant role in idiopathic epilepsy, whereas in PNES, there was no evident trend. However, by examining several studies, the authors put forth evidence that many were subject to several methodological limitations, potentially introducing bias. Specifically, they pointed out that studies using machine-learning methods should report information such as model architecture and training parameters to guarantee reproducibility. Moreover, it is necessary to analyze EEG recordings that did not occur a few hours before a seizure.
Recent studies in the field of seizure classification from EEG data have highlighted the importance of adopting more stringent evaluation criteria. Specifically, Peng et al. [16] proposed the leave-one-patient-out validation method based on the leave-one-out validation criterion. Additionally, Shafiezadeh et al. [17] compared the performance of a XGBoost classifier using two different validation criteria across two distinct EEG datasets. Results indicate a significant decline in accuracy from 80% to 50% when changing the evaluation strategy.
In this paper, we investigate the possibility of distinguishing between epileptic patients, PNES, and healthy subjects by analyzing resting-state EEG data using machine-learning techniques. The objective is to avoid inducing seizures in patients and minimize the need for lengthy EEG recording sessions. By leveraging machine-learning algorithms, we seek to identify specific patterns and features within the resting-state EEG signals that can effectively classify and differentiate between epileptic seizures and PNES events. This approach offers the potential to establish a non-invasive and efficient diagnostic method, reducing patient discomfort and optimizing resource utilization in clinical settings.
This work extends the work of Zucco et al. [18]. Specifically, our paper discusses a semi-automatic pipeline to discriminate between healthy, PNES, and epileptics subjects based on the extraction of spectral features from EEG signals and classification through machine-learning-based approaches. The innovative aspects are highlighted in the following. First, we tested the implemented pipeline on a larger dataset of 225 EEG recordings, equally distributed between the three classes. As pointed out in the previous paragraphs, the datasets are smaller in the literature; moreover, in our study a multi-class problem is addressed. We also implemented an automatic method to discard EEG-corrupted recordings due to the amplifier’s saturation on at least one channel for the entire duration of the EEG recording. Finally, we trained a suite of different machine learning algorithms to classify EEG recordings. In order to highlight the importance of establishing evaluation protocols that ensure the reproducibility and effectiveness of the models, we also aimed to explore how variations in the validation method, notably the use of patient-aware 10-fold cross-validation vs. standard 10-fold cross-validation, affected the average accuracy values of the models trained on our dataset.
In detail, the paper compares three different validation approaches: (i) preprocessing and feature extraction are performed on the EEG recordings without epochs segmentation and a standard 10-fold cross validation strategy; (ii) the preprocessing is performed on the entire EEG recording. The EEG is subsequently segmented in epochs, and patient-aware 10-fold cross-validation strategy evaluates several classifiers; (iii) the preprocessing is performed on the entire EEG recording. Each EEG is subsequently subdivided into epochs, and a standard 10-fold cross-validation evaluates several classifiers.
2. Materials and Methods
Neurologists of the Operative Unit of Neurology, Mater Domini Polyclinic, University of Catanzaro, Italy, had acquired EEG signals from three groups of subjects in a resting condition. The first group includes healthy subjects and is referred to as CNT, the control group. The second group included PNES subjects (PNES) who were diagnosed based on a video-EEG recording of a typical episode, where the EEG did not display any concurrent ictal activity or post-ictal patterns, whereas the last group comprises epileptic subjects. The study followed the Declaration of Helsinki and was formally approved by the local Medical Research Ethics Committee.
2.1. EEG Signals Acquisition
EEG recordings were acquired using 19 Ag/AgCl surface electrodes (C3, C4, O1, O2, Cz, F3, F4, F7, F8, Fz, Fp1, Fp2, P3, P4, Pz, T3, T4, T5 and T6) positioned according to the International 10/20 System (see Figure 1). Recordings were performed with an Xltek Brain Monitor EEG Amplifier with a sampling rate of 256 Hz. A band-pass filter with cut-off frequencies 0.5–70 Hz, and a 50 Hz notch filter were used. Participants were comfortably seated in a semi-darkened room and with open eyes.
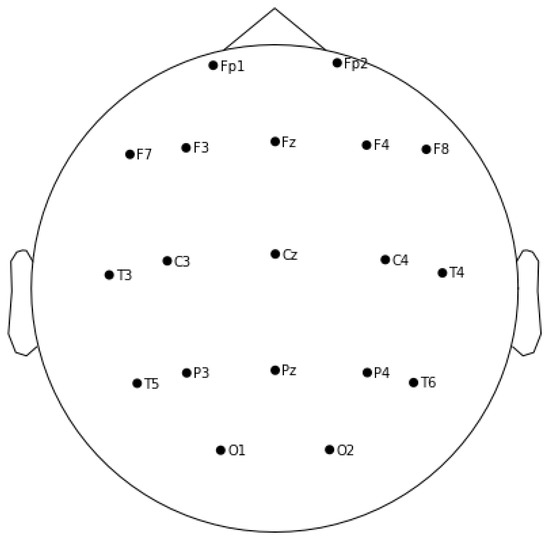
Figure 1.
The figure shows 10/20 International Standard for EEG electrodes positioning.
All the electrode-skin impedance was kept below 5 kΩ. The average duration of EEG acquisition ranged from 10 to 20 min.
2.2. EEG Data Pre-Processing
Generally, acquiring EEG signals is a challenging task due to their weak nature and susceptibility to contamination from environmental noise or distortion caused by physiological artifacts such as ocular and muscle artifacts. Consequently, noise removal plays a crucial role in the processing of EEG signals. Furthermore, proper data cleaning may improve the signal-to-noise ratio and allow for discriminating the most meaningful features from the EEG signals.
In clinical practice, trained neurologists visually detect artefacts by discarding contaminated EEG epochs. Although the pre-processing phase is operator-dependent, monotonous and time-consuming, in clinical practice, no automatic tools are applied in clinical context to detect and eliminate artefacts without eliminating useful signal parts.
In this study, a qualified neurologist examined each EEG recording to identify and annotate epochs corrupted by noise and artifacts (refer to Figure 2). Afterwards, all EEG data were pre-processed using an automatic routine that individuates the EEG portions where the amplifier is in a saturation stage (i.e., electrode malpositioning). The procedure scans all recordings and eliminates the portions of the EEG data (for all channels) where the amplifier is in saturation mode or all EEG recordings, if corrupted for the entire duration. Then, digital filtering techniques were applied. To mitigate high-frequency artifacts and power-line interference, we specifically applied a Butterworth band-pass filter with a frequency range of 0.1 to 70 Hz. Additionally, a notch filter with a cut-off frequency of 50 Hz was utilized. These filtering techniques were employed to minimize the impact of such disturbances during signal processing, as depicted in Figure 3.
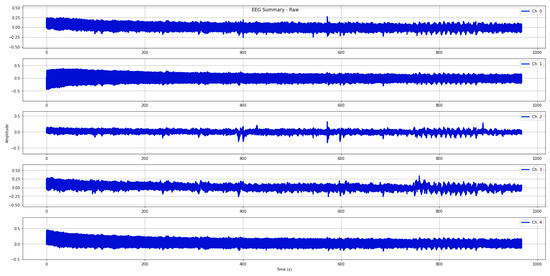
Figure 2.
The plots show EEG signals acquired from electrodes positioned on the frontal and central lobes.
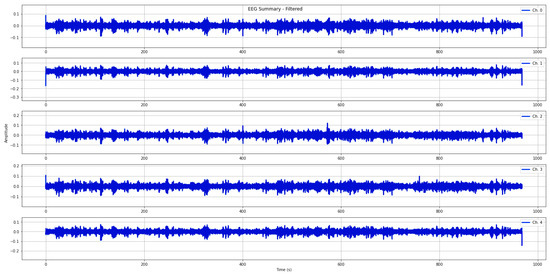
Figure 3.
The plots show filtered EEG signals acquired from electrodes positioned on the frontal and central lobes.
After noise removal, EEG signals were segmented in EEG epochs of 10 s to apply the following feature extraction methods on each epoch. Considering the average duration of each recording, an average number of 111 epochs was obtained for each channel for each subject. Segmenting the EEG signal into epochs is beneficial as it enables a more precise analysis of local variations within the signal. This segmentation approach facilitates a detailed examination of the EEG signal’s fluctuations on a localized level, and also extends the dataset without using artificial methods, enhancing the accuracy of the analysis.
2.3. EEG Feature Extraction
Following the pre-processing step, the subsequent stage in the EEG software pipeline is the feature extraction stage. As previously mentioned, the purpose of feature extraction is to extract pertinent information contained within the signals.
We decided to implement the Power Spectral Density (PSD) analysis because it is a robust extractor largely used for EEG quantitative analysis. Specifically, among the several methods for PSD estimation reported in the literature, Welch’s method was employed in our analysis. Let be the samples from an EEG epoch. The evaluation of PSD by using Welch’s method consists of the following steps:
- the EEG epoch is divided into N sections (possibly overlapped O) of equal lengths M;
- a window is applied to each section, and then the periodogram on the windowed sections is calculated. We can define the periodogram in the following way:
- the periodograms are averaged from the K sections in order to obtain an estimation of the spectral density
In this equation, represents the estimation of the cross power spectral density between two discrete-time signals, x and y. The Welch method eliminates the trade-off between spectral resolution and variance by allowing overlapping segments. When a high-frequency resolution is required, the recorded data can be divided into a small number (N) of segments with a length of L. However, in our analysis, we used non-overlapping segments and applied the Hamming window.After PSD calculation on all EEG epochs, we consider the following EEG sub-bands: delta (1–4 Hz), theta (4–8 Hz), alpha (8–13 Hz), beta (13–30 Hz), gamma (30–70 Hz) and calculated the cumulative power for each sub-band. Moreover, we evaluated the total cumulative power for all EEG frequencies.
The main processing steps of our feature extraction approach can be summarized as:
- Power Spectral Density (PSD) was estimated through Welch method;
- From PSD matrix output, we selected five frequency sub-bands;
- For each band (delta, teta, alpha, beta, gamma) we computed cumulative power.
Figure 4 shows an example of cumulative power in beta band for all epochs extracted from an EEG signal.
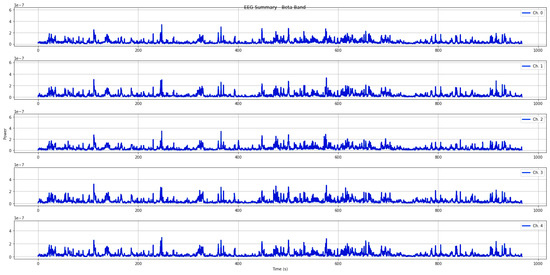
Figure 4.
EEG signals power in the beta band evaluated for all epochs.
We constitute the features vector with the six power cumulative coefficients for all EEG epochs.
The features vector has the following dimension: the number of cumulative power coefficients (six) times the number of epochs (an average of 105 epochs for each EEG) times the number of channels (19) times the number of subjects (225).
2.4. EEG Classification
The PyCaret library (available on https://pycaret.org, accessed on 13 May 2023) was used for the classification of EEG signals. PyCaret is an open-source low-code machine learning library for the Python language that automatically trains and evaluates a suite of different algorithms for classification. The present study considers the classifiers who obtained the best results in the various experiment conducted. In particular different ensemble techniques have been tested to discriminate among PNES, EPI and CNT: AdaBoost, Random Forest, Decision Trees, and Gradient Boosting.
Ensemble learning approaches assume the principle that different base models can be combined to build a more predictive model. Bagging [19] and Boosting [20] are the most popular ensemble learning techniques. In Bagging, the base learners are trained in parallel on bootstrap replicates of the training set and then the final prediction is made by majority voting, while in Boosting, the aggregate model is built in sequential fashion in order to train models that are increasingly more “attentive” to instances misclassified by previous models. AdaBoost [21] is one of the most popular boosting algorithm.
In a decision tree algorithm [22], knowledge is acquired through a collection of rules organized in a tree structure. The Random Forest algorithm, introduced by Breiman in 2001 [23], is an ensemble method that combines multiple decision trees through bagging. On the other hand, the Light Gradient Boosted Machine (LGBM), proposed by Ke et al., in 2017 [24], is a boosting ensemble of decision trees. LGBM constructs the ensemble by minimizing a differentiable loss function using the gradient descent optimization algorithm.
3. Results
This section presents and compares the results of training different machine learning algorithms in three different fashions.
Before presenting the results of the different test batteries in detail, a preliminary exploratory analysis of the data is presented to gain useful insights into the EEG data distribution after their preprocessing and epoching.
The experimental EEG raw data are available from 75 patients with PNES, 75 healthy patients (CNT), and 75 epileptic patients (EPI).
However, 10 EEGs (1 CNT, 3 PNES, 6 EPI) showed the presence of an amplifier in saturation stage on at least one channel for the entire duration of the EEG recording and were excluded from further analyses. As shown in the pie chart in Figure 5a, the exclusion criteria did not produce an imbalance between the classes.
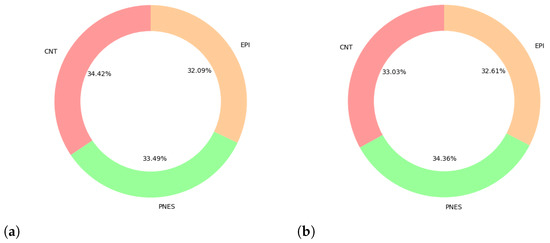
Figure 5.
Class distribution (a) after preprocessing (b) after segmentation. Due to the different duration of the EEGs recordings, the segmentation process does not produce the same number of epochs for each EEG. However, the comparison of the two pie charts shows that the dataset of segmented EEGs epochs does not presents any significant imbalance among classes.
Since the 225 input EEG data do not have the same duration, and the routine that eliminates EEG portions where the amplifier is in a saturation stage further reduces the duration of some EEG recordings, the segmentation process does not produce the same number of epochs for each EEG. Therefore, we also verified that the dataset continued to be balanced even after segmenting the signal into epochs, as shown in the pie chart in Figure 5b.
The violin plots in Figure 6 allow to visually compare the distributions of the EEGs epochs obtained for each class. To formally compare the epochs distribution obtained by class, we first check the three distributions are not normally distributed through the Shapiro–Wilk’s test and a significance level with for the EPI group, for the PNES group, and for the CNT group. Then, a Kruskal–Wallis test showed that there was no statistically significant difference in segmented epochs among the three groups, , , .
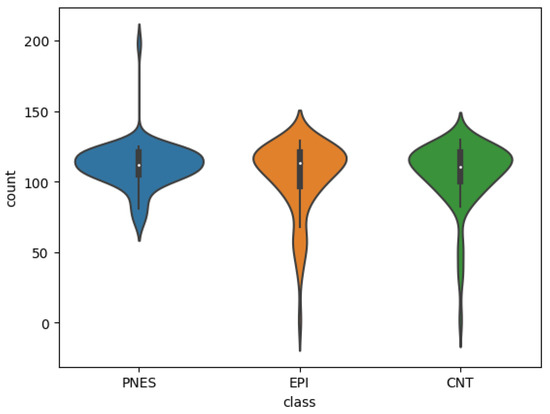
Figure 6.
Violin plot of the EEGs epochs distributions by class. Although the PNES class shows an high outlier and the CNT and EPI classes show low outliers, the three distributions are unimodal and show similar median and interquartile range.
The different test batteries described in Section 2 were conducted through the Python PyCaret library.
To be able to evaluate how much the segmentation of the EEG recordings in epochs affects the performance, three different approaches were considered. In particular:
- B1
- We extracted features as described in Section 2 from the 215 EEG recordings, without EEG epoching, and evaluate the classifiers by a standard 10-fold cross validation.
- B2
- We performed EEG epoching and feature extraction as described in Section 2, then different models built on the 22,261 EEG epochs dataset were evaluated by a patient-aware 10-fold cross validation strategy. Specifically, the 215 patients were randomly divided into 10 sets and all the 10 s epochs extracted from the same EEG recording were constrained to belong to the same fold. Therefore, at every run, the features of the EEG epochs segmented from the same EEG recording, appear either in the training or in the test set.
- B3
- We performed EEG epoching and feature extraction as described in Section 2, then different models built on the 22,261 EEG epochs dataset were evaluated by a standard 10-fold cross validation strategy. Each EEG epoch was considered independent from the others and therefore, while ensuring that the 10-folds were disjointed, we allowed that different EEG epochs segmented from of the same EEG recording could appear in different folds. In other words, the same EEG epoch cannot be present simultaneously in two different folds, while two different EEG epochs segmented from the same patient can appear in different folds and therefore, at a particular 10-fold validation run, one epoch could appear in the training set and the other in the test set.
For each test battery, we compared the performance of different algorithms provided by PyCaret. PyCaret includes a variety of classification algorithms that can be used for machine learning tasks. These algorithms cover a wide range of classification techniques, from traditional algorithms such as, for instance, logistic regression and decision trees, to ensemble methods as for instance random forest and gradient boosting. PyCaret provides a unified interface to work with these algorithms, making it easy to compare and evaluate their performance on different datasets. To streamline the discussion, we focused on the performance of the two top-performing models for each battery, namely, AdaBoost and Decision Tree (the most performant models for Battery [B1]), and Random Forest and Light Gradient Boosting Machine (the top performers for the validation approaches described in [B2] and [B3]). To assess the effectiveness of the algorithms, we examined various evaluation metrics, including accuracy and AUC. The results demonstrated notable variations in performance across the different batteries and algorithms.
Accuracy and AUC (Area Under the Curve) with a One vs. Rest (OvR) strategy [25] were considered for comparison.
Accuracy measures the number of correct prediction made by the algorithm. Denoting with , and and , and , the number of CNT, EPI and PNES subjects well or misclassified by the model, respectively, the model accuracy is:
AUC was chosen as measure to assess the model’s capability of distinguishing among classes. In the multiclass problem formulation, using the One-vs.-Rest (OvR) strategy, the evaluation of the AUC measure was performed individually for each class.
Table 1 presents Accuracy and micro-average Area Under the Curve (AUC) of the classifier models built from the 215 × 116 EEG dataset, where the 116 features described in Section 2 were extracted without epoching the EEG recordings. A 10-fold cross validation was exploited for perfomance evaluation (see the validation approach described in [B1]). Best values are highlighted in bold. The results show that also the Ada Boost model, which was the most-performing, reached low accuracy levels.

Table 1.
Accuracy and AUC of the considered models for test battery B1.
Table 2 presents Accuracy and micro-average Area Under the Curve (AUC) of the classifiers trained on 22,261 × 116 EEG dataset. A patient-aware 10-fold cross validation was exploited for perfomance evaluation (see the validation approach in [B2]). Best values are highlighted in bold. The results obtained in terms of accuracy and AUC summarized in Table 2 show that, especially with regard to the best performing classifiers, i.e., Random Forest and AdaBoost, the EEG epoching combined with a patient-aware 10-fold cross validation strategy, did a slight improvement of performance.

Table 2.
Accuracy and AUC of the considered models for test battery B2.
Table 3 summarizes Accuracy and micro-average Area Under the Curve (AUC) of the classifiers trained on 22,261 × 116 EEG dataset. A 10-fold cross validation was exploited for perfomance evaluation (see the approach described in [B3]). Best values are highlighted in bold. The results obtained in terms of accuracy and AUC show a significant increase in performance with respect to the previous approaches.

Table 3.
Accuracy and AUC of the considered models for test battery [B3].
Figure 7 compares the approaches in [B2] and [B3] in terms of learning curves for Light Gradient Boosting.
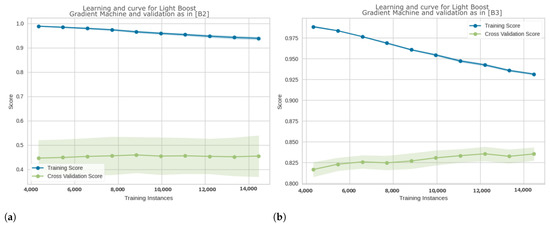
Figure 7.
Comparison of learning curves for Light Gradient Boosting Machine classifier on the same EEG epoched dataset, and (a) with a patient-aware 10-fold cross validation as described in approach [B2], (b) with a Stratified 10-fold cross validation as described in approach [B3]. The y-axis represents accuracy score. In figure (a), the learning curve shows high performance on the training set (in blue) but significantly lower performance on the training set where accuracy is between 40% and 50% (green curve), with a large gap between the two curves. This indicates overfitting on the training dataset and poor generalization on unseen data. In figure (b), the learning curve shows accuracy over 90% on the training set (in blue) and accuracy over 80% on the training set (in green). This indicates that the model generalizes to unseen data.
Learning curves, in the context of machine learning, are graphical representations that depict the relationship between a model’s performance and the amount of training data used. They provide insights into how the model’s performance improves or stabilizes as more data become available for training. A learning curve typically plots a performance metric; in this work, we considered the accuracy on the y-axis against the number of training examples or the training set size on the x-axis. Accuracy is computed on both training and test set. The learning curve shows how the model’s performance evolves as more data points are included in the training process. The learning curve can exhibit different patterns that convey valuable information about the model:
- Overfitting: If the learning curve shows high performance on the training set but significantly lower performance on the validation set, with a large gap between the two curves. This indicates that the model is capturing noise or specific patterns in the training data that do not generalize well to unseen data.
- Underfitting: If the learning curve demonstrates low performance on both the training and validation sets. In this case, more data are needed or the model is probably too simple and is unable to capture the underlying patterns in the data.
- Convergence: Ideally, a learning curve shows that the model’s performance on both the training and validation sets converge or stabilize as more data are included. This suggests that the model is learning from the data and generalizing well to unseen examples.
The results in Figure 7a suggest that the model leads to a poor generalization, with probable model overfitting on the training dataset, while Figure 7b shows a better ability to generalize the model on the data.
To better investigate the performance of the LBGM model and [B3] test battery, in Figure 8 the OvR ROC curves and the micro and macro average ROC curves for each class are compared. In Figure 9, precision-recall curve is displayed for LBGM, illustrating a favorable balance between false positive and false negative rates. Furthermore, Figure 10 provides additional insights into which features have the most influence on the model’s predictions, allowing researchers and practitioners to understand the relative importance of different variables in the model’s decision-making process. Significant features have been selected through PyCaret’s feature importance permutation technique. This method involves randomly permuting the values of a single feature and measuring the impact on the model’s performance. By comparing the model’s performance before and after permuting the feature, the importance of the feature can be determined. Features with larger performance drops after permutation are considered more important.
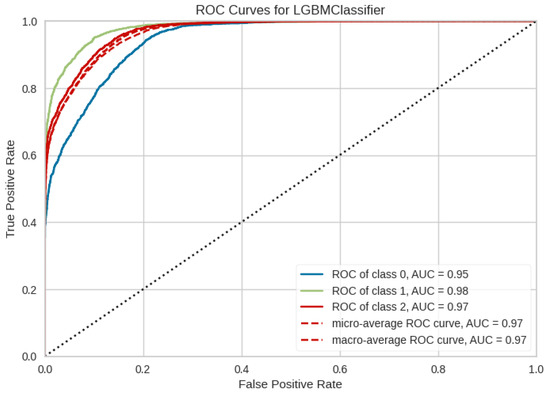
Figure 8.
ROC Curves for LGBM Classifier and [B3] validation approach. Class 0 refers to CNT, class 1 to EPI, and class 2 to PNES.
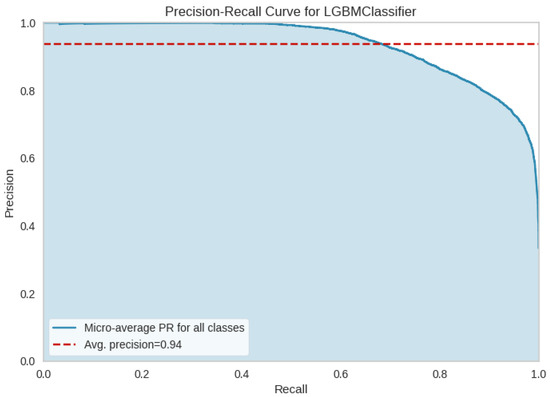
Figure 9.
Precision-Recall curve and [B3] validation approach.
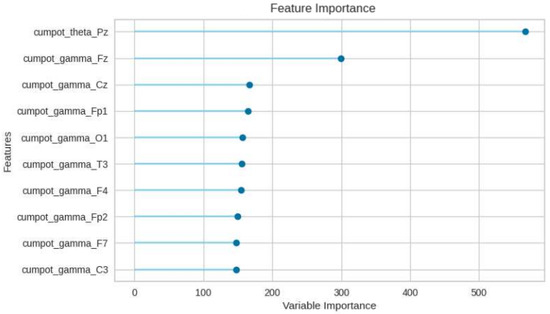
Figure 10.
The ten most important features w.r.t. LGBM and [B3] validation approach.
The analysis reveals that the cumulative power in the theta band at the Pz electrode and the gamma band are particularly influential in predicting the target variable. These findings are consistent with prior research in the field, which has consistently identified the theta and gamma frequency bands as relevant indicators in similar contexts.
In general, the results show a good predictive power of the last discussed model, and the EPI class demonstrated the highest level of discrimination, with an AUC of 98% and an error rate of 11%, see Figure 8, Figure 9, Figure 10 and Figure 11. However, more comprehensive analyses are required to delve deeper into the comparison of different validation approaches across multiple EEG datasets. These analyses would help investigate the underlying factors contributing to the significant difference in performance observed between validation approaches [B2] and [B3].
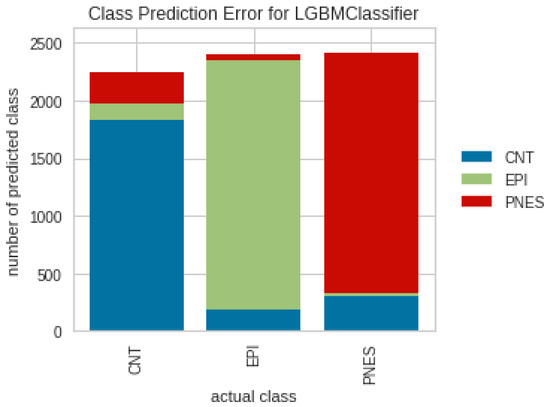
Figure 11.
Class prediction errors for LGBM classifier and [B3] validation approach.
To gain a better understanding of the observed performance disparities, further investigations should be conducted on a larger and more diverse set of EEG datasets. This would enable researchers to explore potential sources of variation, such as differences in data characteristics, recording protocols, or patient populations. By examining the performance across multiple datasets, it would be possible to assess the generalization capabilities of the model and determine whether the observed discrepancies are consistent or dataset-specific.
4. Conclusions
In recent years, the use of machine-learning techniques for EEG analysis for the automatic differentiation of PNES and epileptic subjects has attracted a lot of interest in the scientific community.
This work proposed an analysis pipeline that, starting from raw EEG acquired in resting conditions, combines signal processing and machine learning techniques for the automatic classification of PNES, epileptic and healthy subjects. Our analysis pipeline underwent testing on a large dataset comprising approximately 225 subjects, resulting in classification accuracy exceeding 83% for distinguishing between control subjects, PNES, and epileptics.
The study also focused on the effects of segmenting the EEG in epochs and in particular showed that, under the Kruskal–Wallis test assumptions, we found no statistically significant difference in segmented epochs among the three groups, , , .
Then, different evaluation strategies were compared to quantify how much the segmentation of the EEG recordings in epochs may affect the performance.
The results indicate the presence of overfitting in the best-performing model when validated using patient-based 10-fold cross validation. Specifically, the learning curve of this model exhibits signs of overfitting, with the training curve showing a large gap between the accuracy on training and test set. However, by only changing the validation method on the same dataset, the accuracy is doubled and the learning curve did show good convergence. These findings aligned well with recent studies that highlight the critical importance of carefully selecting an appropriate validation strategy to ensure the reproducibility and applicability of the method. It emphasizes the need to investigate and compare different validation criteria to determine the most effective approach for evaluating the modelsas well as the importance of defining rigorous evaluation protocols to ensure reproducibility and good generalization of the proposed model on unseen data.
Therefore, we plan to conduct more in-depth analyses that extend the comparison of different validation approaches to several EEG datasets, to investigate the cause of the strong difference in performance between different validation approaches.
Author Contributions
Conceptualization, B.C., C.Z. and M.C.; methodology, C.Z., B.C. and M.C.; validation, C.Z. and B.C.; data curation, M.S. writing—original draft preparation, B.C. and C.Z.; writing—review and editing, B.C., C.Z., M.C., R.M., M.S., F.P. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data sharing not applicable The data are not publicly available due to privacy regulation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gasparini, S.; Beghi, E.; Ferlazzo, E.; Beghi, M.; Belcastro, V.; Biermann, K.P.; Bottini, G.; Capovilla, G.; Cervellione, R.A.; Cianci, V.; et al. Management of psychogenic non-epileptic seizures: A multidisciplinary approach. Eur. J. Neurol. 2019, 26, 205-e15. [Google Scholar] [CrossRef]
- Albert, D.V. Psychogenic Nonepileptic Seizures in Children and Adolescents. Semin. Pediatr. Neurol. 2022, 41, 100949. [Google Scholar] [CrossRef]
- Volbers, B.; Walther, K.; Kurzbuch, K.; Erdmann, L.; Gollwitzer, S.; Lang, J.D.; Dogan Onugoren, M.; Schwarz, M.; Schwab, S.; Hamer, H.M. Psychogenic nonepileptic seizures: Clinical characteristics and outcome. Brain Behav. 2022, 12, e2567. [Google Scholar] [CrossRef]
- Rosengard, J.L.; Ferastraoaru, V.; Donato, J.; Haut, S.R. Psychogenic nonepileptic seizures during the COVID-19 pandemic in New York City—A distinct response from the epilepsy experience. Epilepsy Behav. 2021, 123, 108255. [Google Scholar] [CrossRef]
- Valente, K.D.; Alessi, R.; Baroni, G.; Marin, R.; Dos Santos, B.; Palmini, A. The COVID-19 outbreak and PNES: The impact of a ubiquitously felt stressor. Epilepsy Behav. 2021, 117, 107852. [Google Scholar] [CrossRef]
- Gomez-Figueroa, E.; Vargas-Sanchez, Á.; Alvarado-Bolaños, A.; Paredes-Aragón, E.; Alatriste-Booth, V.; Moreno-Avellan, Á.; Fernández, M. The role of short-term video electroencephalogram monitoring for epilepsy and psychogenic seizures. J. Clin. Neurosci. 2020, 82, 105–110. [Google Scholar] [CrossRef]
- Deli, A.; Huang, Y.G.; Toynbee, M.; Towle, S.; Adcock, J.E.; Bajorek, T.; Okai, D.; Sen, A. Distinguishing psychogenic nonepileptic, mixed, and epileptic seizures using systemic measures and reported experiences. Epilepsy Behav. 2021, 116, 107684. [Google Scholar] [CrossRef]
- Magaudda, A.; Laganà, A.; Calamuneri, A.; Brizzi, T.; Scalera, C.; Beghi, M.; Cornaggia, C.M.; Di Rosa, G. Validation of a novel classification model of psychogenic nonepileptic seizures by video-EEG analysis and a machine learning approach. Epilepsy Behav. 2016, 60, 197–201. [Google Scholar] [CrossRef]
- Benoliel, T.; Gilboa, T.; Har-Shai Yahav, P.; Zelker, R.; Kreigsberg, B.; Tsizin, E.; Arviv, O.; Ekstein, D.; Medvedovsky, M. Digital Semiology: A Prototype for Standardized, Computer-Based Semiologic Encoding of Seizures. Front. Neurol. 2021, 12, 711378. [Google Scholar] [CrossRef]
- Ahmadi, N.; Pei, Y.; Carrette, E.; Aldenkamp, A.P.; Pechenizkiy, M. EEG-based classification of epilepsy and PNES: EEG microstate and functional brain network features. Brain Inform. 2020, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Fıçıcı, C.; Telatar, Z.; Eroğul, O. Automated temporal lobe epilepsy and psychogenic nonepileptic seizure patient discrimination from multichannel EEG recordings using DWT based analysis. Biomed. Signal Process. Control 2022, 77, 103755. [Google Scholar] [CrossRef]
- Arıkan, K.; Öksüz, Ö.; Metin, B.; Günver, G.; Laçin Çetin, H.; Esmeray, T.; Tarhan, N. Quantitative EEG Findings in Patients with Psychogenic Nonepileptic Seizures. Clin. EEG Neurosci. 2021, 52, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Lo Giudice, M.; Varone, G.; Ieracitano, C.; Mammone, N.; Tripodi, G.G.; Ferlazzo, E.; Gasparini, S.; Aguglia, U.; Morabito, F.C. Permutation Entropy-Based Interpretability of Convolutional Neural Network Models for Interictal EEG Discrimination of Subjects with Epileptic Seizures vs. Psychogenic Non-Epileptic Seizures. Entropy 2022, 24, 102. [Google Scholar] [CrossRef] [PubMed]
- Varone, G.; Boulila, W.; Lo Giudice, M.; Benjdira, B.; Mammone, N.; Ieracitano, C.; Dashtipour, K.; Neri, S.; Gasparini, S.; Morabito, F.C.; et al. A Machine Learning Approach Involving Functional Connectivity Features to Classify Rest-EEG Psychogenic Non-Epileptic Seizures from Healthy Controls. Sensors 2021, 22, 129. [Google Scholar] [CrossRef] [PubMed]
- Faiman, I.; Smith, S.; Hodsoll, J.; Young, A.H.; Shotbolt, P. Resting-state EEG for the diagnosis of idiopathic epilepsy and psychogenic nonepileptic seizures: A systematic review. Epilepsy Behav. 2021, 121, 108047. [Google Scholar] [CrossRef]
- Peng, P.; Song, Y.; Yang, L.; Wei, H. Seizure prediction in EEG signals using STFT and domain adaptation. Front. Neurosci. 2022, 15, 1880. [Google Scholar] [CrossRef]
- Shafiezadeh, S.; Duma, G.M.; Mento, G.; Danieli, A.; Antoniazzi, L.; Del Popolo Cristaldi, F.; Bonanni, P.; Testolin, A. Methodological issues in evaluating machine learning models for EEG seizure prediction: Good cross-validation accuracy does not guarantee generalization to new patients. Appl. Sci. 2023, 13, 4262. [Google Scholar] [CrossRef]
- Zucco, C.; Calabrese, B.; Mancuso, R.; Sturniolo, M.; Gambardella, A.; Cannataro, M. Resting-State EEG Classification for PNES Diagnosis. In Proceedings of the Computational Science–ICCS 2022: 22nd International Conference, London, UK, 21–23 June 2022; pp. 526–538. [Google Scholar]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Schapire, R.E.; Freund, Y. Boosting: Foundations and algorithms. Kybernetes 2013, 42, 164–166. [Google Scholar] [CrossRef]
- Schapire, R.E. Explaining adaboost. In Empirical Inference: Festschrift in Honor of Vladimir N. Vapnik; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013; pp. 37–52. [Google Scholar]
- Quinlan, J.R. Induction of decision trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).