Development of Artificial Intelligence-Based Dual-Energy Subtraction for Chest Radiography
Abstract
:1. Introduction
- We developed an AI-based DES system to provide soft-tissue- and bone-enhanced images using virtually generated low-energy images;
- The virtual low-energy images were generated through the AI technique from only high-energy images, which can be obtained by routine chest radiography;
- AI-DES has the potential to provide specific tissue-enhanced images while avoiding issues associated with DES systems, such as multiple exposures and noise increments;
- A comparison of the generated images with those produced by a clinically applied system suggests that AI-DES can achieve superior sharpness and noise characteristics.
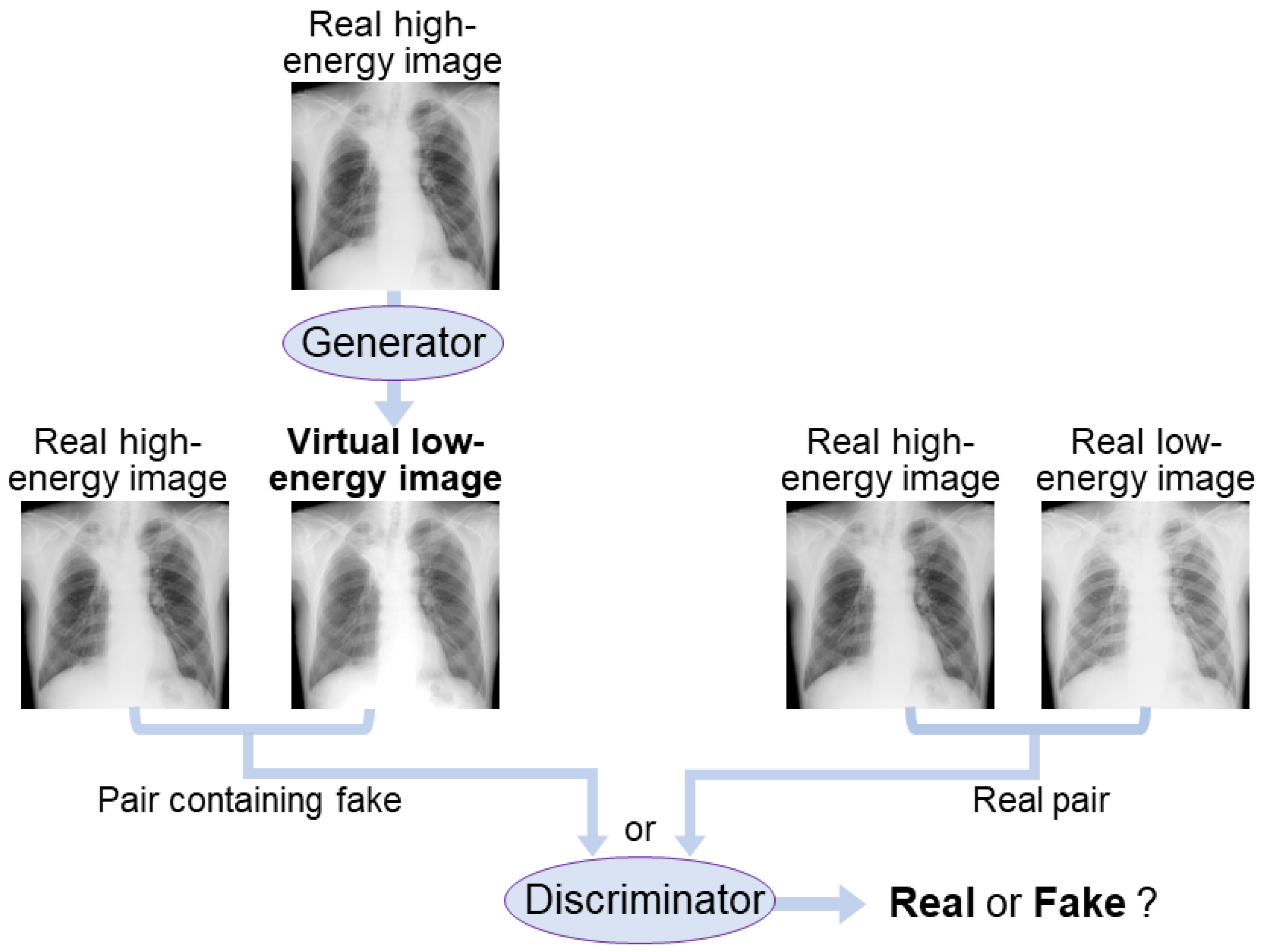
2. Materials and Methods
2.1. AI-DES Development
2.1.1. AI Network
2.1.2. Weighted Image Subtraction
2.2. Dataset Preparation
2.3. Training Environment and Parameter Settings
2.4. Performance Evaluation
3. Results
3.1. Generated Virtual Low-Energy Images
3.2. Soft Tissue and Bone Images
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, J.; Deng, Y.; Tin, M.S.; Lok, V.; Ngai, C.H.; Zhang, L.; Lucero-Prisno, D.E., 3rd; Xu, W.; Zheng, Z.J.; Elcarte, E.; et al. Distribution, risk factors, and temporal trends for lung cancer incidence and mortality: A global analysis. Chest 2022, 161, 1101–1111. [Google Scholar] [CrossRef]
- Panunzio, A.; Sartori, P. Lung cancer and radiological imaging. Curr. Radiopharm. 2020, 13, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Ge, T.; Jiang, M.; Jia, K.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; et al. Early diagnosis of lung cancer: Which is the optimal choice? Aging 2021, 13, 6214–6227. [Google Scholar] [CrossRef]
- Huo, J.; Shen, C.; Volk, R.J.; Shih, Y.T. Use of CT and chest radiography for lung cancer screening before and after publication of screening guidelines: Intended and unintended uptake. JAMA Intern. Med. 2017, 177, 439–441. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.J.; Stone, E.; Baldwin, D.R.; Vliegenthart, R.; Lee, P.; Fintelmann, F.J. Lung cancer screening. Lancet 2022, 401, 390–408. [Google Scholar] [CrossRef] [PubMed]
- Wender, R.; Fontham, E.T.; Barrera, E., Jr.; Colditz, G.A.; Church, T.R.; Ettinger, D.S.; Etzioni, R.; Flowers, C.R.; Gazelle, G.S.; Kelsey, D.K.; et al. American Cancer Society lung cancer screening guidelines. CA Cancer J. Clin. 2013, 63, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Kazerooni, E.A.; Baum, S.L.; Eapen, G.A.; Ettinger, D.S.; Hou, L.; Jackman, D.M.; Klippenstein, D.; Kumar, R.; Lackner, R.P.; et al. Lung cancer screening, Version 3.2018, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2008, 98, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Nakayama, T.; Kusunoki, Y.; Iso, H.; Suzuki, T. Sensitivity and specificity of lung cancer screening using chest low-dose computed tomography. Br. J. Cancer 2018, 16, 412–441. [Google Scholar] [CrossRef]
- Stitik, F.P.; Tockman, M.S. Radiographic screening in the early detection of lung cancer. Radiol. Clin. N. Am. 1978, 16, 347–366. [Google Scholar]
- Li, F.; Engelmann, R.; Doi, K.; MacMahon, H. Improved detection of small lung cancers with dual-energy subtraction chest radiography. AJR Am. J. Roentgenol. 2008, 190, 886–891. [Google Scholar] [CrossRef]
- Gomi, T.; Nakajima, M. Dual-energy subtraction X-ray digital tomosynthesis: Basic physical evaluation. Open J. Med. Imaging 2012, 2, 111–117. [Google Scholar] [CrossRef] [Green Version]
- MacMahon, H.; Li, F.; Engelmann, R.; Roberts, R.; Armato, S. Dual energy subtraction and temporal subtraction chest radiography. J. Thorac. Imaging 2008, 23, 77–85. [Google Scholar] [CrossRef]
- Kuhlman, J.E.; Collins, J.; Brooks, G.N.; Yandow, D.R.; Broderick, L.S. Dual-energy subtraction chest radiography: What to look for beyond calcified nodules. Radiographics 2006, 26, 79–92. [Google Scholar] [CrossRef]
- Oda, S.; Awai, K.; Funama, Y.; Utsunomiya, D.; Yanaga, Y.; Kawanaka, K.; Yamashita, Y. Effects of dual-energy subtraction chest radiography on detection of small pulmonary nodules with varying attenuation: Receiver operating characteristic analysis using a phantom study. Jpn. J. Radiol. 2010, 28, 214–219. [Google Scholar] [CrossRef]
- Oda, S.; Awai, K.; Funama, Y.; Utsunomiya, D.; Yanaga, Y.; Kawanaka, K.; Nakaura, T.; Hirai, T.; Murakami, R.; Nomori, H.; et al. Detection of small pulmonary nodules on chest radiographs: Efficacy of dual-energy subtraction technique using flat-panel detector chest radiography. Clin. Radiol. 2010, 65, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Manji, F.; Wang, J.; Norman, G.; Wang, Z.; Koff, D. Comparison of dual energy subtraction chest radiography and traditional chest X-rays in the detection of pulmonary nodules. Quant. Imaging Med. Surg. 2016, 6, 1–5. [Google Scholar] [PubMed]
- Van der Heyden, B. The potential application of dual-energy subtraction radiography for COVID-19 pneumonia imaging. Br. J. Radiol. 2021, 94, 20201384. [Google Scholar] [CrossRef] [PubMed]
- Fukao, M.; Kawamoto, K.; Matsuzawa, H.; Honda, O.; Iwaki, T.; Doi, T. Optimization of dual-energy subtraction chest radiography by use of a direct-conversion flat-panel detector system. Radiol. Phys. Technol. 2015, 8, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Do, Q.; Seo, W.; Shin, C.W. Automatic algorithm for determining bone and soft-tissue factors in dual-energy subtraction chest radiography. Biomed. Signal Process. Control 2023, 80, 104354. [Google Scholar] [CrossRef]
- Vock, P.; Szucs-Farkas, Z. Dual energy subtraction: Principles and clinical applications. Digit. Radiogr. 2009, 72, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Park, J.; Kim, J.; Kim, H.K. Noise-reduction approaches to single-shot dual-energy imaging with a multilayer detector. J. Instrum. 2019, 14, C01021. [Google Scholar] [CrossRef]
- Shunkov, Y.E.; Kobylkin, I.S.; Prokhorov, A.V.; Pozdnyakov, D.V.; Kasiuk, D.M.; Nechaev, V.A.; Alekseeva, O.M.; Naumova, D.I.; Dabagov, A.R. Motion artefact reduction in dual-energy radiography. Biomed. Eng. 2022, 55, 415–419. [Google Scholar] [CrossRef]
- Hong, G.S.; Do, K.H.; Son, A.Y.; Jo, K.W.; Kim, K.P.; Yun, J.; Lee, C.W. Value of bone suppression software in chest radiographs for improving image quality and reducing radiation dose. Eur. Radiol. 2021, 31, 5160–5171. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, N.; Teramoto, A.; Saito, K.; Fujita, H.; Naumova, D.I. Bone suppression for chest X-ray image using a convolutional neural filter. Phys. Eng. Sci. Med. 2020, 43, 97–108. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, M.; Xi, Y.; Qin, G.; Shen, D.; Yang, W. Generating dual-energy subtraction soft-tissue images from chest radiographs via bone edge-guided GAN. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; pp. 678–687. [Google Scholar]
- Bae, K.; Oh, D.Y.; Yun, I.D.; Jeon, K.N. Bone suppression on chest radiographs for pulmonary nodule detection: Comparison between a generative adversarial network and dual-energy subtraction. Korean J. Radiol. 2022, 23, 139–149. [Google Scholar] [CrossRef]
- Cho, K.; Seo, J.; Kyung, S.; Kim, M.; Hong, G.-S.; Kim, N. Bone suppression on pediatric chest radiographs via a deep learning-based cascade model. Comput. Methods Programs Biomed. 2022, 215, 106627. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhou, L.; Shen, K. Dilated conditional GAN for bone suppression in chest radiographs with enforced semantic features. Med. Phys. 2020, 47, 6207–6215. [Google Scholar] [CrossRef]
- Zarshenas, A.; Liu, J.; Forti, P.; Suzuki, K. Separation of bones from soft tissue in chest radiographs: Anatomy-specific orientation-frequency-specific deep neural network convolution. Med. Phys. 2019, 46, 2232–2242. [Google Scholar] [CrossRef]
- Rajaraman, S.; Cohen, G.; Spear, L.; Folio, L.; Antani, S. DeBoNet: A deep bone suppression model ensemble to improve disease detection in chest radiographs. PLoS ONE 2022, 17, e0265691. [Google Scholar] [CrossRef]
- Rani, G.; Misra, A.; Dhaka, V.S.; Zumpano, E.; Vocaturo, E. Spatial feature and resolution maximization GAN for bone suppression in chest radiographs. Comput. Methods Programs Biomed. 2022, 224, 107024. [Google Scholar] [CrossRef]
- Isora, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar]
- Yoshida, N.; Kageyama, H.; Akai, H.; Yasaka, K.; Sugawara, H.; Okada, Y.; Kunimatsu, A. Motion correction in MR image for analysis of VSRAD using generative adversarial network. PLoS ONE 2022, 17, e0274576. [Google Scholar] [CrossRef]
- Sun, J.; Du, Y.; Li, C.; Wu, T.-H.; Yang, B.; Mok, G.S.P. Pix2Pix generative adversarial network for low dose myocardial perfusion SPECT denoising. Quant. Imaging Med. Surg. 2022, 12, 3539–3555. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- junyanz/pytorch-CycleGAN-and-pix2pix. Available online: https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix (accessed on 5 June 2023).
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Pathak, D.; Krähenbühl, P.; Donahue, J.; Darrell, T.; Efros, A.A. Context encoders: Feature learning by inpainting. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2536–2544. [Google Scholar]
- Hayashi, N.; Taniguchi, A.; Noto, K.; Shimosegawa, M.; Ogura, T.; Doi, K. Development of a digital chest phantom for studies on energy subtraction techniques. Nihon Hoshasen Gijutsu Gakkai Zasshi 2014, 70, 191–198. (In Japanese) [Google Scholar] [CrossRef] [Green Version]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs trained by a two time-scale update rule converge to a local nash equilibrium. In Proceedings of the Advances in Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 6626–6637. [Google Scholar]
- Borji, A. Pros and cons of GAN evaluation measures. arXiv 2018, arXiv:1802.03446. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error measurement to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hóre, A.; Ziou, D. Image Quality Metrics: PSNR vs. SSIM. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2366–2369. [Google Scholar]
- Wang, Z.; Simoncelli, E.P.; Bovik, A.C. Multiscale structural similarity for image quality assessment. In Proceedings of the 37th IEEE Asilomar Conference on Signals, Systems & Computers, Pacific Grove, CA, USA, 9–12 November 2003; Volume 2, pp. 1398–1402. [Google Scholar]
- Umme, S.; Morium, A.; Mohammad, S.U. Image quality assessment through FSIM, SSIM, MSE and PSNR—A comparative study. J. Comput. Commun. 2019, 7, 8–18. [Google Scholar]
- Mudeng, V.; Kim, M.; Choe, S. Prospects of structual similarity index for medical image analysis. Appl. Sci. 2022, 12, 3754. [Google Scholar] [CrossRef]








| Type | Norm 1, Dropout | Activation | Input Shape 2 | Output Shape 2 | ||
|---|---|---|---|---|---|---|
| Encoder | Layer1 | Conv2d (4,2,1) | – | LekyReLU | 1024 × 1024 × 3 | 512 × 512 × 64 |
| Layer2 | BN | 512 × 512 × 64 | 256 × 256 × 128 | |||
| Layer3 | 256 × 256 × 128 | 128 × 128 × 256 | ||||
| Layer4 | 128 × 128 × 256 | 64 × 64 × 512 | ||||
| Layer5 | 64 × 64 × 512 | 32 × 32 × 512 | ||||
| Layer6 | 32 × 32 × 512 | 16 × 16 × 512 | ||||
| Layer7 | 16 × 16 × 512 | 8 × 8 × 512 | ||||
| Layer8 | 8 × 8 × 512 | 4 × 4 × 512 | ||||
| Decoder | Layer9 | Deconv2d (4,2,1) | BN | – | 4 × 4 × 512 | 8 × 8 × 512 |
| Layer10 | ReLU+Deconv2d (4,2,1) | BN+Dropout | 8 × 8 × 512 | 16 × 16 × 512 | ||
| Layer11 | 16 × 16 × 512 | 32 × 32 × 512 | ||||
| Layer12 | 3 2 × 32 × 512 | 64 × 64 × 512 | ||||
| Layer13 | BN | 64 × 64 × 512 | 128 × 128 × 256 | |||
| Layer14 | 128 × 128 × 256 | 256 × 256 × 128 | ||||
| Layer15 | 256 × 256 × 128 | 512 × 512 × 64 | ||||
| Layer16 | – | Tanh | 512 × 512 × 64 | 1024 × 1024 × 3 |
| Type | Normalization | Activation | Input Shape 1 | Output Shape 1 | |
|---|---|---|---|---|---|
| Layer1 | Conv2d (4,2,1) | – | LekyReLU | 1024 × 1024 × 6 | 512 × 512 × 64 |
| Layer2 | BN | 512 × 512 × 64 | 256 × 256 × 128 | ||
| Layer3 | 256 × 256 × 128 | 128 × 128 × 256 | |||
| Layer4 | Conv2d (4,1,1) | 128 × 128 × 256 | 127 × 127 × 512 | ||
| Layer5 | – | – | 127 × 127 × 512 | 126 × 126 × 1 |
| PSNR | SSIM | MS-SSIM | Weight Factor | |
|---|---|---|---|---|
| 60 kV images (virtual and real) | 33.8 ± 5.39 | 0.984 ± 0.00554 | 0.957 ± 0.0514 | – |
| Soft tissue images (AI-DES and Discovery) | 21.1 ± 2.56 | 0.711 ± 0.0551 | 0.794 ± 0.0640 | 2.47 ± 0.159 |
| Bone images (AI-DES and Discovery) | 18.3 ± 1.97 | 0.433 ± 0.0827 | 0.571 ± 0.101 | 1.52 ± 0.102 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamazaki, A.; Koshida, A.; Tanaka, T.; Seki, M.; Ishida, T. Development of Artificial Intelligence-Based Dual-Energy Subtraction for Chest Radiography. Appl. Sci. 2023, 13, 7220. https://doi.org/10.3390/app13127220
Yamazaki A, Koshida A, Tanaka T, Seki M, Ishida T. Development of Artificial Intelligence-Based Dual-Energy Subtraction for Chest Radiography. Applied Sciences. 2023; 13(12):7220. https://doi.org/10.3390/app13127220
Chicago/Turabian StyleYamazaki, Asumi, Akane Koshida, Toshimitsu Tanaka, Masashi Seki, and Takayuki Ishida. 2023. "Development of Artificial Intelligence-Based Dual-Energy Subtraction for Chest Radiography" Applied Sciences 13, no. 12: 7220. https://doi.org/10.3390/app13127220
APA StyleYamazaki, A., Koshida, A., Tanaka, T., Seki, M., & Ishida, T. (2023). Development of Artificial Intelligence-Based Dual-Energy Subtraction for Chest Radiography. Applied Sciences, 13(12), 7220. https://doi.org/10.3390/app13127220






