Optimal Thresholding for Multi-Window Computed Tomography (CT) to Predict Lung Cancer
Abstract
:1. Introduction
2. Proposed Method
2.1. Preprocessing
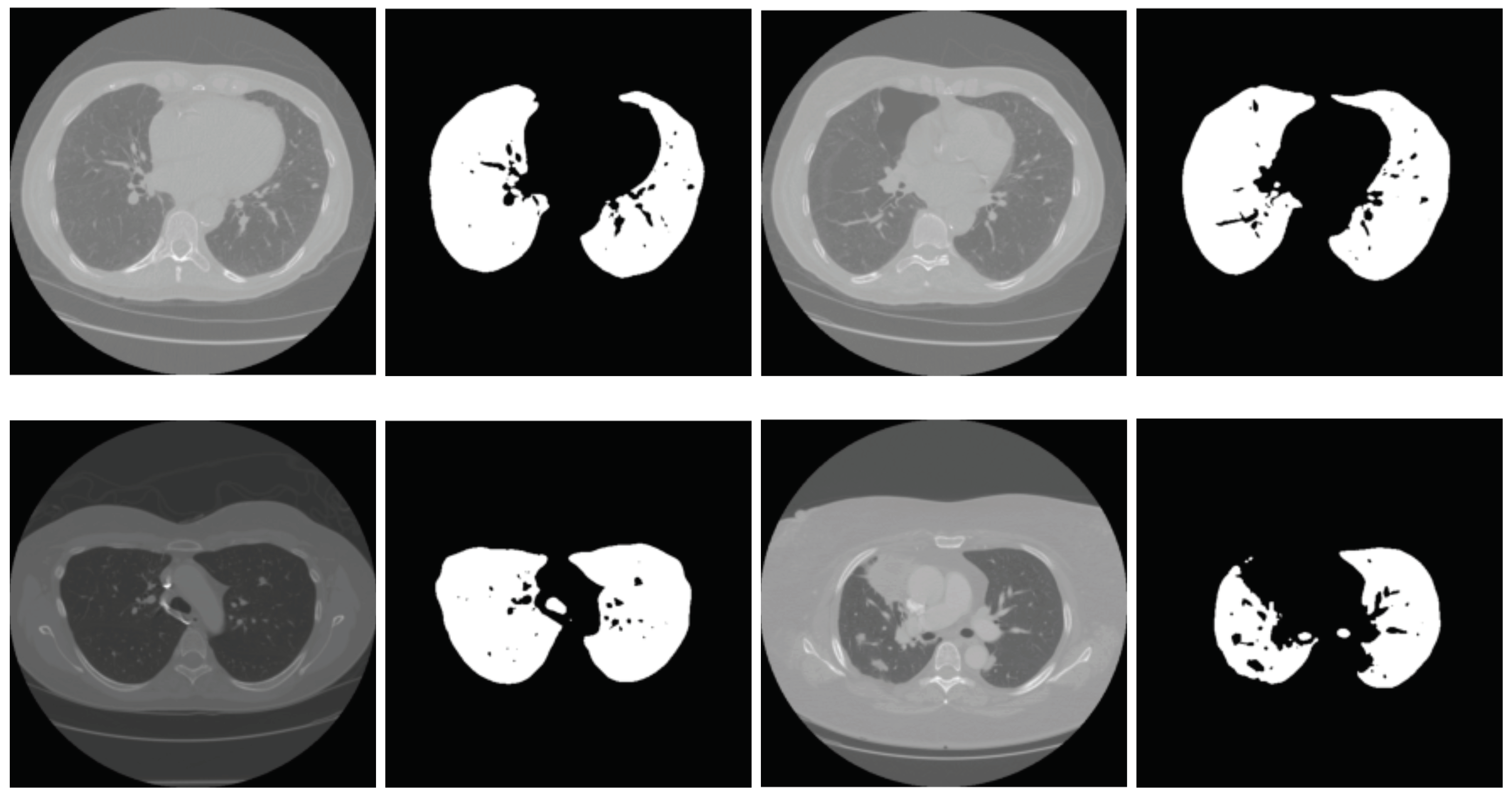
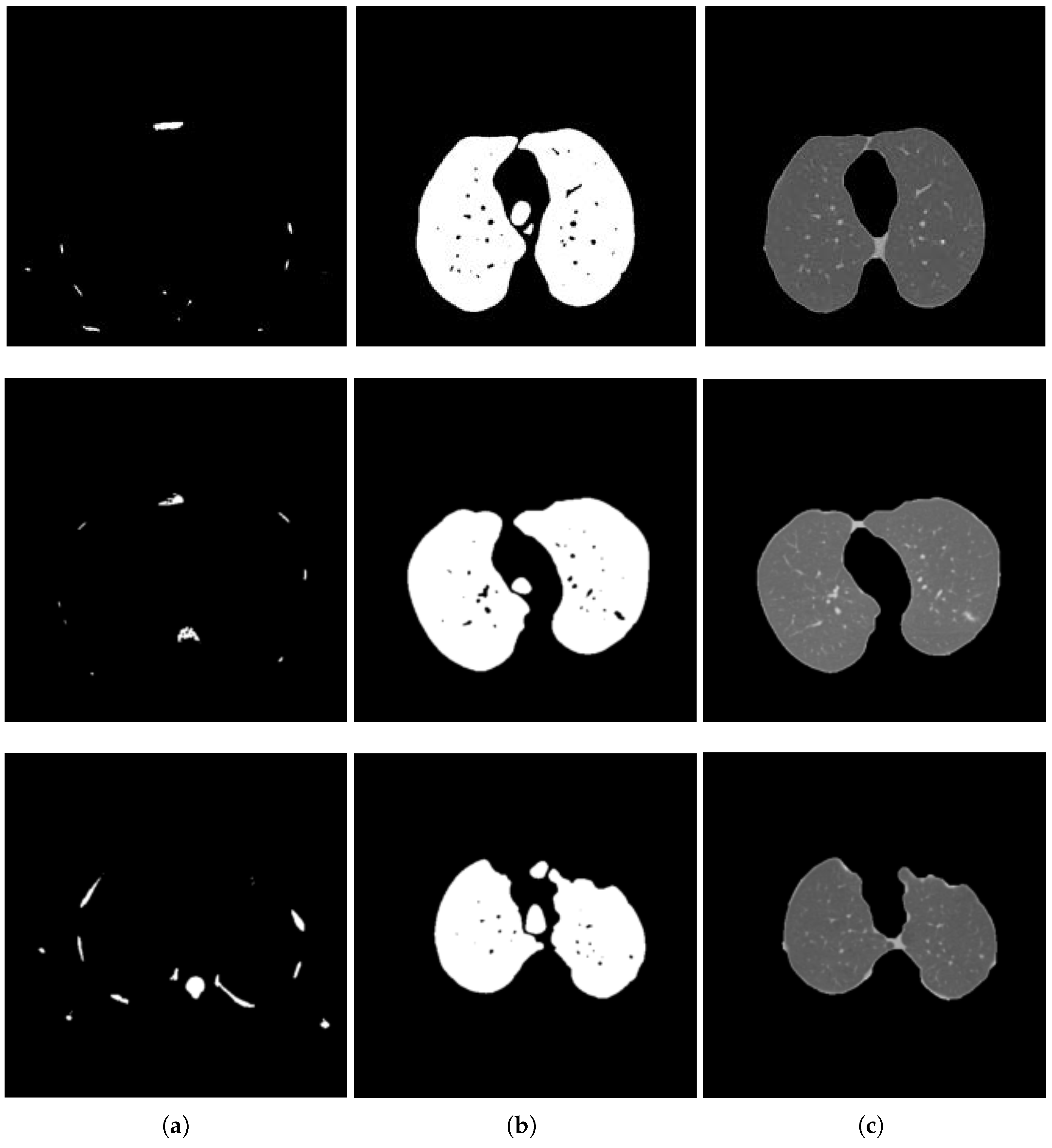
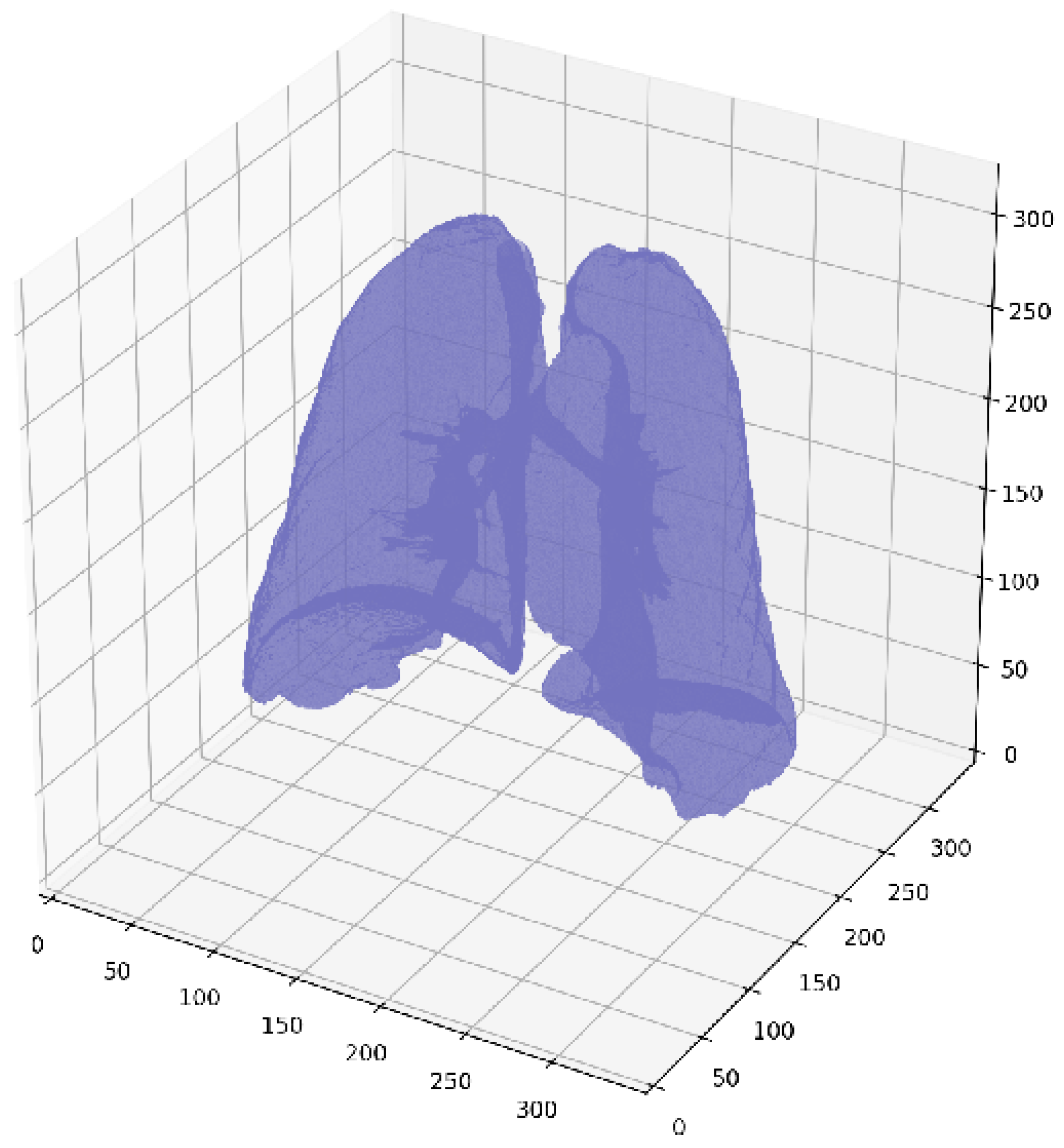
2.2. Lung Segmentation
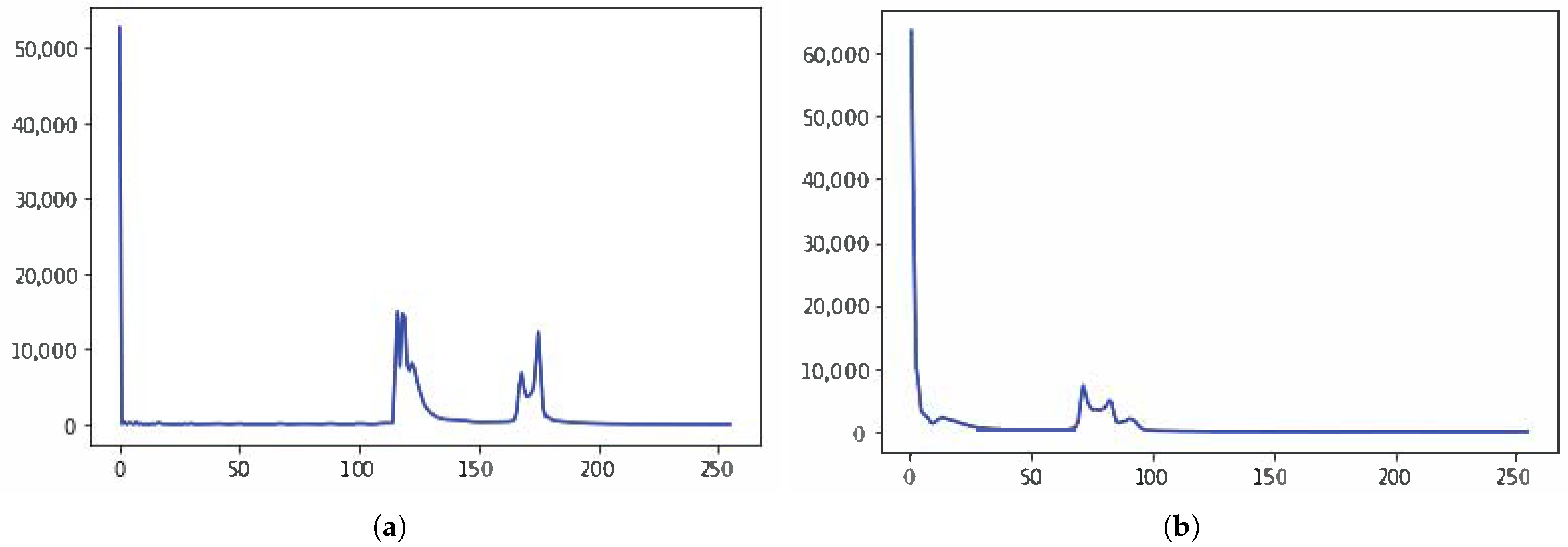
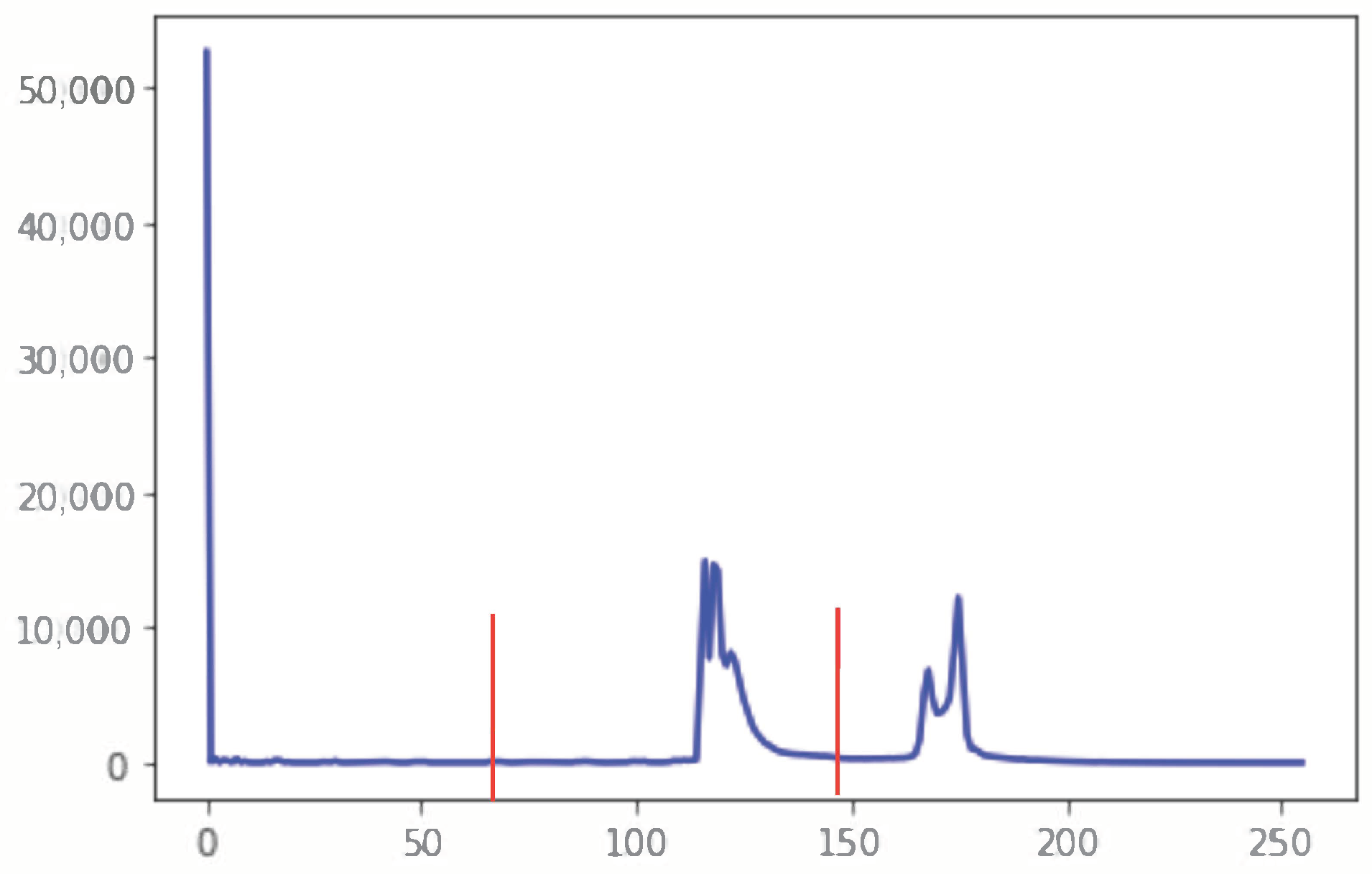
2.2.1. Segmentation in Lung Window
2.2.2. Segmentation in Mediastinal Window
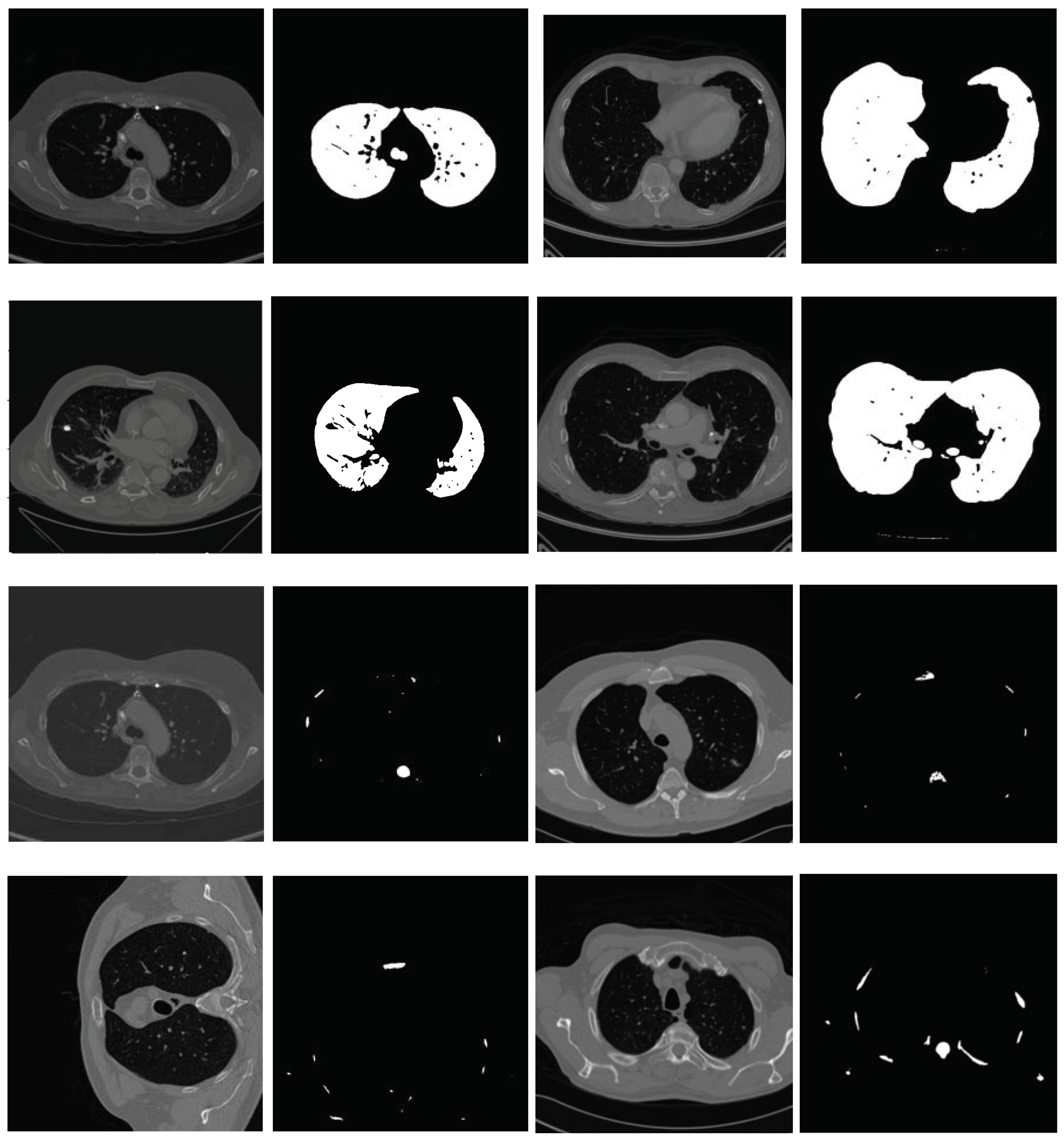
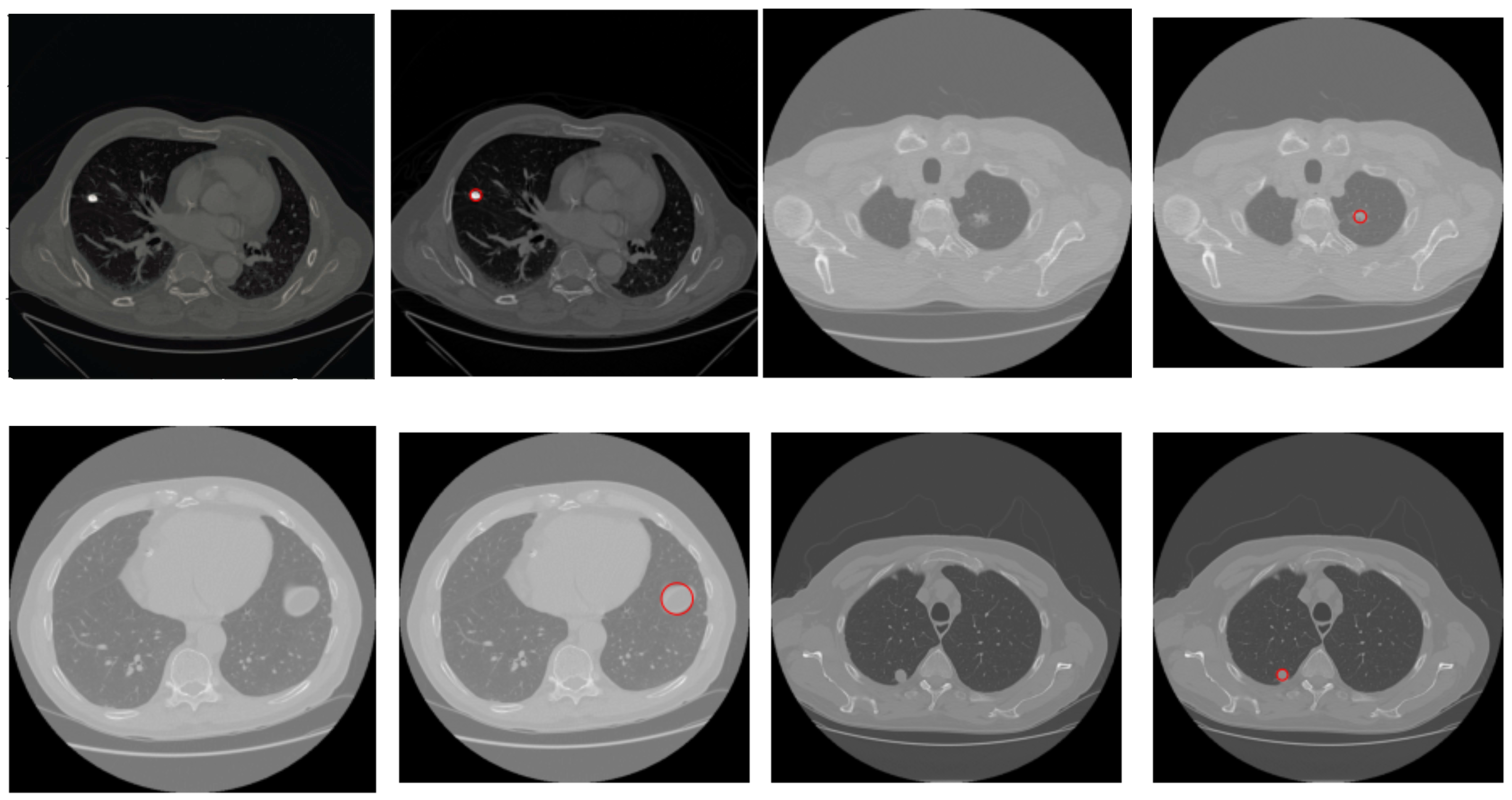
2.3. Nodule Detection
| Algorithm 1 Proposed Nodule Detection Algorithm |
| Require: A CT Image I of size Ensure: Detected nodule(s). 1: Normalize I to bring the intensity values within range 0–255 2: Suppress noise in I using Gaussian blur [] 3: if I is a lung window then 4: Compute histogram H of I 5:
Find the two local minima in H 6: [] ← multi-Otsu() 7: Threshold I using to separate lungs from rest of the lung window image 8: else {I is a mediastinal window} 9: {Compute the global mean of I} 10: Use to roughly separate divide I into two regions, and . 11: {Compute the local mean of the region } 12: {Compute the local mean of the region } 13: {The threshold is then taken as the average of the two local means} 14: Threshold I using to separate lungs from rest of the mediastinal window image 15: end if 16: Threshold I using threshold value This extracts vessels and nodules from the segmented lungs I. 17: I = OPEN (I) {OPEN is morphological binary open operator.} 18: I = CLOSE (I) {CLOSE is morphological binary open operator.} 19: Drop the objects in I that have the elongation feature, i.e., those that are not circular 20: Compute the diameter d of each lesion in I 21: if mm then 22: Mark the region as nodule 23: else mm 24: Mark the region as non-nodule 25: end if 26:Highlight the nodule regions, if any, using red circles |
3. Experiments and Results
3.1. Dataset
3.2. Performance Evaluation and Comparison
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Key Statistics for Lung Cancer. Available online: https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html (accessed on 6 June 2023).
- Lung Cancer—Non-Small Cell: Statistics. Available online: https://www.cancer.net/cancer-types/lung-cancer-non-small-cell/statistics (accessed on 6 June 2023).
- Society, A.C. What Is Lung Cancer? 2019. Available online: https://www.cancer.org/cancer/lung-cancer/about/what-is.html (accessed on 6 June 2023).
- Hacking, C.; Hsu, C. Dual Energy CT. 2014. Available online: https://radiopaedia.org/articles/31353 (accessed on 6 June 2023). [CrossRef]
- System, L.U.H. Technology Developed to Improve Lung Cancer Detection, Treatment. 2014. Available online: https://www.sciencedaily.com/releases/2014/11/141113194950.htm (accessed on 28 June 2022).
- WHO. Preventing Cancer. Available online: https://www.who.int/activities/preventing-cancer (accessed on 6 June 2023).
- Li, Q.; Li, F.; Doi, K. Computerized Detection of Lung Nodules in Thin-Section CT Images by Use of Selective Enhancement Filters and an Automated Rule-Based Classifier. Acad. Radiol. 2008, 15, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdillah, B.; Bustamam, A.; Sarwinda, D. Image processing based detection of lung cancer on CT scan images. J. Phys. Conf. Ser. 2017, 893, 012063. [Google Scholar] [CrossRef]
- Wason, J.V.; Nagarajan, A. Image processing techniques for analyzing CT scan images towards the early detection of lung cancer. Bioinformation 2019, 15, 596–599. [Google Scholar] [PubMed]
- Uzelaltinbulat, S.; Ugur, B. Lung tumor segmentation algorithm. Procedia Comput. Sci. 2017, 120, 140–147. [Google Scholar] [CrossRef]
- Gaikwad, A.; Inamdar, A.; Behera, V. Lung cancer detection using digital Image processing On CT scan Images. Int. Res. J. Eng. Technol. 2016, 3, 2395-0056. [Google Scholar]
- Nardi-Agmon, I.; Abud-Hawa, M.; Liran, O.; Gai-Mor, N.; Ilouze, M.; Onn, A.; Bar, J.; Shlomi, D.; Haick, H.; Peled, N. Exhaled Breath Analysis for Monitoring Response to Treatment in Advanced Lung Cancer. J. Thorac. Oncol. 2016, 11, 827–837. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.H.; Helsper, J.; Farid, M.S.; Grzegorzek, M. A computer vision-based system for monitoring Vojta therapy. Int. J. Med Informatics 2018, 113, 85–95. [Google Scholar] [CrossRef]
- Khan, M.H.; Schneider, M.; Farid, M.S.; Grzegorzek, M. Detection of Infantile Movement Disorders in Video Data Using Deformable Part-Based Model. Sensors 2018, 18, 3202. [Google Scholar] [CrossRef] [Green Version]
- de Kock, R.; van den Borne, B.; Soud, M.Y.E.; Belderbos, H.; Stege, G.; de Saegher, M.; van Dongen-Schrover, C.; Genet, S.; Brunsveld, L.; Scharnhorst, V.; et al. Circulating biomarkers for monitoring therapy response and detection of disease progression in lung cancer patients. Cancer Treat. Res. Commun. 2021, 28, 100410. [Google Scholar] [CrossRef]
- Fu, W.; Tang, D.; Yang, F.; Wang, J.; Wu, Y.; Shen, X.; Gao, W. Short-term home remote monitoring of patients after lung cancer surgery. Clin. Surg. Oncol. 2022, 1, 100004. [Google Scholar] [CrossRef]
- Yeganeh, A.; Shadman, A.; Shongwe, S.C.; Abbasi, S.A. Employing evolutionary artificial neural network in risk-adjusted monitoring of surgical performance. Neural Comput. Appl. 2023, 35, 10677–10693. [Google Scholar] [CrossRef]
- Kadir, T.; Gleeson, F. Lung cancer prediction using machine learning and advanced imaging techniques. Transl. Lung Cancer Res. 2018, 7, 304–312. [Google Scholar] [CrossRef]
- Makaju, S.; Prasad, P.; Alsadoon, A.; Singh, A.; Elchouemi, A. Lung cancer detection using CT scan images. Procedia Comput. Sci. 2018, 125, 107–114. [Google Scholar] [CrossRef]
- Khehrah, N.; Farid, M.S.; Bilal, S.; Khan, M.H. Lung nodule detection in CT images using statistical and shape-based features. J. Imaging 2020, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Maqsood, A.; Farid, M.S.; Khan, M.H.; Grzegorzek, M. Deep Malaria Parasite Detection in Thin Blood Smear Microscopic Images. Appl. Sci. 2021, 11, 2284. [Google Scholar] [CrossRef]
- Bhatia, S.; Sinha, Y.; Goel, L. Lung cancer detection: A deep learning approach. In Soft Computing for Problem Solving; Springer: Singapore, 2019; pp. 699–705. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Asuntha, A.; Srinivasan, A. Deep learning for lung Cancer detection and classification. Multimed. Tools Appl. 2020, 79, 7731–7762. [Google Scholar] [CrossRef]
- Huidrom, R.; Chanu, Y.J.; Singh, K.M. Pulmonary nodule detection on computed tomography using neuro-evolutionary scheme. Signal Image Video Process. 2018, 13, 53–60. [Google Scholar] [CrossRef]
- Masood, A.; Yang, P.; Sheng, B.; Li, H.; Li, P.; Qin, J.; Lanfranchi, V.; Kim, J.; Feng, D.D. Cloud-Based Automated Clinical Decision Support System for Detection and Diagnosis of Lung Cancer in Chest CT. IEEE J. Transl. Eng. Health Med.-JTEHM 2020, 8, 1–13. [Google Scholar] [CrossRef]
- Shakeel, P.M.; Burhanuddin, M.A.; Desa, M.I. Lung cancer detection from CT image using improved profuse clustering and deep learning instantaneously trained neural networks. Measurement 2019, 145, 702–712. [Google Scholar] [CrossRef]
- Chon, A.; Balachandar, N.; Lu, P. Deep Convolutional Neural Networks for Lung Cancer Detection; Standford University: Stanford, CA, USA, 2017; pp. 1–9. [Google Scholar]
- Alakwaa, W.; Nassef, M.; Badr, A. Lung cancer detection and classification with 3D convolutional neural network (3D-CNN). Lung Cancer 2017, 8, 409. [Google Scholar] [CrossRef] [Green Version]
- Kuan, K.; Ravaut, M.; Manek, G.; Chen, H.; Lin, J.; Nazir, B.; Chen, C.; Howe, T.C.; Zeng, Z.; Chandrasekhar, V. Deep learning for lung cancer detection: Tackling the kaggle data science bowl 2017 challenge. arXiv 2017, arXiv:1705.09435. [Google Scholar]
- Lu, H.; Kim, J.; Qi, J.; Li, Q.; Liu, Y.; Schabath, M.B.; Ye, Z.; Gillies, R.J.; Balagurunathan, Y. Multi-Window CT Based Radiological Traits for Improving Early Detection in Lung Cancer Screening. Cancer Manag. Res. 2020, 12, 12225–12238. [Google Scholar] [CrossRef] [PubMed]
- Sung Liao, P.; Sheng Chen, T.; Choo Chung, P. A fast algorithm for multilevel thresholding. J. Inf. Sci. Eng. 2001, 17, 713–727. [Google Scholar]
- Feng, Y.; Zhao, H.; Li, X.; Zhang, X.; Li, H. A multi-scale 3D Otsu thresholding algorithm for medical image segmentation. Digit. Signal Process. 2017, 60, 186–199. [Google Scholar] [CrossRef] [Green Version]
- Malarvel, M.; Sethumadhavan, G.; Bhagi, P.C.R.; Kar, S.; Thangavel, S. An improved version of Otsu’s method for segmentation of weld defects on X-radiography images. Optik 2017, 142, 109–118. [Google Scholar] [CrossRef]
- Liu, H.; Rashid, T.; Habes, M. Cerebral Microbleed Detection Via Fourier Descriptor with Dual Domain Distribution Modeling. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging Workshops (ISBI Workshops), Iowa City, IA, USA, 4 April 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Digabel, H.; Lantuéjoul, C. Iterative algorithms. In Proceedings of the 2nd European Symposium Quantitative Analysis of Microstructures in Material Science, Biology and Medicine; Scientific Research Publishing Inc.: Wuhan, China, 1978; pp. 85–89. [Google Scholar]
- Kornilov, A.S.; Safonov, I.V. An overview of watershed algorithm implementations in open source libraries. J. Imaging 2018, 4, 123. [Google Scholar] [CrossRef] [Green Version]
- Lung Nodule Analysis 2016. 2016. Available online: https://luna16.grand-challenge.org/Data/ (accessed on 6 June 2023).
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.; van den Bogaard, C.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Nasser, I.M.; Abu-Naser, S.S. Lung cancer detection using artificial neural network. Int. J. Eng. Inf. Syst. (IJEAIS) 2019, 3, 17–23. [Google Scholar]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Xiang, H. Automated detection and classification for early stage lung cancer on CT images using deep learning. In Proceedings of the Pattern Recognition and Tracking XXX; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 10995, p. 109950S. [Google Scholar]
- Xie, H.; Yang, D.; Sun, N.; Chen, Z.; Zhang, Y. Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit. 2019, 85, 109–119. [Google Scholar] [CrossRef]
- Jin, X.Y.; Zhang, Y.C.; Jin, Q.L. Pulmonary nodule detection based on CT images using convolution neural network. In Proceedings of the 2016 9th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 10–11 December 2016; Volume 1, pp. 202–204. [Google Scholar]
- Khumancha, M.B.; Barai, A.; Rao, C.R. Lung cancer detection from computed tomography (CT) scans using convolutional neural network. In Proceedings of the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019; pp. 1–7. [Google Scholar]
- Ye, J.C.; Sung, W.K. Understanding Geometry of Encoder-Decoder CNNs. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Chaudhuri, K., Salakhutdinov, R., Eds.; Proceedings of Machine Learning Research. Volume 97, pp. 7064–7073. [Google Scholar]
- Tan, M.; Le, Q.V. Mixconv: Mixed depthwise convolutional kernels. arXiv 2019, arXiv:1907.09595. [Google Scholar]



| Attribute | Property |
|---|---|
| Number of CT scans | 888 |
| Annotated lung nodule | 1186 |
| Format | 3D MHD |
| 2D slice size | 512 × 512 |
| Method | Accuracy | Recall | Precision | Dataset |
|---|---|---|---|---|
| Nasser [43] | 0.96 | — | — | — |
| Sang [44] | — | 0.94 | — | LIDC-IDRI |
| Makaju [19] | 0.92 | — | — | — |
| Jin [46] | 0.84 | 0.82 | 0.86 | — |
| Xie [45] | — | 0.86 | — | LUNA16 |
| Alakwaa [31] | 0.86 | — | — | LUNA16 |
| Khumancha [47] | — | 0.82 | 0.89 | LUNA16 |
| Proposed | 0.94 | 0.97 | 0.92 | LUNA16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasir, M.; Farid, M.S.; Suhail, Z.; Khan, M.H. Optimal Thresholding for Multi-Window Computed Tomography (CT) to Predict Lung Cancer. Appl. Sci. 2023, 13, 7256. https://doi.org/10.3390/app13127256
Nasir M, Farid MS, Suhail Z, Khan MH. Optimal Thresholding for Multi-Window Computed Tomography (CT) to Predict Lung Cancer. Applied Sciences. 2023; 13(12):7256. https://doi.org/10.3390/app13127256
Chicago/Turabian StyleNasir, Muflah, Muhammad Shahid Farid, Zobia Suhail, and Muhammad Hassan Khan. 2023. "Optimal Thresholding for Multi-Window Computed Tomography (CT) to Predict Lung Cancer" Applied Sciences 13, no. 12: 7256. https://doi.org/10.3390/app13127256






